Abstract
RNase P from Escherichia coli cleaves the coenzyme B12 riboswitch from E. coli and a similar one from Bacillus subtilis. The cleavage sites do not occur in any recognizable structure, as judged from theoretical schemes that have been drawn for these 5′ UTRs. However, it is possible to draw a scheme that is a good representation of the E. coli cleavage site for RNase P and for the cleavage site in B. subtilis. These data indicate that transient structures are important in RNase P cleavage and in riboswitch function. Coenzyme B12 has a small inhibitory effect on E. coli RNase P cleavage of the E. coli riboswitch. Both E. coli RNase P and a partially purified RNase P from Aspergillus nidulans mycelia succeeded in cleaving a putative arginine riboswitch from A. nidulans. The cleavage site may be a representative of another model substrate for eukaryotic RNase P. This 5′ UTR controls splicing of the arginase mRNA in A. nidulans. Four other riboswitches in E. coli were not cleaved by RNase P under the conditions tested.
Keywords: arginase, Aspergillus nidulans, Bacillus subtilis, coenzyme B12, Escherichia coli
Riboswitches occur in the untranslated regions at the 5′ side of the mRNA for translated genes (1). In some cases, the riboswitch extends into the ORF of the gene to be transcribed and translated. Small metabolites that are metabolized or transported by the products of the ORFs downstream from the riboswitch interact with the riboswitch without the action of proteins and control the translation, and possibly the transcription, of the riboswitch and the ORF. These results indicate the general importance and antiquity of RNA-based methods in gene regulation.
In this paper, we show that RNase P (2) cleaves some riboswitches once. In fact, we show this for the riboswitch in the 5′ region of the btuB gene in Escherichia coli and a similar gene in Bacillus subtilis (3). The putative arginine riboswitch, the 5′ region before the arginase gene in Aspergillus nidulans (4) is also cleaved, indicating that the unusual RNase P cleavage activity is a phenomenon that is widespread in eukaryotes as well as in prokaryotes. The RNase P cleavage occurs actually in the beginning of the gene coding for the arginase gene. We also assayed four other riboswitches found in E. coli but found no cleavage by RNase P in those regions.
Some genetic experiments indicate that in E. coli, the presence of RNase P cleavage in the btuB riboswitch might promote degradation of the 3′ region downstream from the cleavage site. In operons, this result is a polarity effect and means that the downstream genes after the enzyme cleavage site are usually degraded quickly, and the ORF is not translated as frequently as the upstream genes (5). A polarity effect in riboswitches means a loss of the uncleaved translation efficiency of the downstream gene. It is also certainly the case that RNase P does cleave some substrates that are not operons or mature small stable RNA precursors (2, 6), but the biological function of these latter cleavages is not fully understood.
Although RNase P cleaves hairpin-like substrates with a single-stranded region attached at the 5′ end (2), it had been assumed that such regions would be buried inside many folded mRNAs. We now find that some of the riboswitch substrates that we are currently using have structures recognized by RNase P, at least transiently. Some of the cleavage sites in riboswitches do not correspond to classical RNase P cleavage sites at thermodynamic equilibrium in terms of the general secondary structure according to the programs of Zuker and coworkers (7, 8). We show this is the case below and present explanations of the peculiarity of these sites.
Materials and Methods
Strains. NHY322 and NHY312 (9), competent for transformation, were prepared and transformed with pRS414btuB (10) as described, with the exception that all cells were grown at 30°C (11).
Substrates. The precursor to E. coli tRNATyr [pTyr (12)] and the precursor to a yeast suppressor tRNA [pSup S1 (13)] were used as controls in RNase P assays. For mRNA substrates, the yvrc167 RNA extends from –235 to –86 in the 5′ UTR of the B. subtilis yvrC gene. The btuB202 RNA extends from –240 to –38 in the 5′ UTR of the E. coli btuB gene. mRNA substrates used for RNase P assays were prepared by transcription in vitro by using T7 RNA polymerase (Promega) and 32P-α-GTP by using plasmid DNAs linearized by restriction digests as templates.
mRNA substrates of btuB202 and yvrc167 were transcribed in vitro, radioactively labeled (32P-α-GTP) as indicated, and cleaved with E. coli RNase P holoenzyme. The products were isolated and digested with RNase T1 as described below to determine the cleavage sites. For yvrc167, the experiment was repeated with 32P-α-CTP-labeled substrate to further confirm the exact site of cleavage (14).
