Abstract
Type IV secretion is used by pathogenic microorganisms to transfer effector macromolecules to eukaryotic target cells. The VirB/D4 apparatus of Agrobacterium tumefaciens transfers DNA and proteins to plant cells. We postulated that the cell pole is the site of assembly of the A. tumefaciens type IV apparatus. Using immunofluorescence microscopy, we now demonstrate that 10 of the VirB proteins localized primarily to one cell pole and a macromolecular VirB complex is assembled at the pole. Neither the assembly of the complex nor polar localization of a VirB protein requires ATP utilization by the VirB ATPases. The requirement of other VirB proteins for the polar localization of at least six VirB proteins indicates an essential role of protein–protein interaction in polar targeting. Four proteins (VirB3, VirB4, VirB8, and VirB11) could target themselves to a cell pole independent of a VirB protein. We provide evidence that VirB6–VirB10 are the structural components of the type IV apparatus. Using strains that express defined subsets of the virB genes, we demonstrate that VirB7–VirB10 are the minimum components sufficient for the assembly of a polar VirB complex. VirB6 associates with this complex to form the type IV secretion apparatus. VirB8 functions as the assembly factor and targets the apparatus to the cell pole.
Keywords: cell pole, polar targeting protein, VirB proteins, scaffold protein
Bacteria use type IV secretion (T4S) to deliver macromolecules to plants, humans, animals, and bacteria (1, 2). Substrates transferred include, among others, bacterial conjugative plasmids, Bordetella pertussis toxin, Helicobacter pylori CagA, and several Legionella pneumophila proteins. The T4S system of the phytopathogen Agrobacterium tumefaciens transfers oncogenic DNA (T-DNA) and proteins to plant cells, causing neoplastic crown gall tumor disease. The A. tumefaciens T4S system can also direct conjugative transfer of an IncQ plasmid between bacteria and T-DNA to yeast and other eukaryotes (3–5). In addition to DNA, the system delivers at least three proteins (VirE2, VirE3, and VirF) to target cells (6, 7). Substrate transfer requires virB and virD4, genes conserved in most T4S families.
The VirB proteins are thought to assemble a T4S apparatus that spans the bacterial membranes, forming a conduit for substrate translocation. The coupling factor VirD4 presumably facilitates interaction between a substrate and the apparatus (8). Ten VirB proteins (VirB2–VirB11) are essential for DNA transfer and the assembly of a T-pilus composed primarily of VirB2 (9–11). The VirB2 homolog of the Escherichia coli plasmid RP4, TrbC, assembles a RP4 pilus (12). VirB3, an F plasmid TraL homolog, and VirB5 are minor components of the T-pilus (13, 14). VirB4 and VirB11, the two ATPases, probably provide energy required for the assembly and/or function of the transport apparatus.
The remaining five VirB proteins (VirB6–VirB10) are postulated to be the “core components” of the T4S apparatus (15, 16). VirB6 may serve a dual role, also contributing as an assembly factor for the T-pilus (17). Biochemical, genetic, and cellular analyses have uncovered many interactions among the VirB family of proteins that may be essential for apparatus assembly. VirB9 form a disulfide-linked complex with the lipoprotein VirB7 (18–21). VirB6 interacts with VirB7 and VirB9 and is necessary for the accumulation of a VirB7 homodimer (17, 22). Complexes of VirB6 with VirB7, VirB9, and VirB10 were identified by immunoprecipitation (23). VirB8, VirB9, VirB10, and their homologues interact among themselves and with other VirB proteins (24–26). VirB9 is necessary for the formation of higher-order oligomers of VirB10 (27). In biochemical fractionation and T-DNA immunoprecipitation studies, many of these proteins were found in high-molecular-weight protein complexes associated with the bacterial membranes and to contact the T-DNA during its transit through the T4S apparatus (28, 29).
Subcellular localization of the VirB proteins by immunomicroscopy showed that VirB9 and VirB10 form protein complexes and these complexes localize to a few sites on the cell membranes (30). VirB8 is required for the formation of the VirB9 and VirB10 complexes. Another component essential for T4S in most bacteria, VirD4, localized primarily to a cell pole (31). The cell pole is also the site of assembly of the T-pilus, and A. tumefaciens attaches to plant cells in a polar manner (32, 33). These observations led us to propose that the cell pole is the site of assembly of the T4S apparatus (31). In support of this hypothesis, Atmakuri et al. (34) observed that VirD4 could target a GFP–VirE2 fusion to a cell pole. We recently reported that another component of the T4S apparatus, VirB6, localizes to a cell pole and several VirB proteins are required for its polar localization (16). To further investigate the site of assembly of the A. tumefaciens T4S apparatus and determine the mechanism of its polar targeting, we analyzed the subcellular location of the individual VirB proteins.
In the present study, we demonstrate that 10 VirB proteins localize to a cell pole and polar localization of six depends on other VirB proteins. We demonstrate that assembly of the A. tumefaciens apparatus does not require a VirB energy source. We identified the minimum components necessary for the localization of a VirB protein complex at a cell pole and demonstrate that VirB8 is the scaffold for polar assembly of the A. tumefaciens apparatus.
