Abstract
Misfolded neuronal proteins have been identified in a number of neurodegenerative disorders and have been implicated in the pathogenesis of diseases that include Alzheimer's disease, Parkinson's disease, prion-based dementia, Huntington's disease (HD), and other polyglutamine diseases. Although underlying mechanisms remain the subject of ongoing research, it is clear that aberrant processing, protein degradation, and aggregate formation or spurious protein association of the abnormal neuronal proteins may be critical factors in disease progression. Recent work in these diseases has demonstrated in vitro that specific engineered antibody species, peptides, or other general agents may suppress the formation of aggregates. We have modified an approach with intracellularly expressed single-chain Fv (sFv) antibodies (intrabodies) that bind with unique HD protein epitopes. In cell and tissue culture models of HD, anti-N-terminal huntingtin intrabodies (C4 sFv) reduce aggregation and cellular toxicity. Here, we present the crucial experiment of intrabody-mediated in vivo suppression of neuropathology, using a Drosophila model of HD. In the presence of the C4 sFv intrabody, the proportion of HD flies surviving to adulthood increases from 23% to 100%, and the mean and maximum lifespan of adult HD flies is significantly prolonged. Neurodegeneration and formation of visible huntingtin aggregates are slowed. We conclude from this investigation that engineered intrabodies are a potential new class of therapeutic agents for the treatment of neurodegenerative diseases. They may also serve as tools for drug discovery and validation of sites on mutant neuronal proteins that could be exploited for rational drug design.
Keywords: neurodegeneration, antibody engineering, immunotherapy, nanomedicine
Huntington's disease (HD) is a genetic disorder caused by the pathological expansion of a polyglutamine (polyQ) tract in the huntingtin (htt) protein, resulting in neurodegeneration and premature death of the individual (1). PolyQ stretches of 35 or more residues are associated with pathology. In these cases, mutant htt is found to misfold (2, 3) and form aggregates in the brain and other affected tissues (4, 5). Currently, no effective therapy exists for HD or other polyQ diseases.
In Drosophila, several models of human neurodegenerative diseases (Alzheimer's disease, Parkinson's disease, tau-associated pathologies, and the polyQ diseases SCA1, MJD/SCA3, Kennedy's Disease, and HD) have been created by expression of the relevant human pathogenic protein in the fly nervous system (for reviews, see refs. 6–10). This often results in the faithful recapitulation of many hallmarks of these diseases. Specifically, for the polyQ diseases, expression of pathogenic protein in the nervous system results in neuropathology and cell death, which is progressive and age-dependent, leading to early death of the flies. As in human disease, the severity of pathology is increased with longer polyQ repeats (11–18).
Applying genetic strategies to fly models of polyQ disease has been instrumental in identifying roles for chaperones, motor proteins, RNA processing proteins, glutathione S-transferase, and transcription factors in the pathogenic process (11, 13, 16, 19). This in turn has led to the rational design and testing of a variety of therapeutic interventions, many of which impact both fly and mouse models [e.g., reducing histone deacetylase activity or elevating chaperone proteins (16, 18–21)]. In general, the efficacy of disease therapies exhibits a high degree of concordance between fly and mouse models (8), due to conserved cellular mechanisms, which validates the utility of fly models to rapidly evaluate new therapeutic strategies.
Specific single-chain Fv (sFv) antibody fragments comprise the intact antibody-combining site in a single polypeptide chain composed of the genetically linked heavy- and light-chain variable domains. Very large sFv-phage and/or yeast display libraries can be used to select sFv antibodies with high affinity and specificity for epitopes (22, 23). When genes for these sFvs are transfected into cells and expressed intracellularly, the resulting intrabodies can associate stoichiometrically with the target protein and specifically block the toxic effects of pathogenic agents (24, 25). sFv technology has been applied extensively in studies of cancer and infectious disease, including a phase 1 clinical trial of an anti-Erb-2 intrabody for treatment of ovarian cancer (24–26). However, intrabodies have only recently been tested in cellular models for HD (27–31). To our knowledge, this is the first report to show intrabodies to be efficacious against a neurodegenerative disease in an intact animal model. The anti-HD C4 sFv intrabody binds to an epitope formed by the N-terminal 17 amino acids of htt and has been shown to reduce aggregate formation and to increase turnover of mutant htt fragments in tissue culture models of HD (27, 32). In an organotypic mouse brain-slice model of HD, C4 sFv provided functional protection (33). In experiments described below, htt exon-1 and anti-HD C4 sFv intrabody or a control sFv intrabody are coexpressed exclusively in the fly nervous system. The presence of anti-HD C4 sFv in HD flies increases survival to adulthood from 23% to 100% and significantly prolongs mean, median, and maximum adult lifespan. Furthermore, the progression of neurodegeneration and formation of htt aggregates are slowed. These results provide direct evidence that intrabodies can function as therapeutic agents in vivo against proteins associated with neurodegenerative diseases. As such, they provide a proof of principle for a therapeutic and investigative strategy potentially applicable to some of the most common human neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases, as well as HD.
