Abstract
The T315I mutation poses a significant threat to patients with chronic phase chronic myeloid leukemia (CP-CML). This study aimed to establish a nomogram to predict the risk of T315I mutation in CP-CML patients. The training cohort included 1,466 patients from 24 hematology centers, and the validation cohort included 820 patients from an additional 20 centers. Peripheral blood blast (PBB), additional chromosomal abnormality (ACA), dasatinib use, non-EMR at 3 months, and BCR::ABLIS > 1% at 6 months were identified as independent risk factors through multivariate Cox regression analysis. The performance of the nomogram was assessed via receiver operating characteristic (ROC) curves, calibration curves, and decision curve analysis (DCA). The area under the ROC curve (AUC) values at 5, 10, and 15 years were 0.874, 0.925, and 0.930 for the training cohort, and 0.864, 0.814, and 0.803 for the validation cohort, respectively. The calibration curves for both cohorts were close to the ideal diagonal, and the decision curves indicated clinical net benefit. In conclusion, we developed a nomogram to predict the 5-year, 10-year, and 15-year T315I-free survival probabilities of CP-CML patients. This tool can aid clinicians in the early prediction and timely management of high-risk CP-CML patients with the T315I mutation.
Keywords: Chronic myeloid leukemia, T315I mutation, Prediction nomogram
Subject terms: Predictive medicine, Cancer, Risk factors
Introduction
Chronic myeloid leukemia (CML) is a malignant clonal disorder originating from hematopoietic stem cells in the bone marrow and is characterized by the presence of the Philadelphia chromosome (Ph) and the BCR::ABL1 fusion gene1. The advent of tyrosine kinase inhibitors (TKIs) has revolutionized the treatment of CML, with survival life expectancies approaching those of the general population2. Studies have shown that the 5-year overall survival (OS) rate for chronic phase CML (CP-CML) patients receiving first-line TKI therapy can reach 95%3, and the 10-year OS rate can reach 88%4.
However, many patients develop resistance or intolerance after continuous TKI treatment, leading to a decreased survival probability. A common mechanism of resistance is the emergence of BCR::ABL1 kinase domain (KD) mutations. Over 100 types of amino acid alterations within the BCR::ABL1 KD gene family have been identified to date5. Among these, the T315I gatekeeper mutation has the highest incidence, accounting for approximately 15–20% of all acquired mutations6. As the most aggressive mutation with the poorest prognosis, the T315I mutation is most commonly associated with disease progression and relapse7. The T315I mutation confers high resistance to imatinib, dasatinib, nilotinib, and bosutinib by affecting the affinity of TKIs for the ATP binding site, posing a significant threat to the prognosis of CML patients8.
The median survival time for CP-CML patients with the T315I mutation, accelerated-phase CML (AP-CML) patients, and blast-phase CML (BP-CML) patients is 133 months, 31 months, and 6 months, respectively9. Considering the poor prognosis in AP and BP, clinicians are more inclined to test for BCR::ABL1 KD mutations in AP-CML or BP-CML patients. Additionally, owing to the longer follow-up periods for CP-CML patients, there is a clinical tendency to overlook timely monitoring of BCR::ABL1 KD mutations in CP-CML patients, leading to a lower detection rate of the T315I mutation during CP.
However, research indicates that more than half of T315I mutations occur in the CP-CML rather than in the AP-CML or BP-CMLpatients9. Interestingly, our preliminary study revealed that, the T315I mutation is associated with a poorer prognosis compared with other BCR::ABL1 KD mutations, such as E255K/V, Y253H, and G250E in CP-CML patients. However, in AP-CML and BP-CML patients, the differences in prognosis among patients with various BCR::ABL1 KD mutations were not statistically significant. Therefore, exploring the occurrence of the T315I mutation in CP-CML patients is highly significant. It is crucial to have a tool that identifies high-risk CP-CML patients prone to developing the T315I mutation, facilitating timely switches to TKIs effective against T315I mutations.