Materials and Methods
Strains and Plasmids. A. tumefaciens A136, A348, and A348ΔB contain no Ti-plasmid, the octopine Ti-plasmid pTiA6, and pTiA6 with a deletion in virB, respectively. Strains PC1001–1011 are derivatives of A. tumefaciens A348 with a nonpolar deletion in each virB gene (9). PC1004(virB4K439Q) and PC1011(virB11K175Q) harbor plasmid pBB15 with a K439Q mutation in virB4 and pPCB7113 with a K175Q mutation in virB11, respectively (35, 36).
Mutants of A. tumefaciens that express a subset of the virB genes were constructed by incorporating a virBn::phoA into the Tiplasmid pTiA6 by marker exchange (15, 18). All strains were confirmed by virulence and Western blot assays (31). Strains PJ111–PJ114 and AD997 are derivatives of A. tumefaciens A348 with a polar phoA insertion in virB2 (at codon 121), virB5 (codon 50), virB6 (codon 103), virB10 (codon 91), and virB9 (codon 36), respectively. Strains PJ115 and PJ116 were constructed by incorporating the virB5::phoA insertion into PC1003 (A348ΔB3) and PC1004 (A348ΔB4), respectively. Plasmids pAD1758, pAD1536, pLA17, pAD1746, pAD1746Δ11, and pAD1746Δ8 express virB3, virB8, virB7–virB10, virB6–virB11, virB6–virB10, and virB6, virB7, virB9–virB11, respectively (described in Supporting Text, which is published as supporting information on the PNAS web site). Plasmids were introduced into A. tumefaciens A348ΔB by electroporation (18).
Anti-VirB Antibodies. Antibodies against the VirB proteins were raised in rabbits by injecting a TrpE–VirBn fusion protein purified from overproducing E. coli (see Table 4, which is published as supporting information on the PNAS web site). A synthetic peptide corresponding to 15 C-terminal residues of VirB3 was used as antigen for anti-VirB3 antibodies (Tana Biosystems, Houston). Antigen immobilized by coupling to Affi-gel-10/15 (Bio-Rad) was used as an affinity matrix for antibody purification by using established procedures (37). For colocalization experiments, antibodies against VirB8 were raised in chicken by using the TrpE–VirB8 fusion as an antigen (Cocalico Biologicals, Reamstown, PA). Anti-VirB8 IgY was isolated from egg yolk by precipitation with polyethylene glycol 6000 (3.5–8.5% fraction).
Immunofluorescence Microscopy (IFM). Subcellular location of the Vir proteins was determined by IFM essentially as described by Judd et al. (16). Bacteria were grown overnight in AB (pH 5.8) liquid medium in the absence or presence of 100 μM acetosyringone, as appropriate, to midlog phase (A600 ≈0.5) at 20°C. PJ118 and PJ121 were grown in AB medium containing 1 mM isopropyl β-d-thiogalactoside. Samples were viewed in a Nikon E800 fluorescence microscope equipped with a CoolCam 2000, liquid-cooled, three-chip color video camera. Images were processed with Adobe Systems (San Jose, CA) photoshop 6.0 for presentation.
Colocalization of VirB8 and the other VirB proteins or VirD4 in A. tumefaciens A348 was investigated by probing first with one primary antiserum followed by the cognate secondary antibodies, and then with the second primary antibodies and the cognate secondary antibodies. The order of addition of the primary antibodies had no effect on final results. For quantitative analysis, bacteria labeled with both probes were used. Foci were considered coincident when at least half of each focus overlapped with the other (38).
Results
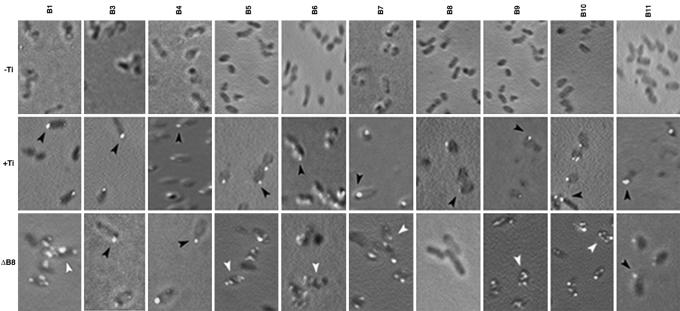
The VirB Proteins Localize to a Cell Pole. To identify the components of the A. tumefaciens T4S apparatus and decipher the mechanism of its assembly, we determined the subcellular location of the VirB proteins by IFM. Polyclonal antibodies against 10 of the 11 VirB proteins were raised and purified by affinity chromatography. Specificity of the antibodies was determined by both Western blot assays and IFM. Specificity of anti-VirB6, -VirB8, -VirB9, -VirB10, and -VirD4 has been demonstrated (16, 30, 31). In Western blot assays, antibodies against the remaining VirB proteins reacted with a single protein present in extracts of induced A. tumefaciens (see Fig. 4, which is published as supporting information on the PNAS web site). In IFM, only bacteria that expressed the cognate antigen were fluorescently labeled (Fig. 1).
Fig. 1.