Materials and Methods
Fly Stocks. Flies were raised on standard media, and all experimental crosses were conducted at 25°C. The P{w[+mW.hs] = GawB}elav[C155] (elav-Gal4) and w[*]; P{w[+mC] = UAS-GFP.S65T}T2 (Uas-GFP) (UAS, upstream activating sequence) flies were obtained from the Bloomington Stock Center, Indiana University. The UAS-htt exon-1-Q20 and UAS-htt exon-1-Q93 flies have been described (16) and contain the entire exon 1 from htt harboring the polyQ region. The UAS-C4 sFv and UAS-D10 sFv flies were made by standard transgenic methods. For each transgene, 5–10 independent insertions were obtained and maintained as homozygous stocks. All were shown to express the UAS-sFv transgene by driving expression with neural-specific elav-Gal4 and detecting the hemagglutinin tag on the sFv in the nervous system. For C4 sFv, three independent lines (C4a, C4b, and C4c) were tested in survival assays.
Plasmids. Both the C4 sFv and D10 sFv clones were subcloned from the pcDNA3.1 (Invitrogen) plasmids into the pUAST plasmid, which had been engineered to contain a hemagglutinin epitope sequence in-frame with the sFv upon ligation (27, 34).
Survival to Adult Emergence. Crosses were set such that 50% of the progeny harbored the UAS-transgene(s) but did not express it, and 50% carried the elav-Gal4 driver, resulting in expression of the transgene(s) in the nervous system. To avoid the use of balancers, which can impact relative survival rates, males carrying the elav-Gal4 driver on the X chromosome were crossed to virgin females homozygous for the UAS-transgene(s). In the progeny, the males carry but do not express the UAS-transgene(s), whereas in the females, the UAS-transgene(s) is driven by the elav-Gal4 on one X chromosome. The percent survival to adult emergence (eclosion) was calculated as follows: (no. of females/no. of males) × 100. Bottles were cleared each day, and the numbers of males and females counted until all of the viable F1 progeny had eclosed. P values were derived from a two-tailed z test, which tests the null hypothesis that the number of individuals expressing the transgene(s) equals the number of individuals not expressing the transgene(s) (i.e., the specified genotype causes no change in survival to emergence). The total number of males and females scored for each genotype was between 243 and 684 (Table 1).
Table 1. C4 sFv completely rescues HD adult emergence.
| Transgene(s) | Percent survival to emergence | n | P value |
|---|---|---|---|
| htt exon-1-Q93 | 23 | 243 | <0.00002 |
| htt exon-1-Q20 | 100 | 307 | NS |
| htt exon-1-Q93 & C4a sFv | 94 | 388 | NS |
| htt exon-1-Q93 & C4b sFv | 116 | 313 | NS |
| htt exon-1-Q93 & C4c sFv | 103 | 378 | NS |
| htt exon-1-Q93 & D10 sFv | 21 | 684 | <0.00002 |
| C4 sFv | 96 | 399 | NS |
Three separate lines of C4 sFv flies (a, b, and c) were tested. Crosses were set such that 50% of the progeny express and 50% do not express the indicated transgene(s) (see Materials and Methods for details of crosses). Percent survival to emergence = (no. of emerging adults expressing the transgene(s)/no. of emerging adults not expressing the transgene(s)) × 100. Between 243 and 684 flies were tested for each genotype. NS, not significant at the P = 0.05 level.
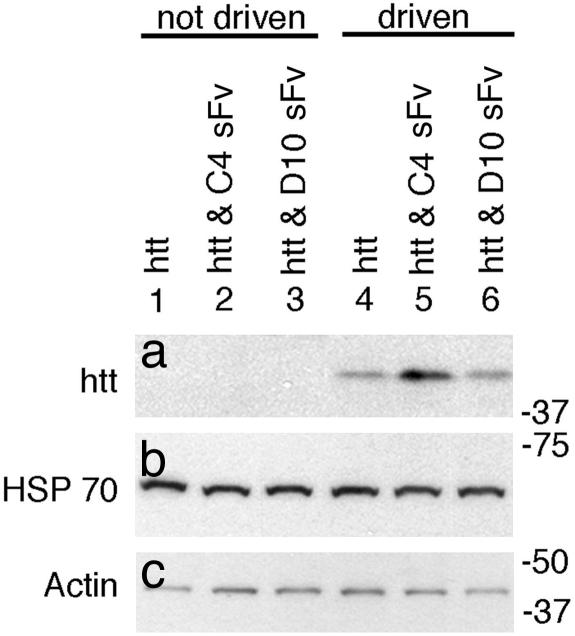
Western Blotting. Fly heads were homogenized in ice-cold lysis buffer [1% Triton X-100/150 mM NaCl/50 mM Tris/1 mM EDTA/protease inhibitor mixture (Boehringer), pH 7.2.] and incubated on ice for 20 min. Insoluble material was cleared by centrifugation. Ten micrograms of protein from each sample was separated by SDS/PAGE, and immunoblots from duplicate gels were probed with either sheep-anti-htt exon-1 S830 (1/2,000) or antihemagglutinin (Covance), stripped, and reprobed with antiactin (Sigma) or anti-HSP70 (Santa Cruz Biotechnology). A representative blot from the three independent experiments is shown in Fig. 2.
Fig. 2.