Currently, no clinical predictive model for the T315I mutation in CML patients has been developed. As simple statistical visualization tools, nomograms have been widely used in recent years to predict disease occurrence, development, prognosis, and survival10. This retrospective study aimed to identify risk factors for the T315I mutation in CP-CML patients and to establish a nomogram to predict the 5-year, 10-year, and 15-year T315I-free survival probability in these patients. This work could support clinicians in conducting early assessments, implementing proactive measures, and improving patient outcomes.
Materials and methods
Study design and participants
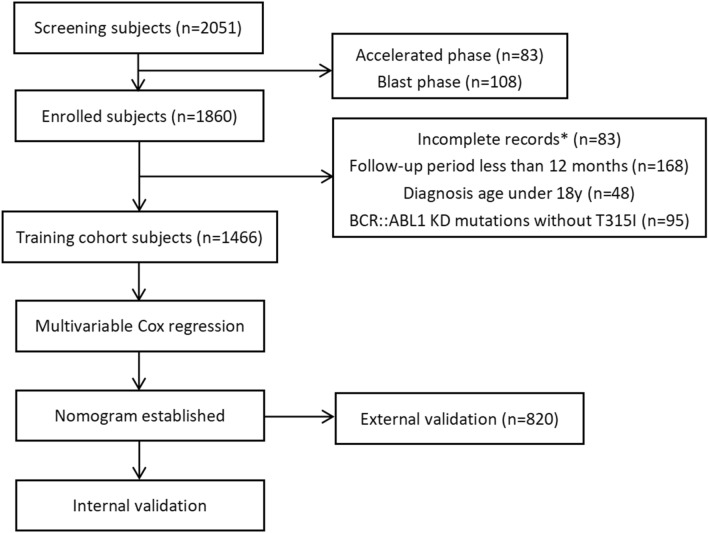
This multicenter retrospective investigation was conducted for the CML Cooperation Group of Zhejiang Hematology in China. From January 2010, to May 2023, a total of 1,956 CML patients were treated and followed up at 24 hematology centers. After 83 AP-CML patients and 108 BP-CML patients were excluded, a total of 1,860 CP-CML patients were included in the study. Among them, 83 patients had incomplete records, 168 patients had a follow-up period of less than 12 months, 48 patients were diagnosed younger than 18 years, and 95 patients had BCR::ABL1 KD mutations other than T315I. Ultimately, 1,466 CP-CML patients were included in the training cohort. The external validation cohort comprised 820 patients from 20 hematologic oncology centers between January, 2010, and May, 2023. The inclusion criteria were as follows: (1) met the diagnostic criteria for CML11; (2) were aged older than 18 years; and (3) had a follow-up duration exceeding 12 months. The exclusion criteria were (1) having experienced AP or BP, (2) missing critical clinical records, and (3) having other BCR::ABL1 KD mutations excluding T315I. The criteria for BCR::ABL1 KD mutation testing in CP-CML patients are based on the NCCN guidelines7. BCR::ABL1 KD mutation analysis were conducted for patients who do not achieve response milestones, for those with any sign of loss of response, and if there is a 1-log increase in BCR::ABL1 level with loss of MMR. The detection of kinase domain mutations in patients strictly followed the standards outlined in the "Chinese Expert Consensus on the Laboratory Practice of BCR-ABL Tyrosine Kinase Domain Mutation Detection"12, and all patients in this study underwent testing using the same criteria.
Data collection and definition
The data collected included the patient’s age, sex, additional chromosomal abnormality (ACA), spleen size below the costal margin, white blood cell count (WBC), hemoglobin level (HB), platelet count (PLT), eosinophil count (EOS), basophil count (BAS), peripheral blood blast (PBB), time from CML diagnosis to the start of first TKI, choice of TKI, history of TKI discontinuation, number of TKI lines used, number of lines of dasatinib used, duration of dasatinib use, and molecular response at 3, 6, and 12 months.