Subcellular location of the VirB proteins was determined by IFM (16). Induced bacteria were probed with rabbit anti-VirBn (1:10–600 dilution) or chicken anti-VirB8 (1:20,000) antibodies at 4°C followed by Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes). Strains analyzed were A. tumefaciens A136 (-Ti), A348 (+Ti), and A348ΔB8 (ΔB8). Polar and nonpolar foci are identified by black and white arrowheads, respectively. (Magnifications: ×1,500–2,500.)
We previously reported that three VirB proteins (VirB8, VirB9, and VirB10) localized to several sites on the cell membrane (30). Our subsequent study on VirD4, another component essential for T4S, showed that VirD4 localizes to a cell pole (31). These observations and a report on a temperature-sensitive phenotype of VirB protein accumulation and tumor formation (39) prompted us to examine all of the VirB proteins under conditions that support tumorigenicity. At a growth temperature of 28°C or above, a condition routinely used for bacterial growth in the laboratory, many VirB proteins are unstable (39). At this temperature, the T-pilus does not elaborate and virulence of most A. tumefaciens strains is strongly inhibited. At the lower growth temperature of 20°C, all VirB proteins accumulated at a similar level and no defect in either T-pilus assembly or virulence was observed (39). To maximize assembly of a functional T4S apparatus, bacteria grown to midlog phase at 20°C were used for the analysis of the subcellular location of the VirB proteins.
Analysis of WT A. tumefaciens A348 by IFM showed that 10 VirB proteins localized primarily to a cell pole (Fig. 1). In control experiments, the antibodies exhibited little or no nonspecific labeling of bacteria. Only anti-VirB2 antibodies, although highly specific in Western blot assays (11), showed significant nonspecific fluorescence in IFM. They were not used in further studies. Quantitative analysis showed that 77–89% of foci in bacteria probed with all antibodies except for anti-VirB4 antibodies localized to the cell poles (Table 1). The frequency of VirB4 foci at a cell pole was somewhat lower (57%) but is significantly above that expected from a random occurrence. To determine whether the VirB proteins are unipolar, we analyzed bacteria that contained polar foci. In those cells, 72–88% of the foci mapped to one pole, strongly suggesting that the VirB proteins are unipolar (Table 1).
Table 1. Subcellular localization of the VirB proteins.
| Probe antibodies | No. of foci (no. of cells) | No. of foci at a pole | Percent polar foci | No. of cells with polar foci | No. of cells with foci at one pole | Percent unipolar |
|---|---|---|---|---|---|---|
| VirB1 | 242 (195) | 201 | 83 | 164 | 141 | 86 |
| VirB3 | 243 (200) | 213 | 88 | 177 | 145 | 82 |
| VirB4 | 245 (194) | 139 | 57 | 112 | 90 | 80 |
| VirB5 | 186 (156) | 143 | 77 | 127 | 107 | 84 |
| VirB7 | 232 (203) | 178 | 77 | 158 | 139 | 88 |
| VirB8 | 238 (196) | 209 | 88 | 172 | 139 | 81 |
| VirB9 | 270 (200) | 239 | 89 | 177 | 127 | 72 |
| VirB10 | 193 (162) | 168 | 87 | 153 | 133 | 87 |
| VirB11 | 223 (180) | 175 | 78 | 151 | 118 | 78 |
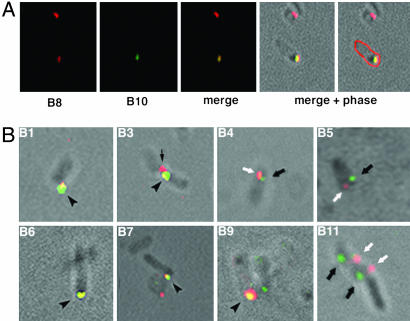
Many VirB Proteins Colocalize with VirB8. Polar localization of the VirB proteins suggests that these proteins can form a protein complex. To test this possibility and identify the components of the complex, colocalization experiments were performed. We examined whether VirB8, a core component of the T4S apparatus, colocalizes with the other components of the transport apparatus. In an analysis of VirB8–VirB10 colocalization, bacteria probed with chicken anti-VirB8 antibodies exhibited polar red fluorescence and those probed with the rabbit anti-VirB10 antibodies fluoresced green (Fig. 2A). When the two images were superimposed, some cells showed polar yellow foci. The yellow foci color, resulting from the mixture of green and red, indicated that VirB8 and VirB10 do colocalize. Similar analyses indicated that five other VirB proteins (VirB1, VirB3, VirB6, VirB7, and VirB9) colocalized with VirB8 (Fig. 2B). Quantitative analysis showed that 60–90% of the foci were coincident (Table 2). Three VirB proteins (VirB4, VirB5, and VirB11) did not colocalize with VirB8 (Table 2 and Fig. 2B). Bacteria labeled with these antibody pairs exhibited noncoincident green and red foci predominantly at the same pole.
Fig. 2.