Plasmids. Plasmids were prepared by inserting PCR products into pUC19 at the SmaI site. To make the pUC19/yvrC167 clone, pDG1661/yvrc (3) was used as the DNA template and primer A, which contains a T7 promoter (TAATACGACTCACTATAGGAAAAACGGATACG), and primer B (GAAUUCUGCGAGGACAGAUGAUG) were used. To make the pUC19/btuB 202 clone, pRS 414 (–70–450) (10) was used as the DNA template and primer C, which contains a T7 promoter (TAATACGACTCACTATAGGGCCGGTCCTGTGAGTT AATA), and primer D (TCATCAATATTACGCGATGATGAGA) were used. pUC19/yvrC167 was digested with BamHI and used for transcription in vitro reactions. pUC19/btuB202 was digested with SacI, treated with Klenow, and used for transcription in vitro reactions.
RNase P Activity Assays. Reactions were carried out at 37°C in 10 μl of 1×PA (50 mM Tris·HCl, pH 8.0/100 mM NH4Cl/10 mM MgCl2) with RNasin (0.1 units/μl; Promega). Either E. coli RNase P holoenzyme [M1 RNA/C5 protein (15, 16)] or A. nidulans RNase P purified through the glycerol gradient step was used with internally labeled mRNA substrates (32P-α-GTP). Substrates were transcribed in vitro by using T7 or SP6 RNA polymerase (Promega). Reactions were stopped by adding 10 μl 10 M urea/10% phenol dye and then analyzed on an 8% PAGE denaturing gel.
RNase P Activity Assays in the Presence of Coenzyme B12. For assays with coenzyme B12, E. coli RNase P holoenzyme in 1×PA reaction buffer was aliquoted into reaction tubes. In a darkroom illuminated by red light, either coenzyme B12 (Sigma), to a final concentration of 0.1 mM, or water was added to the tubes. Labeled btuB 202 and yvrc167 substrates were then added to the respective reactions, and assays were completed as described above. A different protocol of additions to the reaction tubes gave slightly different results.
Preparation of RNase P from A. nidulans. Five grams of frozen Aspergillus mycelia were crushed in a mortar under liquid nitrogen to obtain the consistency of fine powder. The crushed cells were transferred to a 50-ml polypropylene centrifuge tube and resuspended in 25 ml of Buffer A (25 mM Tris·HCl, pH 7.5/10 mM MgCl2/100 mM NaCl/10% glycerol/0.1% Igepal CA360/5 mM 2-mercaptoethanol) to which Complete (Roche Diagnostics) was added (1 tablet/50 ml). The mixture was left on ice for 15 min, inverted occasionally for mixing, until the cells defrosted. The cells were then sonicated for 2 min with a half-inch probe (Branson Sonifier 450) and centrifuged in an SS34 Sorvall rotor at 7,000 × g to prepare a crude extract. The supernatant was then centrifuged at 30,000 × g to prepare an S30 extract. The S30 extract was loaded onto a DEAE Sepharose Fast Flow column (Amersham Pharmacia) equilibrated with Buffer A, and the enzyme was eluted with a NaCl gradient of 100–300 mM in the same buffer. Fractions were assayed for RNase P activity, and the resulting active fractions were pooled and concentrated by using Amicon Ultra filter devices (10K molecular weight cutoff). One ml of concentrated extract was layered onto 12 ml of a 15%–30% glycerol gradient in Buffer A with 200 mM NaCl. Centrifugation was carried out in a Beckman SW41 rotor for 22 h at 2°C and 40,000 rpm. Fractions (400 μl) were collected by puncturing the bottom of the tube with an 18-gauge needle. These were assayed for RNase P activity. The peak of RNase P activity was pooled and concentrated by using Amicon Ultra filter devices (10K molecular weight cutoff). All enzyme extracts were stored on ice at 4°C.
Assays for β-Galactosidase Activity and Cloning of Mutations in btuB DNA. Assays were carried out as described (17, 18). Single-base mutations of G175 to C and G175 to A were made separately via PCR at the site of RNase P cleavage in the 5′ UTR region of the btuB gene. PCR inserts were prepared by using pUC19/btuB 202 as the DNA template and primer C (described above) and primer E [AATTCATCAATATTACGCGATGATGAGAACGA(G/T)ATGCGAC GTTGGCC]. The PCR products were then inserted into pUC19 DNA that had been digested with SmaI. Transformed clones were sequenced, and the appropriate mutant clones were selected for further study. The mutated DNA of the clones selected were first digested with SacI and treated with Klenow to make blunt 3′ ends before transcription reactions in vitro. The mRNA was then used for RNase P assays as described above.