ACAs were defined as the major route ACA + 8, + Ph, i[17q], + 19, + 21, + 17, the minor route ACA 3q26.2, 11q23, − 7/7q − , and complex karyotypes13,14. The choice of TKI was categorized into patients who had used dasatinib and those who had not used dasatinib. In our preliminary analysis, we found that among the various TKIs, patients using dasatinib were more likely to develop the T315I mutation (Table S1a, S1b). This is the basis for the classification of the choice of TKI in our study. The number of TKI lines was classified into 1-line TKI use (no TKI switch) and ≥ 2-line TKI use (one or more TKI switches). Number of lines of dasatinib used was classified as none (no prior use of dasatinib), 1st-line (dasatinib used as first-line therapy), and ≥ 2nd-line (dasatinib used as second-line or third-line therapy). The response definitions were as follows: (1) early molecular response (EMR): BCR::ABLIS ≤ 10% at 3 months; (2) BCR::ABLIS ≤ 1% at 6 months; and (3) major molecular response (MMR): BCR::ABLIS ≤ 0.1% at 12 months.
Development and assessment of the nomogram
Univariate Cox analysis was used to identify clinical parameters significantly associated with the occurrence of T315I. Variables with p values < 0.05 were included in the multivariate Cox analysis. Independent risk factors were used to construct a nomogram to predict the 5-year, 10-year, and 15-year T315I-free survival probabilities in CP-CML patients. Each predictive factor is assigned a line according to its weight, corresponding to different scores. The total score is the sum of the points for all variables in the nomogram. The T315I-free survival rates at different time points were obtained on the basis of the total score.
The model was validated via the concordance index (C-index), receiver operating characteristic (ROC) curve, area under the ROC curve (AUC), calibration curve, and decision curve analysis (DCA). The C-index reflects the consistency between the risk ranking predicted by the model and the actual observations. The value of the C-index ranges from 0.5 to 1.0, with a value closer to 1.0 indicating stronger predictive ability of the model. For each time point, ROC curves were calculated based on the predicted probabilities from the model. It generates a series of points by computing the True Positive Rate (TPR) and False Positive Rate (FPR) at different thresholds, which together form the ROC curve for that specific time point. The C-index and AUC were both used to evaluate the discriminative ability of the model15,16. The accuracy of the model was tested via calibration curves, which compare the observed values with the actual values via 1000 bootstrap resamples17. DCA was used to assess the clinical potential application value of the nomogram by quantifying the net benefit at different threshold probabilities18. Additionally, we classified patients into low-risk and high-risk groups on the basis of their nomogram scores. K‒M curves and log-rank tests were used to compare survival differences between the risk groups. The model’s discriminative ability, accuracy, and clinical net benefit in the external validation cohort were validated via the same methods.
Statistical analysis
All eligible patients in the CML Cooperation Group of Zhejiang Hematology from January, 2010 to May, 2023, were included in this analysis, and no additional sample size calculations were performed. All the data were analyzed via IBM SPSS Statistics Version 28.0 and R software version 4.3.0 with the rms, time‒ROC, survival, ggplot2 and dcurves packages. We used SPSS for descriptive statistical analysis. The “rms” package was employed to construct the nomogram model and plot the calibration curve. The “timeROC” package was used for time-dependent ROC curve analysis and to calculate the AUC values at different time points. The “dcurves” package was utilized for DCA. The “survival” package was used for Kaplan–Meier curve analysis, and the “ggplot2” package was applied for graphical plotting and visualization of results. Prior to performing the Cox regression analysis, we tested the proportional hazards assumption using the Schoenfeld residuals test. The overall P-value and the P-values for each variable were all > 0.05 (Fig. S1), and the R code used for the analysis was provided in the Appendix 1. The derivation of the scoring scale was based on the regression coefficients (β values) from the Cox regression model and R code was provided in the Appendix 2. Categorical covariates were reported as percentages and counts. Continuous variables were reported as medians and interquartile ranges (IQRs). For comparisons among these groups, the Pearson chi-square test was used for categorical factors, and the Mann‒Whitney U test was used for continuous variables. P < 0.05 was considered to indicate statistical significance.