C4 sFv increased the relative amount of soluble htt exon-1-Q93 present in fly head homogenates. Equal amounts of detergent soluble protein from heads were subjected to Western blot analysis. a probed for htt; b probed for HSP 70; and c probed for actin. Lane 1, undriven htt; lane 2, undriven htt and C4 sFv; lane 3, undriven htt and D10 sFv; lane 4, driven htt; lane 5, driven htt and C4 sFv; and lane 6, driven htt and D10 sFv. HSP70 and actin serve as loading controls. Note the increased relative abundance of htt exon-1-Q93 in the presence of C4 sFv (lane 5, Top). Molecular weight standards in kDa are shown at the right.
Survival Analysis. Flies of the desired genotype were generated as described above (see Survival to Adult Emergence). Female progeny were collected within 24 h of emergence. For each genotype tested, 10–20 female flies per vial were maintained at 25°C, transferred every other day to fresh food, and the number of dead flies counted each day. Survival curves were generated and data analyzed by using the Kaplan–Meier survival analysis method, and statistical significance was tested by using log rank statistics software (SPSS, Chicago). Two separate cohorts were analyzed for mean, median, and maximum survival and % change from flies expressing htt alone. The total number of flies followed for each genotype ranged from 79 to 194 (see Table 2).
Table 2. C4 sFv increases HD adult survival time.
| Cohort | Transgene(s) | Mean | Median | Maximum | P value | n |
|---|---|---|---|---|---|---|
| 1 | htt | 10 | 10 | 17 | NA | 145 |
| 1 | htt and C4b | 13 (30%) | 14 (40%) | 19 (12%) | <0.0001 | 142 |
| 1 | htt and C4c | 15 (50%) | 15 (50%) | 22 (29%) | <0.0001 | 132 |
| 1 | htt and 2× C4a | 13 (30%) | 14 (40%) | 19 (12%) | <0.0001 | 134 |
| 1 | htt and D10 | 10 (0%) | 10 (0%) | 16 (-6%) | NS | 142 |
| 1 | htt exon-1-Q20 | 71 | 75 | 95 | * | 108 |
| 1 | C4a | 73 | 77 | 88 | * | 121 |
| 1 | htt no driver | 67 | 74 | 88 | * | 79 |
| 2 | htt | 9 | 10 | 19 | NA | 177 |
| 2 | htt and C4a | 12 (30%) | 13 (30%) | 22 (16%) | <0.0001 | 191 |
| 2 | htt and D10 | 8 (-11%) | 8 (-20%) | 15 (-23%) | 0.0001 | 194 |
Three separate lines of C4 sFv flies (a, b, and c) were tested. “htt” refers to the pathogenic htt exon-1-Q93 transgene. htt exon-1-Q20 is the nonpathogenic form of the HD transgene. 2× C4 indicates that the fly carried two doses of the C4 sFv transgene. Mean, median, and maximum survival times in days were calculated from survival curves by using Kaplan–Meier analysis. Numbers in parentheses are percent change in survival time for flies expressing htt exon-1-Q93 and the sFv from flies expressing htt exon-1-Q93 alone in the cohort. P values were calculated by using log rank statistics and compare the genotype with the htt genotype for that cohort. Selected growth curves for cohort 1 are shown in Fig. 1. The two cohorts were followed at different times. The three C4 lines are derived from three independent C4 transgene insertions. The D10 sFv data sets are from the same fly line but were done at different times. NS, not significant; NA, not applicable. *, not shown.
Histology. Histology is described in Supporting Text, which is published as supporting information on the PNAS web site.
Results
The Drosophila HD model used in these studies expresses a human htt fragment, htt exon-1, similar to the fragment that was initially shown to cause HD-like pathology in mice (35). The Drosophila express htt exon-1 containing either 20 glutaminyl residues (htt exon-1-Q20), which is not pathogenic, or 93 residues (htt exon-1-Q93), which causes neurodegeneration and early death (16). Expression of htt exon-1 is regulated by the UAS/GAL4 system in which the htt exon-1 transgene lies downstream of the yeast Gal4 UAS (36). In this system, HD flies are produced by crossing UAS-htt exon-1 flies to flies engineered to express the yeast Gal4 protein exclusively in the nervous system from early embryonic stages onward (elav-Gal4) to produce progeny that express htt exon-1 in the same pattern. To achieve coexpression of htt and an sFv, crosses were constructed so that in progeny, both UAS-htt and UAS-sFv transgenes were coordinately regulated by Gal4 protein. Experiments used anti-htt C4 sFv or anti-α-synuclein D10 sFv as a negative control.