Colocalization of VirB8 and other Vir proteins. Induced A. tumefaciens A348 were processed for IFM and probed successively with chicken anti-VirB8 antibodies, Alexa Fluor 594 (red)-conjugated goat anti-chicken antibodies, rabbit anti-VirBn antibodies, and Alexa Fluor 488 (green)-conjugated goat anti-rabbit antibodies. (A) Colocalization of VirB8 and VirB10. Shown (left to right) are cells probed with anti-VirB8, anti-VirB10, merged image of anti-VirB8 and anti-VirB10, and merged plus phase image. In the far right, the outline of the labeled cell was artificially colored red. (B) Merged images of localization of VirB8 and the indicated VirB protein are shown. Yellow coincident foci are indicated by arrowheads, green VirBn foci are indicated by black arrows, and red VirB8 foci are indicated by white arrows. The red focus in B3 (thin arrow) is from a second bacterium. (Magnifications: ×1,500–2,500.)
Table 2. Colocalization of VirB8 and other Vir proteins.
| Vir protein | No. of cells analyzed | No. of pairs counted | Percentage coincidence |
|---|---|---|---|
| VirB1 | 66 | 66 | 89 |
| VirB3 | 61 | 63 | 60 |
| VirB4 | 97 | 120 | 12 |
| VirB5 | 50 | 52 | 35 |
| VirB6 | 52 | 52 | 88 |
| VirB7 | 101 | 109 | 65 |
| VirB9 | 108 | 132 | 83 |
| VirB10 | 54 | 58 | 86 |
| VirB11 | 76 | 76 | 29 |
| VirD4 | 58 | 62 | 87 |
In addition to the VirB proteins, substrate transfer from A. tumefaciens requires VirD4. To determine whether VirD4 localizes with or near VirB8, we examined the subcellular location of both proteins in the same bacterium. Like most VirB proteins, VirD4 colocalized with VirB8 (Table 2), suggesting that VirD4 is either associated with or in close proximity to the VirB protein complex. In another study, we observed that VirD4 colocalizes with a second component of the T4S apparatus, VirB6 (16).
Polar Localization of Most VirB Proteins Requires VirB8. The polar location of the VirB proteins and colocalization of several proteins suggest that the VirB proteins form a complex at the cell pole. How does a protein find the cell pole? A protein can be targeted to the cell pole because of its intrinsic affinity for the pole or through interaction with a polar protein. A candidate for an interacting protein is VirB8 because it participates in many interactions with other VirB proteins and is indispensable in the formation of VirB complexes (24, 25, 30). To determine the role of VirB8 in polar localization of other VirB proteins, we determined their subcellular location in a nonpolar virB8 deletion mutant. Six proteins (VirB1, VirB5–VirB7, VirB9, and VirB10) failed to localize to the cell pole in PC1008, indicating that VirB8 is required for their polar localization (Fig. 1, white arrowhead). The mutation had no effect on polar localization of VirB3, VirB4, and VirB11. Because the virB8 mutant is likely to be defective in the assembly of the apparatus, polar localization of VirB3, VirB4, and VirB11 does not require T4S apparatus assembly.
Polar Localization Does Not Require the VirB ATPases. VirB4 and VirB11, the two VirB proteins with nucleotide-binding motif, can bind and probably hydrolyze ATP (35, 36). ATP utilization may be required for the assembly and/or function of the T4S apparatus. To determine whether the T4S apparatus assembly is an energy-dependent process, we determined the subcellular location of VirB proteins in two well characterized null mutants defective in ATP utilization, virB4K439Q and virB11K175Q (35, 36). All VirB proteins, including the mutant proteins, localized to the cell pole in both strains (Table 3). Therefore, polar assembly of the VirB protein complex does not require ATP hydrolysis.
Table 3. Subcellular location of the VirB proteins in Agrobacterium strains.
| Strain | VirB expressed | VirB protein, subcellular location* |
|---|---|---|
| PC1004K439Q | VirB1–VirB3, VirB4K439Q, VirB5–VirB11 | All, polar |
| PC1011K175Q | VirB1–VirB10, VirB11K175Q | All, polar |
| AD997 | VirB1–VirB8, VirB936–PhoA | VirB3, VirB4, VirB8, polar |
| VirB1, VirB5–VirB7, distributed | ||
| PC1002 | VirB1, VirB3–VirB11 | All, polar |
| PC1009 | VirB1–VirB8, VirB10, VirB11 | VirB3, VirB8, polar |
| VirB1, VirB5, VirB7, VirB10, distributed | ||
| PC1011 | VirB1–VirB10 | VirB7–VirB10, polar |
| VirB6, distributed | ||
| PJ111 | VirB1, VirB2121–phoA | VirB1, distributed |
| PJ112 | VirB1–VirB4, VirB550–PhoA | VirB3, VirB4, polar |
| PJ114 | VirB1–VirB9, VirB1091–PhoA | VirB3, VirB8, polar |
| VirB7, VirB9, distributed | ||
| PJ115 | VirB1, VirB2, VirB4, VirB550–phoA | VirB4, polar |
| PJ116 | VirB1–VirB3, VirB550–phoA | VirB3, polar |
| PJ117 | VirB8 | VirB8, polar |
| PJ118 | VirB11K175Q | VirB11K175Q, polar |
| PJ120 | VirB3 | VirB3, polar |
| PJ121 | VirB4K439Q | VirB4K439Q, polar |
| PJ122 | VirB7–VirB10 | VirB7–VirB10, polar |
| PJ123 | VirB6–VirB10 | VirB7–VirB10, polar |
| VirB6, distributed |
Subcellular location of VirB2 was not determined
Four VirB Proteins Can Target Themselves to a Cell Pole. Studies with A348ΔB8 (PC1008) suggested that assembly of the T4S apparatus is not required for the polar localization of VirB3, VirB4, and VirB11 (Fig. 1). To determine the requirements for polar localization of these and other VirB proteins, we analyzed mutants that express a defined subset of the virB genes. Analysis of AD997, a strain with a polar virB9::phoA insertion, indicated that VirB8 also localizes to the cell pole in the absence of the T4S apparatus. In this strain, VirB3, VirB4, and VirB8 localized to the cell poles (Table 3). The remaining VirB proteins were found distributed throughout the cell surface. A similar analysis of PC1002, a mutant with a nonpolar deletion in the virB2 pilin gene, indicated that T-pilus assembly is not required for polar localization of any VirB protein.