Results
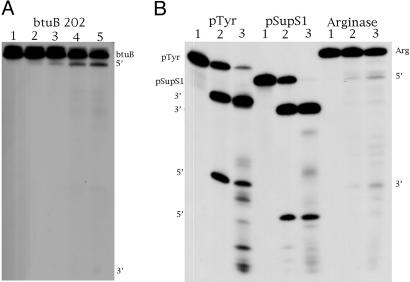
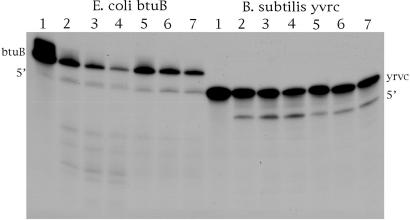
Assays of RNase P Activity. Prokaryotes. Five riboswitches from E. coli were assayed. These are TPP, FMN, SAM, lysine, and btuB (1). Of these five riboswitches, only the btuB (coenzyme B12) riboswitch was cleaved by E. coli RNase P. One cleavage site was observed in the relevant experiment (Fig. 1). A similar riboswitch (coenzyme B12) was also assayed from B. subtilis and it, too, was cleaved once. Some clones were made that encode longer mRNAs than the ones we tried in the coenzyme B12 riboswitch, and these clones, which contained sequences from the ORF, were cleaved in a minor way at several sites. The minor cleavages could be other RNase P sites that were unveiled by using a longer test sequence but were still cleaved at extremely minor rates. These RNAs transcribed from longer clones were not studied further at this point. The sequences of the cleavage product were investigated by using complete RNase T1 digestion and electrophoresis to determine the size and composition of some cleavage products (see Fig. 7, which is published as supporting information on the PNAS web site, and Materials and Methods).
Fig. 1.
Activity of RNase P from E. coli and A. nidulans on various substrates. (A) E. coli btuB 202 mRNA. Reactions were carried out as described in Materials and Methods by using E. coli RNase P holoenzyme with 12,500 cpm of labeled E. coli btuB 202 mRNA substrate. Lane 1, mRNA alone; lanes 2 and 3, mRNA with E. coli RNase P holoenzyme (20 nM M1 RNA/200 nM C5) incubated for 15 and 30 min, respectively, at 37°C; lanes 4 and 5, mRNA with E. coli RNase P holoenzyme (100 nM M1 RNA/1 μM C5) incubated for 15 and 30 min, respectively, at 37°C. (B) A. nidulans 5′ UTR of the arginase gene with controls. Reactions were carried out as described in Materials and Methods by using E. coli RNase P holoenzyme (20 nM M1 RNA/200 nM C5 protein) or A. nidulans concentrated glycerol gradient extracts (2 μl/10 μl reaction) with 10,000 cpm of labeled mRNA substrate as indicated. Reactions were incubated for 20 min at 37°C. Substrates were pTyr, the precursor to Saccharomyces cerevisiae tRNASer (pSupS1), and the A. nidulans mRNA: lane 1, mRNA alone; lane 2, mRNA with E. coli RNase P holoenzyme; and lane 3, mRNA with A. nidulans RNase P extract.
A variety of reaction buffer conditions were attempted in the RNase P assay; only the buffer given in Materials and Methods and the same buffer with 400 mM NH4Cl instead of 100 mM were optimal.
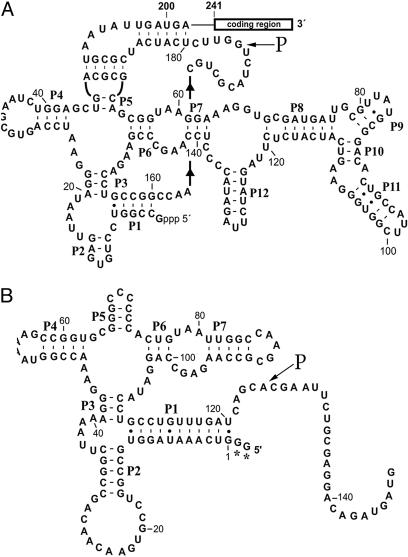
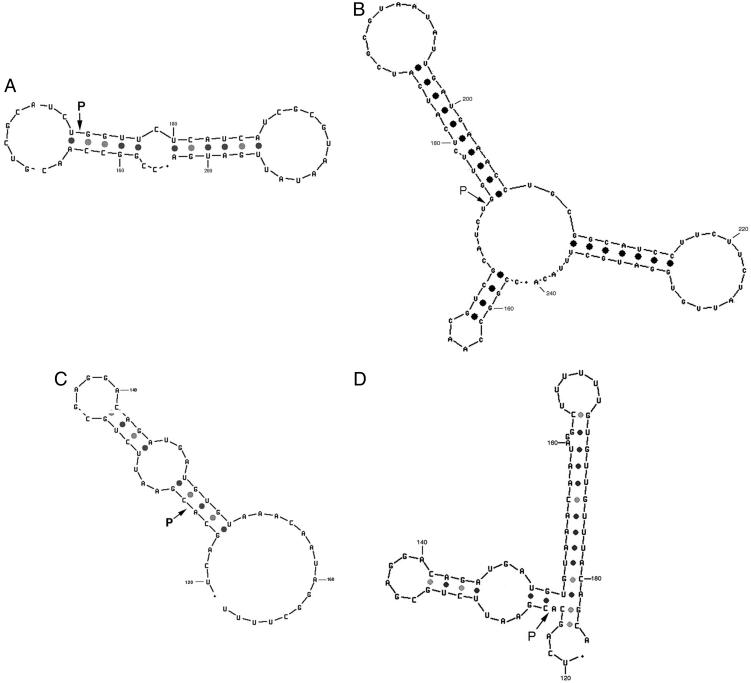
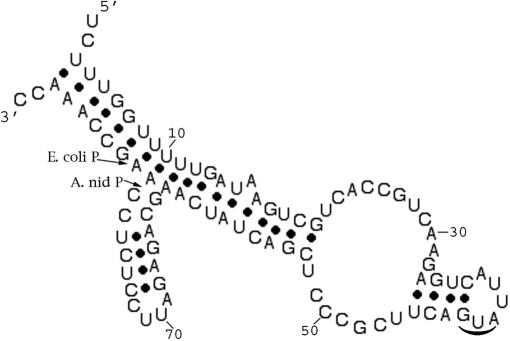