Results
Baseline characteristics
Out of 2,051 CML patients from 24 hematology centers, 585 patients were excluded. Ultimately, 1,466 patients were included in the training cohort (Fig. 1). A total of 898 (61.3%) patients were male, with a median age of 50 years (IQR 37–60 years). The median follow-up time was 3.6 years (range 1–24.9 years), and 30 patients (2.0%) developed the T315I mutation. The 5-year, 10-year, and 15-year T315I-free survival probabilities were 98.3% (95% CI 97.6–99.1%), 97.1% (95% CI 95.6–98.7%), and 91.3% (95% CI 86.1–96.8%), respectively (Fig. S2). The external validation cohort included 820 patients who met the inclusion and exclusion criteria, with a median follow-up time of 3.7 years (range 1–29.5 years), and 23 patients (2.8%) who developed the T315I mutation. The 5-year, 10-year, and 15-year T315I-free survival probabilities were 97.9% (95% CI 96.5–99.2%), 92.4% (95% CI 88.8%–96.1%), and 88.3% (95% CI 81.6%–95.5%), respectively (Fig. S2). There were no statistically significant differences between the training and validation cohorts across the 19 listed parameters, including age, sex, WBC, HB, PLT, EOS, BAS, PBB, spleen size below the costal margin, ACA, time from diagnosis to first TKI, TKI discontinuation, TKI lines, TKI choice, lines of dasatinib, duration of dasatinib, EMR at 3 months, BCR::ABLIS ≤ 1% at 6 months, and MMR at 12 months (Table 1).
Fig. 1.
Flow chart of the study. *Incomplete hematologic parameters (n = 10), missing chromosomal data (n = 12), missing spleen data (n = 8), unclear TKI usage (n = 15), missing molecular response data (n = 38).
Table 1.
Baseline characteristics of the training cohort and validation cohort.
| Variables | Training cohort (n = 1466) M (P25, P75)/N (%) | External validation cohort (n = 820) M (P25, P75)/N (%) | P-value |
|---|---|---|---|
| Age (years) | 50 (37, 60) | 49 (38, 61) | 0.466 |
| Sex | 0.351 | ||
| Male | 898 (61.3) | 486 (59.3) | |
| Female | 568 (38.7) | 334 (40.7) | |
| White blood cell (× 10ˆ9/L) | 92.80 (38.5, 159.5) | 93.00 (38.0, 159.5) | 0.623 |
| Hemoglobin (g/L) | 111.00 (98.0, 127.0) | 110.00 (98.0, 128.0) | 0.509 |
| Platelet (× 10ˆ9/L) | 419.50 (262.0, 591.0) | 424.00 (261.0, 579.3) | 0.71 |
| Eosinophils (%) | 2.25 (1.1, 3.0) | 2.33 (1.2, 3.3) | 0.216 |
| Basophils (%) | 4.00 (2.0, 5.3) | 4.00 (1.8, 5.1) | 0.591 |
| Peripheral blood blast (%) | 0.00 (0.0, 2.0) | 0.00 (0.0, 2.0) | 0.092 |
| Spleen size below costal margin (cm) | 3.50 (0.0, 6.0) | 3.50 (0.0, 5.7) | 0.974 |
| ACA (%) | 0.517 | ||
| Yes | 77 (5.3) | 38 (4.6) | |
| No | 1389 (94.7) | 782 (95.4) | |
| Time from diagnosis to first TKI (months) | 0.00 (0.0, 0.0) | 0.00 (0.0, 0.0) | 0.44 |
| TKI discontinuation (%) | 0.666 | ||
| Yes | 133 (9.1) | 70 (8.5) | |
| No | 1333 (90.9) | 750 (91.5) | |
| TKI lines (%) | 0.821 | ||
| 1-line | 1122 (76.5) | 631 (77.0) | |
| ≥ 2-line | 344 (23.5) | 189 (23.0) | |
| TKI choice (%) | 0.424 | ||
| Dasatinib | 168 (11.5) | 85 (10.4) | |
| Other | 1298 (88.5) | 735 (89.6) | |
| Lines of dasatinib | 0.627 | ||
| None | 1298 (88.5) | 735 (89.6) | |
| 1st-line | 27 (1.8) | 16 (2.0) | |
| ≥ 2nd-line | 141 (9.6) | 69 (8.4) | |
| Duration of dasatinib (months) | 0.00 (0.0, 0.0) | 0.00 (0.0, 0.0) | 0.418 |
| EMR at 3 months | 0.51 | ||
| Yes | 1107 (75.5) | 609 (74.3) | |
| No | 359 (24.5) | 211 (25.7) | |
| BCR::ABLIS ≤ 1% at 6 months | 0.651 | ||
| Yes | 1067 (72.8) | 604 (73.7) | |
| No | 399 (27.2) | 216 (26.3) | |
| MMR at 12 months | 0.668 | ||
| Yes | 1044 (71.2) | 577 (70.4) | |
| No | 422 (28.8) | 243 (29.6) | |
ACA additional chromosomal abnormalities, EMR early molecular response, MMR major molecular response.