Four VirB proteins (VirB3, VirB4, VirB8, and VirB11) do not require the T4S apparatus for polar localization. To determine the requirement of VirB3 and VirB4 localization, we analyzed mutants that express virB1–virB4 or their subset. Results presented in Table 3 suggested that no other VirB protein is required for the polar localization of VirB3 or VirB4. To verify this conclusion, we determined the subcellular location of VirB3 and VirB4K439Q in bacteria that express no additional virB gene. Each protein was competent in self-targeting to the cell pole. Similar analysis of bacteria that expressed virB8 or virB11K175T showed that these two proteins were also competent in targeting themselves to the cell pole (Table 3). Therefore, no other VirB protein is required for polar localization of VirB3, VirB4, VirB8, and VirB11.
Polar localization of several VirB proteins required VirB8 (Fig. 1). Additional VirB proteins are also required for the polar localization of these proteins. Polar localization of VirB6 required VirB7–VirB11, VirB7 required VirB8–VirB10, VirB9 required VirB8 and VirB10, and VirB10 required VirB7–VirB9 (Table 3, ref. 16, and data not shown). The complete requirements for VirB1 and VirB5 localization are not yet known, but at least VirB8 and VirB9 are required for their polar localization (Fig. 1 and Table 3).
VirB8 Is the Scaffold for the Assembly of the T4S Apparatus at a Cell Pole. VirB6–VirB11 are the putative core constituents of the T4S apparatus (16). If these proteins are sufficient for targeting the T4S apparatus to a cell pole, all six proteins will localize to the pole in bacteria expressing virB6--virB11. VirB8 and VirB11 are the only two among the six proteins that could localize to a cell pole in the absence of another VirB protein (Table 3). This property makes them candidates for the “polar targeting factor,” a protein that targets the T4S apparatus to a cell pole. Because VirB8 is required for the formation of VirB9 and VirB10 protein complexes and a complex of VirB7–VirB10 or their homologues is functional in importing DNA to bacteria (3, 30, 40–42), we strongly favor VirB8 to be the polar targeting factor.
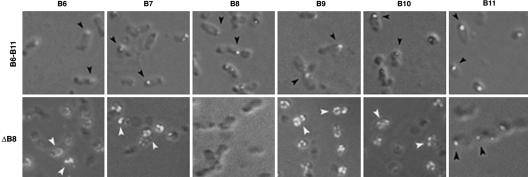
To test this hypothesis, we first determined the role of VirB11 in polar localization of the other VirB proteins. In the virB11 deletion mutant PC1011, VirB7–VirB10 localized to the cell pole (Table 3). VirB11 is therefore not required for the polar localization of these proteins. We then determined whether VirB6–VirB11 are sufficient for their polar localization. All six VirB proteins localized to the cell pole in A. tumefaciens PJ119 that express virB6–virB11 (Fig. 3). None of the proteins, except for VirB11, was found at the cell pole in an isogenic strain with a nonpolar deletion in virB8. Therefore, VirB8 is required for polar targeting of the VirB protein complex and VirB11 is not sufficient for this process. In Western blot assays, no significant effect of VirB8 on the level of the other proteins was observed (data not shown). To identify the minimum subset of proteins required for the formation of a polar VirB complex, we analyzed A. tumefaciens strains that express virB6--virB10 or virB7–virB10 (Table 3). VirB7–VirB10 localized to the cell pole in both strains. VirB6, however, exhibited no preference for the pole in the absence of VirB11. These results indicate that VirB8, but not VirB6 or VirB11, is required to target VirB7, VirB9, and VirB10 to a cell pole.
Fig. 3.
Minimum components required for the polar assembly of a VirB protein complex. Subcellular localization of the VirB proteins in bacteria expressing virB6–virB11 or virB6–virB11 minus virB8 (ΔB8) was determined by IFM. Polar foci, black arrowhead; nonpolar, white arrowhead. (Magnifications: ×1,500–2,500.)