C4 sFv Improves Survival of HD Flies. Previous studies using this HD fly model have shown 30% survival during development to adult emergence when flies express htt exon-1-Q93 in the nervous system with a substantially shortened adult lifespan for those flies that do eclose (16). In our studies, only 23% of flies expressing mutant htt exon-1-Q93 alone at 25°C emerge from the pupal case, but 100% emerge when the nonpathogenic htt exon-1-Q20 is expressed (Table 1). With three independent insertion lines of anti-HD C4 sFv (C4a, C4b, and C4c), coexpression of an anti-HD intrabody with htt exon-1-Q93 completely rescued survival to adult emergence (Table 1). Our control intrabody, anti-α-synuclein D10 sFv, was unable to rescue survival of the HD flies. C4 sFv did not affect survival to adult emergence in non-HD flies (Table 1). Rescue by the C4 sFv transgene was not unique to a particular genomic site of insertion and did not result from disruption of another gene by the insert, because the three independent lines produced complete rescue. Additional controls showed that rescue is not a function of C4 sFv down-regulation of the Gal4-dependent expression of our UAS-regulated htt transgene (data not shown). Levels of GFP in flies expressing a UAS-GFP transgene in the nervous system with the same driver (elav-Gal4) are equivalent with or without C4 sFv coexpression by using quantitative Western blot analyses. We conclude that C4 sFv completely rescues adult emergence in HD flies.
HD flies that complete metamorphosis and emerge as viable adults die prematurely (16, 37). To assess the effect of the C4 sFv intrabody on lifespan, we generated survival curves for cohorts of flies expressing either htt exon-1-Q93 alone, htt exon-1-Q93 plus C4 sFv, or htt exon-1-Q93 plus D10 sFv, htt exon-1-Q20 (nonpathogenic) alone (selected survival curves for cohort 1 are shown in Fig. 1, and all data are shown in Table 2). For each individual in the cohort, the number of days survived after adult emergence was recorded. The mean and median survival time for HD flies with C4 sFv compared with those in the cohort without C4 sFv increased significantly (P < 0.0001), with increases ranging from 30% to 50% in four independent experiments (Fig. 1 and Table 2, cohorts 1 and 2). Maximum lifespan was also significantly improved. Genetically doubling the dose of C4 sFv produced no further improvement in survival (2× C4; Table 2). Increased survival was specific to C4 sFv, because our control intrabody, D10 sFv, produced either no change (cohort 1) or a modest reduction in mean, median, and maximum survival time (cohort 2) (Fig. 1, Table 2). C4 sFv was not toxic, as shown by the survival curves and times similar to other controls expressing the nonpathogenic htt exon-1-Q20, or flies in which the htt exon-1-Q93 is harbored but silent (Fig. 1, Table 2). Depending on the comparison made, control survival times were about five to six times longer than experimental flies in the cohort coexpressing htt exon-1-Q93 and C4 sFv. From these results, we conclude that C4 sFv significantly improved survival of HD flies after adult emergence but did not restore lifespan to control ages.
Fig. 1.
C4 sFv increased the survival time of adult HD flies. Selected survival curves for cohort no. 1 are shown (see Table 2 for characterization of curves of all of the genotypes examined including cohort no. 2). HD (htt exon-1-Q93) flies without intrabody (open circle, dashed line); HD flies with C4b sFv intrabody (closed circle, dashed line); HD flies with C4c sFv intrabody (closed triangle, dash-dot line); and HD flies with control intrabody D10 sFv (open triangle, solid line) (a). Survival curves for control flies expressing C4a sFv alone (solid triangle, solid line); expressing nonpathogenic htt exon-1-Q20 (solid circle, dashed line); and flies harboring but not expressing an htt exon-1-Q93 transgene (open circle, dotted line) (b). The number of flies that survived was determined for each cohort once per day. In a, asterisks indicate survival curves that were significantly different from flies expressing htt exon-1-Q93 alone (P < 0.0001). The different range and scale for the x axis in a and b are to accommodate the large difference in survival times for the two sets of data. Htt, htt exon-1-Q93 and htt Q20, htt exon-1-Q20. n = between 79 and 145 animals for each genotype.
C4 sFv Slows Appearance of htt Aggregates. In studies using human postmortem brain tissue, as well as in mouse and fly models of HD, one manifestation of aberrant protein–protein interactions is the accumulation of microscopically detectable intracellular aggregates of htt (3–5, 12, 19, 37, 38). To determine whether C4 sFv could block or reduce these aberrant protein interactions, we examined brains of HD adult flies, with or without C4a sFv, for the presence of soluble and aggregated htt. In flies expressing htt exon-1-Q93, immunolocalization of htt exon-1 in newly emerged adult brain sections revealed diffuse staining throughout the brain and numerous visible htt aggregates in the cell bodies of optic lobe neurons (Fig. 4a, which is published as supporting information on the PNAS web site). Few or no aggregates in other regions of the brain were detected (data not shown). Significantly aggregate density in HD flies was reduced by 44% in the optic lobe lamina when anti-HD C4 sFv was coexpressed with htt exon-1-Q93 (Table 3, Fig. 4b) but was unaffected by control D10 sFv intrabody (Table 3, Fig. 4c) (the observed 10% reduction is not significant). In 6-day-old flies, C4 sFv-dependent reduction in aggregate formation was no longer evident. Htt aggregates were never detected in flies expressing the nonpathogenic htt exon-1-Q20 transgene. We conclude that C4 sFv slows but does not block accumulation of visible aggregates.