We note that cleavage of btuB mRNA occurs at the 5′ side of position G175 and the B. subtilis homologous riboswitch at the 5′ side of position A125, as shown in Fig. 2. An examination of these cleavage sites does not indicate a classical RNase P recognition site or even a cleavable model site (2), as shown in theoretical structures drawn to reflect the phylogenetic conserved regions of riboswitches (3). As such, these schemes should be ultimately redrawn to reflect structures in solution that are recognized by RNase P. Some possibilities are shown in Fig. 3, which are redrawn sites for E. coli and B. subtilis cleavage of RNase P of btuB mRNA. In these cases, ≈45 nucleotides were taken upstream (5′) and ≈25 nucleotides downstream (3′) of the cleavage site for the E. coli btuB riboswitch, and structures were drawn according to Zuker's methods (5, 6). As shown in Fig. 3A, the RNase P cleavage site in E. coli btuB mRNA has a structure that might be expected for a RNase P model substrate. [No cleavages occur at the 5′ side of a pyrimidine (2).] We would expect such a site to map at the junction of a single- and double-stranded region. This apparently transient substrate structure can be formed as an intermediate during kinetic transitions between different, more stable, conformations of the btuB riboswitch. Northern blots in experiments designed to search for effects of this cleavage in strains that are temperature-sensitive for RNase P indicate that this cleavage site does exist in vivo (see below). The B. subtilis RNase P cleavage site is located in a region that has a short single-stranded region next to a putative double-stranded region in Fig. 3C; although we included various combinations of nucleotides from the actual sequence in searching for new structures, we could not draw even a transient structure for this substrate for E. coli RNase P, even though cleavage occurs in a double-stranded region. We rearranged a structure to indicate a theoretical possible RNase P cleavage site (Fig. 3D). We note also that no 3D structural effects or pseudoknots are shown in the schemes we present.
Fig. 2.
Schemes of the E. coli (A) and B. subtilis (B) coenzyme B12 5′ UTR riboswitches as determined by phylogenetic considerations (1). The RNase P cleavage sites are marked. In A, some inherent uncertainty in the analysis of the RNase T1 digests indicates that the cleavage site might be at G176 rather than at G175. In B. subtilis yvrC, G132 has been changed to C132 to correct the sequence published in ref. 3. The schemes are reprinted by permission from ref. 1.
Fig. 3.
Redrawn schemes of btuB regions in E. coli and B. subtilis. Different sequences were chosen, as described in the text, to redraw structures to determine RNase P cleavage sites (7, 8). (A) E. coli btuB 202. (B) The same as in A, except more nucleotides are at the 3′ end, and these give a theoretical structure that has not been tested. (C) B. subtilis yvrc167(btuB). (D) Redrawn from a Zuker structure that showed no RNase P cleavage sites. This has more nucleotides at the 3′ end than does C and represents a possible RNase P cleavage site. The nucleotide numbers refer to those in Fig. 2.
Some aberrant cleavages by E. coli RNase P, i.e., not at the first position of the junction between a single- and a double-stranded region, were also observed in cleavages of TYMV derived substrates (19, 20) and a model substrate for human RNase P (21). This latter substrate had some resemblance to the simple hairpin substrates that are attacked by E. coli RNase P.