Discussion
T4S involves the assembly of a multiprotein complex at the bacterial membranes. The mechanism of complex assembly, the components of the complex, and its distribution on the cell surface are largely unknown. The cell pole has recently been implicated in T4S (16, 31, 34). In the present study, we demonstrate that the cell pole is the site of assembly of the A. tumefaciens T4S apparatus and report the mechanism of its assembly. Ten of the 11 VirB proteins, the primary constituents of the T4S apparatus, localized to a single cell pole (Fig. 1 and Table 1). Polar localization of four proteins (VirB3, VirB4, VirB8, and VirB11) did not require other VirB proteins (Table 3). The remaining proteins localized to the cell pole in a VirB-dependent manner and form a protein complex at the cell pole. Neither complex assembly nor polar localization of a VirB protein required the ATP-binding motifs of the VirB4 and VirB11 ATPases.
All proteins in a multiprotein complex will be near one another and localize to the same site on the cell membrane. Colocalization studies indicated that VirD4 and six VirB proteins (VirB1, VirB3, VirB6, VirB7, VirB9, and VirB10) are in close proximity of VirB8, a core component of the T4S apparatus (Table 2 and Fig. 2). Of these, VirB3 and VirD4 can independently target themselves to the cell pole in the absence of another Vir protein (Table 3 and ref. 31). Colocalization of these two proteins with VirB8 can therefore be caused by their localization to adjacent subcellular sites in the polar region. The remaining five proteins (VirB1, VirB6, VirB7, VirB9, and VirB10), however, did not localize to the cell pole in the absence of other VirB proteins and required more than one VirB protein for their polar localization (Table 3 and ref. 16). The requirement for multiple proteins that do not share a common target strongly suggests that polar localization of these proteins results from an interaction with another protein present in a protein complex, and the six proteins (VirB1 and VirB6–VirB10) are components of a polar VirB complex. A recent report indicating that a VirB6–GFP fusion localized to a cell pole in the absence of other VirB proteins is in disagreement with our observations (23). We demonstrated that VirB6 did not localize to the cell pole when expressed from the native Ti-plasmid carrying a nonpolar deletion in any of five virB genes (virB7–virB11) as well as when expressed in the absence of other virB genes (16). Our studies identified that the C-terminal end of VirB6 contains signals for polar localization. It is likely that in the other study the addition of GFP to the C terminus of VirB6 induced a structural alteration, leading to its mislocalization to the cell pole.
Reconstruction experiments presented here demonstrated that coexpression of VirB7–VirB10 is sufficient for their polar localization. Among the four, only VirB8 can localize to a cell pole in the absence of another VirB protein; therefore, it must function in targeting the others to the cell pole through protein–protein interactions. Interactions of VirB8 with VirB9 and VirB10 and that of VirB7 with VirB9 have been reported (18, 21, 24, 25). VirB1 and VirB6, two proteins that colocalized with VirB8 and required other VirB proteins for polar localization, must interact with the VirB7–VirB10 complex, making them members of the polar VirB complex. Because VirB1 is not an absolute requirement for DNA transfer, our results strongly suggest that the T4S core structure is composed of five proteins, VirB6–VirB10.
Three VirB proteins (VirB4, VirB5, and VirB11) did not colocalize with VirB8. The lack of colocalization can be a result of experimental constraints caused by an inherent difference in the relative strength of antigen recognition by different antibodies. However, this explanation is unlikely because both VirB8 and VirB4/5/11 were found at the same pole in a bacterium but at different regions of the pole. An alternate explanation that we favor is that these three proteins are not structural components of the T4S apparatus. Two of the proteins (VirB4 and VirB11) interact with VirD4 and other VirB proteins (43). These observations raise the questions of whether the interactions take place in vivo and how they can occur when the interacting proteins are >200 nm (typical resolution of fluorescence microscope) away from each other. Although all interactions observed in solution may not take place in the native membrane environment, a condition in which the present study was conducted, a more likely explanation is these proteins are functional, and not structural, components of T4S and their interaction with the T4S apparatus is dynamic in nature. Subtle changes in the subcellular location of the components in a substrate/recipient-dependent process can facilitate interactions necessary for substrate delivery through the secretion channel. Dynamic localization of bacterial proteins and origins of replication has been observed (44). In addition, dissimilar localization of two subunits of an enzyme, topoisomerase IV, has been observed in several bacterial systems (45). Although the ParC subunit colocalized with the replisome at a cell pole, the ParE subunit of the ParC2ParE2 tetramer did not. ParE was dispersed throughout the cell cytoplasm or was restricted to the nucleoid-free region. A functional ParE subunit is essential for polar ParC association with the replisome.
VirB8 directs assembly of the VirB complex at the cell pole. How does this protein (and VirB3, VirB4, VirB11, and VirD4) find the cell pole? In Caulobacter, PodJ serves as a localization factor that targets pilus and signaling proteins to the cell pole (46). A homolog of podJ exists in A. tumefaciens and may serve a similar role for T4S apparatus assembly. Several observations suggest that polar location of the T4S apparatus is probably a common theme in α-proteobacteria. For example, the T-pilus elaborates at a cell pole, and A. tumefaciens attaches to plant cells in a polar manner (32, 33). A filamentous H. pylori surface organelle, part of the T4S system, is often located in one bacterial pole (47). The possible exception is the T4S system of bacterial conjugative plasmids where donor and recipient cells form a conjugation junction over a large area of the cell surfaces (48). The small size of a bacterium restricts the surface area available for cell-to-cell contact necessary for conjugation. A random distribution of the conjugation apparatus will ensure maximal availability of the cell surface for a high efficiency of plasmid transfer. The target for other bacteria that possess a T4S system is an eukaryotic cell. The large size of the target would permit simultaneous attachment of many bacteria to a single cell. The efficiency of substrate delivery in this case is maximized by mass effect.