Table 3. C4 sFv reduces htt aggregates.
| Expressed transgene(s) | Mean aggregate density, aggregates/μm2 ± SE × 10-3 | n | P value |
|---|---|---|---|
| htt exon-1-Q93 | 7.9 ± 0.51 | 6 | NA |
| htt exon-1-Q93 and C4a sFv | 4.4 ± 0.36 | 6 | 0.0003 |
| htt exon-1-Q93 and D10 sFv | 7.1 ± 0.45 | 5 | 0.2906 |
n, number of sections counted. P values were derived from an unpaired t test and test the null hypothesis that aggregate density is equal to that of htt exon-1-Q93 flies. NA, not applicable.
To assess the relative abundance of soluble htt exon-1-Q93 fragment in these flies, equal amounts of protein from fly head lysates were subjected to Western blot analysis. Flies harboring but not expressing either the htt exon-1-Q93 or sFv transgenes (lanes 1–3) show no htt signal (Fig. 2a). In flies in which the transgene(s) are driven (lanes 4–6), the soluble htt exon-1 protein fragment was readily detected in both C4 sFv (lane 5) and D10 sFv (lane 6) coexpressing flies (Fig. 2a). Interestingly, the level of soluble monomeric htt exon-1-Q93 fragment was increased in the presence of C4 sFv compared with either htt exon-1-Q93 alone or when our control D10 sFv was present. HSP 70 (Fig. 2c) and actin (Fig. 2d) controls confirm that equal amounts of protein are being assayed in each lane. Thus, the presence of C4 sFv clearly alters processing of htt exon-1-Q93 in neurons such that soluble but presumably complexed htt exon-1-Q93 in the brain is elevated (Fig. 2a), and aggregation is slowed.
C4 sFv Slows Neurodegeneration in HD Flies. As in the human form of the disease, mutant htt causes neurodegeneration in flies (12, 16, 19, 37). This phenomenon is readily observed and quantified in the compound eye, using the pseudopupil technique to visualize and count seven of the eight photoreceptor cell rhabdomeres that comprise each ommatidium (16, 39). In representative pseudopupil images of 1-day-old flies (Fig. 3a), photoreceptors are preserved in HD flies when C4 sFv is present. This effect is quantified for 1- and 6-day-old flies as a histogram of the distribution of the percent of ommatidia containing one to seven photoreceptors (Fig. 3 b and c). In this assay, HD flies show an age-dependent loss of photoreceptor cells (Fig. 3 b and c) (16). At 1 day of adult life, the distribution of the percent of ommatidia containing one to seven photoreceptors was significantly shifted toward those containing seven photoreceptors in HD flies when C4 sFv (P < 0.0001) but not D10 sFv (P > 0.4) was present (Fig. 3a). The percentage of ommatidia that contained seven rhabdomeres, indicating no neurodegeneration, in HD flies expressing C4 sFv was 88%, which is almost double that in HD flies (49%) or in HD flies expressing the control intrabody (47%) (Fig. 3a). Furthermore, when C4 sFv was present, ommatidia having fewer than six rhabdomeres were never observed in 1-day-old flies, whereas in HD flies alone or with control intrabody, as few as three were found. At 6 days of adult life, C4 sFv HD flies showed a further loss of photoreceptors. Nonetheless, the distribution of the percent of ommatidia containing one to seven photoreceptors was still significantly shifted toward seven photoreceptors in HD flies when C4 sFv was present (P < 0.0001) but not with D10 sFv (P > 0.9). We observed 15% of the C4 sFv HD ommatidia contained seven photoreceptors, three times more than flies expressing htt exon-1-Q93 either alone (5%) or with the control intrabody (6%). Control flies that express only the nonpathogenic htt exon-1-Q20, C4 sFv, or D10 sFv showed no loss of photoreceptors (data not shown). Flies that harbor but do not express htt exon-1-Q93 also have no photoreceptor degeneration. We conclude that the C4 sFv intrabody significantly slows neurodegeneration in HD flies.
Fig. 3.
Neurodegeneration of photoreceptor cells is slowed in HD flies expressing C4 sFv. (a) Pseudopupil images from 1-day-old HD flies without C4 sFv (htt) or with C4 sFv (htt and C4 sFv). In the HD flies coexpressing C4 sFv, most ommatidia contain seven visible photoreceptors in contrast to flies expressing htt alone. We quantified these data as the distribution of the percent of ommatidia, containing the specified number of photoreceptors, in 1- (b) and 6-day-old (c) adult flies. For both days, the distribution is shifted significantly toward a greater numbers of photoreceptors in HD flies when C4 sFv was present (P < 0.0001). There was no statistical difference between the distribution in HD and D10 sFv HD flies. White bar, htt exon-1-Q93 expressing flies; gray bar, C4 sFv and htt exon-1-Q93; black bar, D10 sFv and htt exon-1-Q93. Error bars represent 95% confidence interval. Significance was calculated by using the nonparametric Mann–Whitney test. For each genotype, five animals were assayed with a total number of ommatidia scored for each animal between 137 and 272. Htt, htt exon-1-Q93.