To determine absolutely that the cleavages we studied were made by RNase P, single base substitution mutations were made at the 5′ terminal nucleotide of the 3′ cleavage product (see Materials and Methods). These mutations should produce unpaired bases at the expected stem of the cleavage product and should alter the way in which RNase P cleaved these sites. In fact, a G to C change at nucleotide 175 and G to A change at the same position yielded RNase P cleavages in which in each case a strong band appeared as the new cleavage product (data not shown). An analysis of the cleavage products indicated that strong cleavage in the G to C change occurred at C179, and for the G to A change, cleavage occurred at U174, confirming changes of cleavage site. The model shown in Fig. 3B resembles the new folded structures very closely.
We also investigated the effect of coenzyme B12 on the cleavage by RNase P of E. coli btuB mRNA. These experiments were carried out, illuminated with a red light in a darkroom. The amount of absolute cleavage by RNase P decreased by 20% in the presence of coenzyme B12 (Fig. 4). It is important to note that the degree of this effect depended on the exact order of addition of B12 to the reaction mixture. Procedures other than those reported in Materials and Methods, i.e., order of addition of the components of the reaction, did not yield the result we reported here. At present, we do not know whether this effect can account for a decrease in transcription or translation of btuB. Wickiser et al. (22) suggested that concentrations of the small metabolite might be required to be 50- to 100-fold higher than the KD to achieve a different conformation in the riboswitch. In our case, the concentrations of coenzyme B12 we used achieved that supposed effect. An overall valuation of the effect of the concentration of coenzyme B12 on RNase P cleavage showed a small decrease, ≈20%, at 0.1 mM.
Fig. 4.
Effect of coenzyme B12 on cleavage of E. coli btuB 202 and B. subtilis yvrc167 mRNAs by E. coli RNase P holoenzyme. Reactions were carried out in the dark as described in Materials and Methods by using E. coli RNase P holoenzyme (100 nM M1 RNA/1 μM C5 protein) with 25,000 cpm of labeled E. coli btuB202 or B. subtilis yvrc167 mRNA substrates. For each substrate, lane 1, mRNA alone; lanes 2–4, mRNA with E. coli RNase P holoenzyme, incubated for 10, 20, and 30 min, respectively; and lanes 5–7, same as lanes 2–4 but with the addition of 0.1 mM coenzyme B12 (Sigma).
We used a clone that encodes the btuB riboswitch that lay upstream of a mutant lacZ gene, which expresses β-galactosidase only when an upstream promoter and the riboswitch sequence are in place (10, 23), to investigate the effects of the cleavage of the riboswitch on the expression of lacZ and to determine the amount of the total mRNA produced in the presence and absence of the RNase P function. The plasmid pRS414 btuB, which contains the btuB riboswitch next to lacZ, was transformed into E. coli NHY322, which is temperature-sensitive for RNase P function (mutant A49; ref. 9). Measurements of lacZ function in both a control (NHY312 pRS414 btuB) and experimental strains (NHY322 pRS414 btuB) indicate there is more absolute lacZ function in E. coli A49 at 43°C (75% compared with 100% at 0 min), whereas the amount made in the control strain is much less at 43°C (16% compared with 100% at 0 min; Table 1) compared with NHY322 btuB. Note that A49 is a temperature-sensitive mutation in the protein subunit for RNase P. This result shows that reduced success in cleaving at the RNase P site leads to more lacZ product made at the higher temperature, as expected from the mutation that affects RNase P. We also note that the A49 mutation immediately knocks out RNase P function, but there still is a residue of mature tRNA in the cell that allows protein synthesis for some period until the supply of mature tRNAs is exhausted.
Table 1. lacZ (β-galactosidase) function in E. coli with the btuB 5′ UTR.
| Strain | 0 min | 30 min | 60 min | 90 min |
|---|---|---|---|---|
| 43°C | ||||
| NHY312 btuB | 0.6 | 0.7 | 0.3 | 0.1 |
| NHY322 btuB | 1.6 | 0.9 | 1.7 | 1.2 |
| 30°C | ||||
| NHY312 btuB | 0.9 | 1.7 | 2.0 | |
| NHY322 btuB | 1.8 | 1.7 | 1.6 |
The strains are listed in Materials and Methods, as are the assay methods for β-galactosidase. Numbers shown are for 43°C in the first two lines and for 30°C in the last two lines. NHY322 contains the A49 mutation. NHY312 produced β-galactosidase at 16%, the control 0-min value, whereas NHY322 produced β-galactosidase at 75%, the amount in the control at 0 min at 43°C. This measurement was one of several taken in various cultures and is represented in a general definition of enzyme units.