Two DNA transfer proteins that colocalized with VirB8 and are independently targeted to the cell pole are VirB3 and VirD4. Polar location of VirB3 may be required for its role in the biogenesis of the T-pilus. The coupling protein VirD4 presumably targets a substrate to the T4S apparatus. Structural analysis of TrwB, a VirD4 homolog, showed that the hexameric protein contains a central cavity that can accommodate the transferred DNA (49). It is proposed to form a channel through the cytoplasmic membrane to allow substrate export. Another possibility is that VirD4 delivers a substrate to the T4S apparatus. In this alternative model a VirB protein, e.g., VirB6, would form the cytoplasmic membrane channel. To serve either function, VirD4 needs to be in close proximity with the T4S apparatus, therefore, at a cell pole. Our results agree well with a recent study that outlined the contact of T-DNA with the proteins required for T4S (29). Using T-DNA immunoprecipitation assays the T-DNA was found to directly interact sequentially with VirD4, VirB11, VirB6, VirB8, VirB9, and VirB2.
How is the T4S apparatus assembled? At least four VirB proteins (VirB7–VirB10) are required for complex formation at a cell pole. This complex is biologically active in DNA import into A. tumefaciens in a vir-mediated plasmid transfer assay (3). A similar complex composed of the Com proteins, the VirB7–VirB10 homologues in H. pylori and Campylobacter jejuni, probably functions in DNA uptake in these naturally competent bacteria (40–42). The next step is the VirB11-dependent recruitment of VirB6 into the complex to form the export-competent T4S apparatus. VirB1 associates with this complex facilitating apparatus assembly. The other VirB proteins most likely participate in T-pilus biogenesis and substrate translocation. Whether the T-pilus caps the VirB complex to form a channel through the outer membrane is not known. Biochemical studies identified subcomplexes composed predominantly of the pilus components and the core complex (28). Association of these complexes may lead to channel formation across both the inner and outer membranes.
The VirB proteins are sufficient for substrate export across the bacterial membranes and to eukaryotic targets. The assembly of the A. tumefaciens T-pilus does not require VirD4 but requires all of the VirB proteins (32). Brucella suis and B. pertussis genomes do not encode a VirD4 homolog, but both bacteria are fully pathogenic. A review of substrates that differ in VirD4 requirement shows that the VirD4 requiring substrates, e.g., H. pylori CagA, A. tumefaciens T-DNA, VirD2, VirE2, VirE3, and VirF, and bacterial plasmids, are located in the cell cytoplasm, whereas the VirD4-independent ones, namely pertussis toxin subunits S1–S5, are periplasmic proteins. VirD4 is probably required for the export of a cytoplasmic substrate and functions in bringing the substrate to the transport apparatus. It is possible that VirB3 serves a similar coupling role in delivering the pilin proteins to the T4S apparatus for T-pilus assembly. Consistent with this hypothesis is the recent observation of a Bartonella henselae VirB3–VirB5 interaction in two-hybrid assays in yeast (26).
Supplementary Material
Acknowledgments
We thank Peter Christie (University of Texas, Houston) for the A. tumefaciens mutants; Lorraine Anderson, N. Rao Madamanchi, Yong-Hong Xie, Shane Caine, and Tony Johnson for generation and purification of anti-VirB antibodies; and Duncan Clarke, Mark R. McClellan, and Mark Sanders for assistance with microscopy. This work was supported by grants from the University of Minnesota Agricultural Experiment Station and the National Science Foundation. P.K.J. was an Arnold H. Johnson Fellow of the Biochemistry, Molecular Biology, and Biophysics Graduate Program.
Author contributions: P.K.J., R.B.K., and A.D. designed research; P.K.J. and R.B.K. performed research; P.K.J. and A.D. contributed new reagents/analytical tools; P.K.J., R.B.K., and A.D. analyzed data; and P.K.J. and A.D. wrote the paper.
Abbreviations: T4S, type IV secretion; IFM, immunofluorescence microscopy.