Discussion
Our results demonstrate that anti-htt sFv intrabodies can protect against the neurotoxic effects of mutant htt protein in an intact functioning nervous system. C4 sFv increased survival from 23% to 100% in HD flies through the larval and pupal stages to adult emergence (Table 1). If untreated, most HD flies die as externally normal adults (pharate adults) trapped in the pupal case unable to act out the stereotypic eclosion behavior required to emerge. Remarkably, C4 sFv completely rescues the elaborate eclosion behavior, which requires precisely timed and coordinated motor output, triggered through the complex interaction of the nervous system with both steroid and neuropeptide hormones (40). Importantly, therapeutic levels of C4 sFv alone had no apparent toxicity (Tables 1 and 2, Fig. 1) and were safely expressed at high levels throughout the life of the fly. Thus, C4 sFv is highly effective at protecting the nervous system through the larval and pupal stages without apparent adverse side effects.
Increased adult emergence was associated with increased levels of detergent-soluble htt protein and decreased levels of visible htt aggregates (Figs. 2 and 3). Although the relationship between detergent-soluble htt exon-1 and visible aggregates in the brain is unknown, the observed changes are consistent with a model in which aggregation is slowed, perhaps by intrabody interference with a nucleation step. Detergent-soluble forms of htt may be stabilized by C4 sFv, allowing them to undergo sFv-dependent turnover, as demonstrated in a HD neuronal cell culture system (32). Recently, it was shown in a cell culture model of HD that the presence of visible htt aggregates increased the lifespan of individual neurons by lowering the level of diffuse htt inside the cell (41). In view of the fact that we see elevated soluble htt associated with protection, we would suggest that binding of the intrabody may change the folding and aggregation kinetics by altering the context of the polyQ protein and/or reducing the rate of formation of some toxic oligomeric intermediates (42–45) formed before the appearance microscopically observable aggregates.
The intrabody also increased mean adult lifespan by 30–50% and delayed neurodegeneration in photoreceptor cells (Figs. 1 and 4 and Table 2). Increased survival was as good as or better than any single agent tested to date in this fly model of HD (16, 46–49). The decreased rate of photoreceptor degeneration seen in our experiments was similar to that observed in a number of fly studies that used the same model and assay to test various therapeutic approaches (16, 47, 48, 50). However, neither life-span nor neuronal survival has been rescued to the wild-type norm. Further intrabody engineering, testing of new intrabodies, increasing intrabody expression levels, or the application of combination therapy with either multiple intrabodies or intrabodies plus small-molecule therapeutics may be needed to optimize treatment (30, 31, 46, 47, 51). The genetic accessibility, ease of rearing and handling, and proven correlations with mammalian models of the fly will allow multiple therapeutic approaches to be evaluated rapidly and cost effectively (8).
Other library-based screens and rational design strategies have identified novel small peptide molecules as potential therapeutics for polyQ diseases (15, 52). The QBP1 peptide, which was selected to bind polyQ sequences, rescues survival of a Machado–Joseph Disease (MJDtr-Q78W) fly model (15). However, its efficacy in HD fly models, which die more rapidly, has not been reported. The impact of this complementary approach in fly HD models would be of interest, because intrabodies against the polyQ region have proven toxic in cells and consequently have not been tested in flies. Small intrabody proteins have the advantage that they are selected from human sFv libraries and are therefore less likely to provoke an immune response in clinical applications compared with novel peptides. Compared with reagents selected to bind to polyQ tracts, the very high specificity of intrabodies should also minimize off-target effects due to the presence of polyQ tracts in other mammalian proteins.
The pathology in a number of neurological disorders appears to result from abnormal folding and/or interactions of proteins (3). Intrabodies can be selected that bind with high specificity to conformational epitopes within these proteins, providing powerful analytical tools for drug development and very targeted therapeutics (24, 25). Intrabodies selected against monomeric-α-synuclein (α-syn) have been shown to specifically bind α-syn in cell cultures, altering its aggregation properties and reducing cellular pathogenic effects (34, 53). Safety of a reagent that binds to both mutant and wild type will need particular care, although this intrabody appears to preferentially target the proteolytic fragment that is pathogenic (32). Delivery of intrabodies will also require gene therapy. Fortunately, the technology for delivery of such reagents to the brain is progressing rapidly, with human clinical trials for gene therapy of Parkinson's and Alzheimer's diseases currently in progress (54, 55). Our current results provide a compelling rationale for aggressively pursuing intrabodies as direct therapeutics and as drug development tools against neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank Ms. Danielle Lebrecht for assistance in the production of transgenic flies; Dr. H. Payami and Ms. Lina M. Moses for the survival curve analysis; Dr. T. L. Shirley for helpful discussions; and Drs. R. Glaser, S. D. Hanes, and H. Payami for critical reading of the manuscript. Technical support was also provided by the Wadsworth Center Molecular Genetics Core and Histology Core Facilities and the David Axelrod Institute Core Microscope Facility. This research was supported in part by the Huntington's Disease Society of America, the Hereditary Disease Foundation, the Cure Huntington's Disease Initiative, and the National Institutes of Health (A.M.), and by the National Institutes of Health, the Hereditary Disease Foundation, the Cure Huntington's Disease Initiative, the Huntington's Disease Society of America Coalition for the Cure, and the Human Frontiers in Science Program (L.M.T. and J.L.M.).
Author contributions: W.J.W. and A.M. designed research; W.J.W., T.W.M., and J.M.W. performed research; A.M., J.S.H., L.M.T., and J.L.M. contributed new reagents/analytic tools; W.J.W., A.M., T.W.M., and J.M.W. analyzed data; W.J.W. and A.M. wrote the paper; and T.W.M., J.M.W., J.S.H., L.M.T., and J.L.M. edited the paper.