We also assayed the survival of the mRNA in these strains by using probes for Northern blots that hybridized to the btuB-lacZ gene junction and one that hybridized to 10 nucleotides on both sides of the RNase P cleavage site in the 5′ UTR (Fig. 5). The amount of the particular RNAs we were probing for was very limited, and the results as observed were above background level (Table 2). Regardless, we found that there were two regions of the Northern blots that were relevant. The data indicate that at 43°C in the A49 strain, there is more intact mRNA (lacBZ) in the riboswitch–lacZ construct, as one might expect (Table 2), and virtually none of this product at 30°C, where there is ample RNase P activity. The data also show some RNA that contained the cleavage site for RNase P at 43°C, but this amount decreases with time after 30 min and, consequently, the accumulation of the cleavage products also increases with time until 30 min and then remains constant.
Fig. 5.
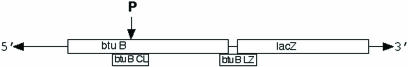
Schematic diagram of pRS414btuB (–70 in the DNA to +450 in frame with the ninth codon of lacZ, a gift of R. Breaker, Yale University) showing locations of the two primers used for Northern analyses. Primer btuB CL covers nucleotides 163–189 of btuB, which includes the site of RNase P cleavage (5′-GCGATGATGAGAACCAGATGCG ACGTTGGC-3′). Primer btuB LZ covers nucleotides 445–450 of btuB (the AUG is located at 241 nucleotides), a linker region between the two genes, and 14 nucleotides of the lacZ gene (5′-ACGTTGTAAAACGACGGG ATCCCGCGGAAG-3′).
Table 2. Extracts that contained RNA of NHY322 grown at two different temperatures were probed in Northern blots for the relevant species indicated.
| Time point, min | lacCL* | lacBZ† | Cleavage products‡ |
|---|---|---|---|
| 30°C | |||
| 0 | 0 | 0 | 0 |
| 30 | 11 | 0 | 24 |
| 60 | 12 | 0 | 49 |
| 90 | 13 | 0 | 17 |
| 43°C | |||
| 0 | 0 | 0 | 0 |
| 30 | 32 | 10 | 44 |
| 60 | 30 | 10 | 75 |
| 90 | 22 | 9 | 78 |
Cells were grown overnight at 30°C, diluted 50-fold, and grown for 1 hr at 30°C. The cultures were then split and one part grown at 30°C and the other at 43°C. Absolute amounts of radioactivity measured in bands shown in the Northern blots were subtracted from 0-min time points to give a true zero, and the same amounts were subtracted from the other values to give normalization. The results shown are for one experiment; three have been performed and the standard error in total counts was a maximum of ≈ 10%. The amount of 5S RNA was used also as a standard value for each time point.
Probed with an oligonucleotide complementary to sequences across the RNase P cleavage site.
As above, the probe contained sequences complementary to the site between the btuB 5′UTR and the lacZ reporter gene.
As above; the probe contained an oligonucleotide on either side of the RNase P cleavage site, respectively. The number listed is the sum of both cleavage products that were detected.
The Michaelis–Menten kinetic constants for RNase P cleavage of the E. coli btuB 202 riboswitch were measured in vitro. The kcat is 5 min–1, and the Km = 4.8 μM. The appropriate numbers for pTyr are kcat = 29 min–1 and Km = 33 nM (2). For btuB, kcat/Km is ≈0.1% of that for pTyr. However, in assays of the kinetics under conditions that may be more appropriate to the situation in vivo, e.g., one molecule substrate per one molecule enzyme, the kobs is 0.02 min–1. Results from Oh et al. (24) indicate that wild-type tRNA precursors are cleaved with a kobs of ≈0.5 min–1 in a relative comparison to our data. This last result indicates a low rate of cleavage when the enzyme is approximately the same concentration of the btuB substrate.
Eukaryotes. Recently, a putative riboswitch has been characterized from A. nidulans that affects splicing of the downstream sequences (4). We also tested this riboswitch, which is a long region at the 5′ side of and extends into the arginase gene, and which has a region that corresponds to an l-arginine-binding site (P. Weglenski, personal communication), for cleavage by both E. coli RNase P and a partially purified extract of RNase P from A. nidulans mycelia (Materials and Methods; P. Weglenski, personal communication). Classical procedures for partially purifying RNase P were used (25) and were successful in yielding fractions with enhanced RNase P activity from A. nidulans mycelia (data not shown).
The A. nidulans extract, which has a small amount of contaminants but contains primarily RNase P in function (see Fig. 1), cleaves the substrate two nucleotides 5′ to the E. coli cleavage site at the 5′ side of position of A79 (Fig. 6). Position 79 is at the first position of a double-stranded region preceded by a single-stranded region of RNA. E. coli RNase P cut the adjacent cleavage site in the ORF for arginase at the 5′ side of position G81 (Fig. 6). We did not investigate the very minor species produced with this extract. Fig. 7 also shows the cleavage site as determined by complete RNase T1 digestion of the cleavage product made by purified E. coli RNase P (Supporting Text, which is published as supporting information on the PNAS web site). We do not understand the differences in cleavage sites, except to say that the eukaryote enzyme is generally more selective in the structure it will recognize for cleavage (7).