References
- 1.Cascales, E. & Christie, P. (2003) Nat. Rev. Microbiol. 1, 137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Covacci, A., Telford, J., Del Giudice, G., Parsonnet, J. & Rappuoli, R. (1999) Science 284, 1328-1333. [DOI] [PubMed] [Google Scholar]
- 3.Liu, Z. & Binns, A. N. (2003) J. Bacteriol. 185, 3259-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bundock, P., den Dulk-Ras, A., Beijersbergen, A. & Hooykaas, P. (1995) EMBO J. 14, 3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piers, K., Heath, J., Liang, X., Stephens, K. & Nester, E. (1996) Proc. Natl. Acad. Sci. USA 93, 1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vergunst, A., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C., Regensburg-Tuink, T. & Hooykaas, P. (2000) Science 290, 979-982. [DOI] [PubMed] [Google Scholar]
- 7.Schrammeijer, B., den Dulk-Ras, A., Vergunst, A., Jurado, J. & Hooykaas, P. (2003) Nucleic Acids Res. 31, 860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabezon, E., Sastre, J. & de la Cruz, F. (1997) Mol. Gen. Genet. 254, 400-406. [DOI] [PubMed] [Google Scholar]
- 9.Berger, B. & Christie, P. (1994) J. Bacteriol. 176, 3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fullner, K., Lara, J. & Nester, E. (1996) Science 273, 1107-1109. [DOI] [PubMed] [Google Scholar]
- 11.Lai, E. & Kado, C. (1998) J. Bacteriol. 180, 2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbrandt, R., Kalkum, M., Lai, E., Lurz, R., Kado, C. & Lanka, E. (1999) J. Biol. Chem. 274, 22548-22555. [DOI] [PubMed] [Google Scholar]
- 13.Shirasu, K. & Kado, C. (1993) FEMS Microbiol. Lett. 111, 287-294. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Eisenlohr, H., Domke, N., Angerer, C., Wanner, G., Zambryski, P. & Baron, C. (1999) J. Bacteriol. 181, 7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, A. & Xie, Y.-H. (1998) Mol. Microbiol. 27, 405-414. [DOI] [PubMed] [Google Scholar]
- 16.Judd, P., Kumar, R. & Das, A. (2005) Mol. Microbiol. 55, 115-124. [DOI] [PubMed] [Google Scholar]
- 17.Hapfelmeier, S., Domke, N., Zambryski, P. & Baron, C. (2000) J. Bacteriol. 182, 4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson, L., Hertzel, A. & Das, A. (1996) Proc. Natl. Acad. Sci. USA 93, 8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farizo, K., Cafarella, T. & Burns, D. (1996) J. Biol. Chem. 271, 31643-31649. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez, D., Dang, T. A., Spudich, G., Zhou, X.-R., Berger, B. & Christie, P. (1996) J. Bacteriol. 178, 3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spudich, G., Fernandez, D., Zhou, X.-R. & Christie, P. (1996) Proc. Natl. Acad. Sci. USA 93, 7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakubowski, S., Krishnamoorthy, V. & Christie, P. (2003) J. Bacteriol. 185, 2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubowski, S., Krishnamoorthy, V., Cascales, E. & Christie, P. (2004) J. Mol. Biol. 341, 961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das, A. & Xie, Y.-H. (2000) J. Bacteriol. 182, 758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward, D., Draper, O., Zupan, J. & Zambryski, P. (2002) Proc. Natl. Acad. Sci. USA 99, 11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shamaei-Tousi, A., Cahill, R. & Frankel, G. (2004) J. Bacteriol. 186, 4796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaupre, C., Bohne, J., Dale, E. & Binns, A. (1997) J. Bacteriol. 179, 78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krall, L., Wiedemann, U., Unsin, G., Weiss, S., Domke, N. & Baron, C. (2002) Proc. Natl. Acad. Sci. USA 99, 11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cascales, E. & Christie, P. (2004) Science 304, 1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, R., Xie, Y.-H. & Das, A. (2000) Mol. Microbiol. 36, 608-617. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, R. & Das, A. (2002) Mol. Microbiol. 43, 1523-1532. [DOI] [PubMed] [Google Scholar]
- 32.Lai, E.-M., Chesnokova, O., Banta, L. & Kado, C. (2000) J. Bacteriol. 182, 3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthysse, A. (1987) J. Bacteriol. 169, 313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atmakuri, K., Ding, Z. & Christie, P. (2003) Mol. Microbiol. 49, 1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger, B. & Christie, P. (1993) J. Bacteriol. 175, 1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashkova, S., Spudich, G. & Christie, P. (1997) J. Bacteriol. 179, 583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harlaw, E. & Lane, D. (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 38.Kahng, L. S. & Shapiro, L. (2003) J. Bacteriol. 185, 3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baron, C., Domke, N., Beinhofer, M. & Hapfelmeier, S. (2001) J. Bacteriol. 183, 6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofreuter, D., Odenbreit, S. & Haas, R. (2001) Mol. Microbiol. 41, 379-391. [DOI] [PubMed] [Google Scholar]
- 41.Bacon, D., Alm, R., Burr, D., Hu, L., Kopecko, D., Ewing, C., Trust, T. & Guerry, P. (2000) Infect. Immun. 68, 4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen, J. C., Szymanski, C. & Guerry, P. (2004) J. Bacteriol. 186, 6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atmakuri, K., Cascales, E. & Christie, P. J. (2004) Mol. Microbiol. 54, 1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gitai, Z., Dye, N. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, S. C. & Shapiro, L. (2004) Proc. Natl. Acad. Sci. USA 101, 9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viollier, P. H., Sternheim, N. & Shapiro, L. (2002) Proc. Natl. Acad. Sci. USA 99, 1383-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohde, M., Puls, J., Buhrdorf, R., Fischer, W. & Haas, R. (2003) Mol. Microbiol. 49, 219-234. [DOI] [PubMed] [Google Scholar]
- 48.Samuels, A., Lanka, E. & Davies, J. (2000) J. Bacteriol. 182, 2709-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomis-Ruth, F., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637-641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.