Abbreviations: htt, huntingtin; HD, Huntington's disease; sFv, single-chain Fv; UAS, upstream activating sequence; polyQ, polyglutamine.
References
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 2.Bossy-Wetzel, E., Schwarzenbacher, R. & Lipton, S. A. (2004) Nat. Med. 10, Suppl., S2-S9. [DOI] [PubMed] [Google Scholar]
- 3.Ross, C. A. & Poirier, M. A. (2004) Nat. Med. 10, Suppl. 1, S10-S17. [DOI] [PubMed] [Google Scholar]
- 4.Gutekunst, C. A., Li, S. H., Yi, H., Mulroy, J. S., Kuemmerle, S., Jones, R., Rye, D., Ferrante, R. J., Hersch, S. M. & Li, X. J. (1999) J. Neurosci. 19, 2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- 6.Bonini, N. M. & Fortini, M. E. (2003) Annu. Rev. Neurosci. 26, 627-656. [DOI] [PubMed] [Google Scholar]
- 7.Driscoll, M. & Gerstbrein, B. (2003) Nat. Rev. Genet. 4, 181-194. [DOI] [PubMed] [Google Scholar]
- 8.Marsh, J. L. & Thompson, L. M. (2004) BioEssays 26, 485-496. [DOI] [PubMed] [Google Scholar]
- 9.Rubinsztein, D. C. (2002) Trends Genet. 18, 202-209. [DOI] [PubMed] [Google Scholar]
- 10.Zoghbi, H. Y. & Botas, J. (2002) Trends Genet. 18, 463-471. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature 408, 101-106. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., MacDonald, M. E. & Zipursky, S. L. (1998) Neuron 21, 633-642. [DOI] [PubMed] [Google Scholar]
- 13.Kazemi-Esfarjani, P. & Benzer, S. (2000) Science 287, 1837-1840. [DOI] [PubMed] [Google Scholar]
- 14.Marsh, J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9, 13-25. [DOI] [PubMed] [Google Scholar]
- 15.Nagai, Y., Fujikake, N., Ohno, K., Higashiyama, H., Popiel, H. A., Rahadian, J., Yamaguchi, M., Strittmatter, W. J., Burke, J. R. & Toda, T. (2003) Hum. Mol. Genet. 12, 1253-1259. [DOI] [PubMed] [Google Scholar]
- 16.Steffan, J. S., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B. L., Kazantsev, A., Schmidt, E., Zhu, Y. Z., Greenwald, M., et al. (2001) Nature 413, 739-743. [DOI] [PubMed] [Google Scholar]
- 17.Takeyama, K., Ito, S., Yamamoto, A., Tanimoto, H., Furutani, T., Kanuka, H., Miura, M., Tabata, T. & Kato, S. (2002) Neuron 35, 855-864. [DOI] [PubMed] [Google Scholar]
- 18.Warrick, J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L. & Bonini, N. M. (1999) Nat. Genet. 23, 425-428. [DOI] [PubMed] [Google Scholar]
- 19.Gunawardena, S., Her, L. S., Brusch, R. G., Laymon, R. A., Niesman, I. R., Gordesky-Gold, B., Sintasath, L., Bonini, N. M. & Goldstein, L. S. (2003) Neuron 40, 25-40. [DOI] [PubMed] [Google Scholar]
- 20.Hockly, E., Richon, V. M., Woodman, B., Smith, D. L., Zhou, X., Rosa, E., Sathasivam, K., Ghazi-Noori, S., Mahal, A., Lowden, P. A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay, D. G., Sathasivam, K., Tobaben, S., Stahl, B., Marber, M., Mestril, R., Mahal, A., Smith, D. L., Woodman, B. & Bates, G. P. (2004) Hum. Mol. Genet. 13, 1389-1405. [DOI] [PubMed] [Google Scholar]
- 22.Feldhaus, M. J., Siegel, R. W., Opresko, L. K., Coleman, J. R., Feldhaus, J. M., Yeung, Y. A., Cochran, J. R., Heinzelman, P., Colby, D., Swers, J., et al. (2003) Nat. Biotechnol. 21, 163-170. [DOI] [PubMed] [Google Scholar]
- 23.Huston, J. S., Margolies, M. N. & Haber, E. (1996) Adv. Protein Chem. 49, 329-450. [DOI] [PubMed] [Google Scholar]
- 24.Lobato, M. N. & Rabbitts, T. H. (2004) Curr. Mol. Med. 4, 519-528. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler, Y. Y., Chen, S. Y. & Sane, D. C. (2003) Mol. Ther. 8, 355-366. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez, R. D., Barnes, M. N., Gomez-Navarro, J., Wang, M., Strong, T. V., Arafat, W., Arani, R. B., Johnson, M. R., Roberts, B. L., Siegal, G. P., et al. (2000) Clin. Cancer Res. 6, 3081-3087. [PubMed] [Google Scholar]
- 27.Lecerf, J. M., Shirley, T. L., Zhu, Q., Kazantsev, A., Amersdorfer, P., Housman, D. E., Messer, A. & Huston, J. S. (2001) Proc. Natl. Acad. Sci. USA 98, 4764-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, R. C. & Messer, A. (2001) Mol. Ther. 3, 113-121. [DOI] [PubMed] [Google Scholar]