Fig. 6.
Scheme of the structure of the arginase mRNA from A. nidulans that shows the RNase P cleavage site. The procedures used were adapted from ref. 20 and show part of the 5′ UTR and of the ORF for arginase. The AUG at the beginning of the ORF is marked, as well as the RNase P cleavage sites [E. coli and A. nidulans (A. nid)].
Discussion
RNase P Cleaves a Riboswitch in Bacteria. RNase P from E. coli cleaves the coenzyme B12 riboswitch in E. coli and B. subtilis. We do not understand why the btuB mRNA and its analog in B. subtilis are cleaved by RNase P, unless there is a need to have mRNA degradation initiated by RNase P or there are polarity effects on the first gene 3′ adjacent to the riboswitch. The cleavages are not of the same magnitude of those for the cleavage of tRNA precursors, which have to occur at a much faster rate to fulfill the cell's need for mature tRNAs or of the cleavage in the lacYA region in the lac operon. The action of RNase P on the btuB riboswitch adjacent to a lacZ reporter gene indicates that btuB–lacZ mRNA (intact mRNA) is in higher amounts in the A49 mutant strain compared with the intact mRNA that contains the cleavage site at a lower permissive temperature. Although there may be a polarity effect on the btuB riboswitch as a consequence of the presence of a “transient” structure that contains an RNase P cleavage site, the possibility exists that this phenomenon is a vestige of a more potent regulatory effect.
The cleavage by RNase P of the btuB riboswitch occurs in a transient structure that is not one of the thermodynamic structures that one can draw with the complete nucleotide sequence of the RNA. Mutations at the 3′ nucleotide next to the site of cleavage are consistent with the notion that it is RNase P that performs the cleavage. The sites of cleavage are changed within a few nucleotides of the site in the wild-type btuB riboswitch, as is expected for newly folded partial structures of the btuB riboswitch.
RNase P Cleavage in the Arginase Riboswitch. The cleavage by an A. nidulans extract that contains RNase P in the putative arginase riboswitch is a further indication that this cleavage phenomenon may not be as trivial as a vestige of ancient times. There is, however, no influence of arginine on the rate of cleavage (data not shown). Two shorter constructs of the 5′ UTR were also cleaved by RNase P at another site (G-35), and these cleavages were slightly inhibited by 1 or 5 mM arginine (≈20%; unpublished results). The modes of cleavage by RNase P may be an important reminder of a more ancient time of gene regulation, even if their function is not fully understood at the moment. Although it is not certain that the 5′ UTR next to the arginase is formally a riboswitch, it seems to share many of the same characteristics, i.e., tight binding of arginine to the 5′ UTR in the absence of protein and the phenotype that shows the effect of the arginine binding on splicing of this region (P. Weglenski, personal communication).
RNase P Cleavage in Operons and Riboswitches. Some operons in E. coli sustain a single or possibly two RNase P cleavages in intergenic regions (26). The presence of transient sites might be true for RNase P cleavage sites in operons where the kcat/Km is very low, <1% of that for pTyr (15). However, for the lac operon or lacYA (5), where the kcat/Km is ≈25% of that for pTyr, the amount of cleavage and the stability of the RNase P cleavage site might be more efficacious, and that effect leads to polarity of the downstream gene, lacA. Transient sites have low half-lives because structural conformations change in vitro. Clearly, the same must occur in vivo during transcription. These transient sites could determine polarity effects.
The results of experiments with the lacZ operon indicate that these cleavages can guarantee polarity in translation effects (5). That may not seem to be the case for riboswitches with respect to RNase P cleavage. In the latter case, those effects on regulation that may affect translation are apparent: (i) conformation of the riboswitch, (ii) the binding of a particular small molecule metabolite, and (iii) cleavage by RNase P. The first two effects on riboswitches are interconnected. It is not clear that conformational changes can occur without the small metabolite. The presence of the latter yields either inhibition of transcription, by promoting the formation of a transcription termination signal or of translation. The presence of RNase P cleavage sites, again variable in different riboswitch substrates, indicates an additional mRNA function, most probably lower transcription and/or translation after RNase P cleavage. We also note that the presence of an RNase P site downstream from the phylogenetically identical “switch” site in the 5′ UTR of btuB suggests that a more distal part of the 5′ UTR, aside from the “switch” site, is involved in conformational switches and should be analyzed as part of the biologically active focus.
Another explanation of our phenomenon is that the downstream gene adjacent to an RNase P cleavage site is just translated at its normal constitutive level without the effect of a small metabolite, inducing some structural change in the riboswitch. The data from our lacZ mRNA and product formation indicate that this may not be the case.
RNase P seems to be involved in regulatory functions, aside from its usual role in the biosynthesis of small stable molecules such as tRNAs (2), but we cannot tell whether this function originated during the RNA world (27) or subsequently during the further evolution of the RNA and protein worlds.
Supplementary Material
Acknowledgments
We thank Drs. Ron Breaker, Ali Nahvi, Don Crothers, and Piotr Weglenski and Mr. Jeffrey Barrick, as well as our laboratory colleagues, for helpful discussions. We also thank Dr. Nahvi (Yale University, New Haven, CT) and Dr. Weglenski (Warsaw University, Warsaw) for the gifts of relevant plasmids. We acknowledge assistance from Yale University (S.A.).
Author contributions: S.A. designed research; D.W. and C.G.-T. performed research; S.A. and Y.L. analyzed data; and S.A. wrote the paper.
Abbreviation: pTyr, precursor to E. coli tRNATyr.
References
- 1.Winkler, W. C. & Breaker, R. R. (2003) ChemBioChem 4, 1024–1032. [DOI] [PubMed] [Google Scholar]
- 2.Altman, S. & Kirsebom, L. (1999) in The RNA World, eds. Gesteland, R., Cech, T. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Woodbury, NY), 2nd Ed., pp. 351–380.
- 3.Nahvi, A., Barrick, J. E. & Breaker, R. R. (2004) Nucleic Acids Res. 32, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsuk, P., Dzikowska, A., Empel, J., Grzelak, A., Grzeskowiak, R. & Weglenski, P. (1999) Acta Biochim. Polon. 46, 391–403. [PubMed] [Google Scholar]
- 5.Li, Y. & Altman, S. (2004) J. Mol. Biol. 339, 31–39. [DOI] [PubMed] [Google Scholar]
- 6.Lyons, A. J. & Robertson, H. D. (2003) J. Biol. Chem. 278, 26844–26850. [DOI] [PubMed] [Google Scholar]
- 7.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 8.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirsebom, L. & Altman, S. (1988) J. Mol. Biol. 204, 879–888. [DOI] [PubMed] [Google Scholar]
- 10.Nahvi, A., Sudarsan, N., Ebert, M. S., Zou, X., Brown, K. L. & Breaker R. R. (2002) Chem. Biol. 9, 1043–1049. [DOI] [PubMed] [Google Scholar]
- 11.Chung, C. T., Niemela, S. L. & Miller, R. H. (1989) Proc. Natl. Acad. Sci. USA. 86, 2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrier-Takada, C., Lumelsky, N. & Altman, S. (1989) Science 246, 1578–1584. [DOI] [PubMed] [Google Scholar]
- 13.Drainas, D., Zimmerly, S., Willis, I. & Soll, D. (1989) FEBS Lett. 251, 84–88. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada, C., McClain, W. H. & Altman, S. (1984) Cell 38, 219–224. [DOI] [PubMed] [Google Scholar]
- 15.Gopalan, V., Baxevanis, A. D., Landsman, D. & Altman, S. (1997) J. Mol. Biol. 267, 818–829. [DOI] [PubMed] [Google Scholar]
- 16.Guerrier-Takada, C. & Altman, S. (1999) Methods Enzymol. 313, 442–456. [DOI] [PubMed] [Google Scholar]
- 17.Guerrier-Takada, C., Li, Y. & Altman, S. (1995) Proc. Natl. Acad. Sci. USA 92, 11115–11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H., in A Short Course in Bacterial Genetics (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 72–80.
- 19.Guerrier-Takada, C., von Belkum, A., Pleij, C. W. A. & Altman, S. (1988) Cell 53, 267–272. [DOI] [PubMed] [Google Scholar]
- 20.Mans, R. M. W., Guerrier-Takada, C., Altman, S. & Pleij, C. W. A. (1990) Nucleic Acids Res. 18, 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan, Y. & Altman, S. (1995) EMBO J. 14, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickiser, J. K., Winkler, W. C., Breaker, R. R. & Crothers, D. M. (2005) Cell 18, 49–60. [DOI] [PubMed] [Google Scholar]
- 23.Simons, R. W., Houman, F. & Kleckner, N. (1987) Gene 53, 85–96. [DOI] [PubMed] [Google Scholar]
- 24.Oh, B.-K., Frank, D. N. & Pace, N. R. (1998) Biochemistry 37, 7277–7283. [DOI] [PubMed] [Google Scholar]
- 25.Bartkiewicz, M., Gold, H. & Altman, S. (1989) Genes Dev. 3, 488–499. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y. & Altman, S. (2003) Proc. Natl. Acad. Sci. USA 100, 13213–13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert, W. (1986) Nature 319, 618. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.