- 29.Khoshnan, A., Ko, J. & Patterson, P. H. (2002) Proc. Natl. Acad. Sci. USA 99, 1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colby, D. W., Garg, P., Holden, T., Chao, G., Webster, J. M., Messer, A., Ingram, V. M. & Wittrup, K. D. (2004) J. Mol. Biol. 342, 901-912. [DOI] [PubMed] [Google Scholar]
- 31.Colby, D. W., Chu, Y., Cassady, J. P., Duennwald, M., Zazulak, H., Webster, J. M., Messer, A., Lindquist, S., Ingram, V. M. & Wittrup, K. D. (2004) Proc. Natl. Acad. Sci. USA 101, 17616-17621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, T. W., Zhou, C., Gines, S., MacDonald, M. E., Mazarakis, N. D., Bates, G. P., Huston, J. S. & Messer, A. (2005) Neurobiol. Dis. 19, 47-56. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, R. C. & Messer, A. (2004) Brain Res. Mol. Brain Res. 121, 141-145. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, C., Emadi, S., Sierks, M. R. & Messer, A. (2004) Mol. Ther. 10, 1023-1031. [DOI] [PubMed] [Google Scholar]
- 35.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W., et al. (1996) Cell 87, 493-506. [DOI] [PubMed] [Google Scholar]
- 36.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 37.Lee, W. C., Yoshihara, M. & Littleton, J. T. (2004) Proc. Natl. Acad. Sci. USA 101, 3224-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 39.Franceschinin, N. (1972) Information Processing in the Visual Systems of Arthropods (Springer, Berlin).
- 40.Baker, J. D., McNabb, S. L. & Truman, J. W. (1999) J. Exp. Biol. 202 (Pt 21.), 3037-3048. [DOI] [PubMed] [Google Scholar]
- 41.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 42.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486-489. [DOI] [PubMed] [Google Scholar]
- 43.Thakur, A. K., Yang, W. & Wetzel, R. (2004) Faseb J. 18, 923-925. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez, I., Mahlke, C. & Yuan, J. (2003) Nature 421, 373-379. [DOI] [PubMed] [Google Scholar]
- 45.Poirier, M. A., Li, H., Macosko, J., Cai, S., Amzel, M. & Ross, C. A. (2002) J. Biol. Chem. 277, 41032-41037. [DOI] [PubMed] [Google Scholar]
- 46.Apostol, B. L., Kazantsev, A., Raffioni, S., Illes, K., Pallos, J., Bodai, L., Slepko, N., Bear, J. E., Gertler, F. B., Hersch, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5950-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollitt, S. K., Pallos, J., Shao, J., Desai, U. A., Ma, A. A., Thompson, L. M., Marsh, J. L. & Diamond, M. I. (2003) Neuron 40, 685-694. [DOI] [PubMed] [Google Scholar]
- 48.Ravikumar, B., Vacher, C., Berger, Z., Davies, J. E., Luo, S., Oroz, L. G., Scaravilli, F., Easton, D. F., Duden, R., O'Kane, C. J., et al. (2004) Nat. Genet. 36, 585-595. [DOI] [PubMed] [Google Scholar]
- 49.Agrawal, N., Pallos, J., Slepko, N., Apostol, B. L., Bodai, L., Chang, L. W., Chiang, A. S., Thompson, L. M. & Marsh, J. L. (2005) Proc. Natl. Acad. Sci. USA 102, 3777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., Smith, D. L., Meriin, A. B., Engemann, S., Russel, D. E., Roark, M., Washington, S. L., Maxwell, M. M., Marsh, J. L., Thompson, L. M., et al. (2005) Proc. Natl. Acad. Sci. USA.
- 51.Beal, M. F. & Ferrante, R. J. (2004) Nat. Rev. Neurosci. 5, 373-384. [DOI] [PubMed] [Google Scholar]
- 52.Kazantsev, A., Walker, H. A., Slepko, N., Bear, J. E., Preisinger, E., Steffan, J. S., Zhu, Y. Z., Gertler, F. B., Housman, D. E., Marsh, J. L., et al. (2002) Nat. Genet. 30, 367-376. [DOI] [PubMed] [Google Scholar]
- 53.Emadi, S., Liu, R., Yuan, B., Schulz, P., McAllister, C., Lyubchenko, Y., Messer, A. & Sierks, M. R. (2004) Biochemistry 43, 2871-2878. [DOI] [PubMed] [Google Scholar]
- 54.Crystal, R. G., Sondhi, D., Hackett, N. R., Kaminsky, S. M., Worgall, S., Stieg, P., Souweidane, M., Hosain, S., Heier, L., Ballon, D., et al. (2004) Hum. Gene Ther. 15, 1131-1154. [DOI] [PubMed] [Google Scholar]
- 55.Mandel, R. J. & Burger, C. (2004) Curr. Opin. Mol. Ther. 6, 482-490. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.