Nomogram model development
According to the univariate Cox regression analysis, WBC (P = 0.019), HB (P = 0.017), PBB (P = 0.001), ACA (P = 0.011), TKI lines (P = 0.01), TKI choice (P < 0.0001), lines of dasatinib (P = 0.001), EMR at 3 months (P < 0.0001), and BCR::ABLIS ≤ 1% at 6 months (P < 0.0001) differed significantly between the T315I mutation group and the non-T315I mutation group (Table 2). These variables were then included in the multivariable Cox regression analysis. PBB (P = 0.042, hazard ratio (HR) = 1.171, 95% confidence interval (CI) 1.006–1.364), ACA (P = 0.020, HR = 3.712, 95% CI 1.224–11.255), TKI choice of dasatinib (P = 0.011, HR = 7.769, 95% CI 1.594–37.868), non-EMR at 3 months (P = 0.033, HR = 5.692, 95% CI 1.146–28.262), and BCR::ABLIS > 1% at 6 months (P = 0.005, HR = 22.794, 95% CI 2.529–205.450) were found to be independent risk factors for the occurrence of the T315I mutation in CP-CML patients (Table 2).
Table 2.
General characteristics of the patients and multivariate Cox regression analyses for the training group.
| Variables | T315I mutation (n = 30) M (P25, P75)/N (%) |
Non-T315I mutation (n = 1436) M (P25, P75)/N (%) |
P-value | HR 95% CI |
|---|---|---|---|---|
| Age (years) | 44 (31, 51) | 50 (37, 61) | 0.177 | |
| Sex | 0.674 | |||
| Male | 19 (63.3) | 879 (61.2) | ||
| Female | 11 (36.7) | 557 (38.8) | ||
| White blood cell (× 10ˆ9/L) | 151.6 (57.6, 244.7) | 92.8 (38.2, 159.5) | 0.019 | |
| Hemoglobin (g/L) | 108.0 (87.8, 118.0) | 112.0 (98.0, 127.0) | 0.017 | |
| Platelet (× 10ˆ9/L) | 400.0 (246.5, 540.0) | 423.0 (263.0, 591.0) | 0.937 | |
| Eosinophils (%) | 2.0 (1.25, 3.4) | 2.3 (1.1, 3.0) | 0.574 | |
| Basophils (%) | 3.9 (1.8, 5.9) | 4.0 (2.0, 5.3) | 0.411 | |
| Peripheral blood blast (%) | 1.0 (0.0, 4.0) | 0.0 (0.0, 1.8) | 0.001 | 1.171 (1.006,1.364) |
| Spleen size below costal margin (cm) | 4.7 (0.5, 7.0) | 3.5 (0.0, 6.0) | 0.306 | |
| ACA (%) | 0.011 | 3.712 (1.224,11.255) | ||
| Yes | 4 (13.3) | 73 (5.1) | ||
| No | 26 (86.7) | 1363 (94.9) | ||
| Time from diagnosis to first TKI (months) | 0.0 (0.0, 1.0) | 0.0 (0.0, 0.0) | 0.912 | |
| TKI discontinuation (%) | 0.253 | |||
| Yes | 7 (23.3) | 126 (8.8) | ||
| No | 23 (76.7) | 1310 (91.2) | ||
| TKI lines (%) | 0.010 | |||
| 1-line | 14 (46.7) | 1108 (77.2) | ||
| ≥ 2-line | 16 (53.3) | 328 (22.8) | ||
| TKI choice (%) | < 0.0001 | 3.128 (1.016,9.631) | ||
| Dasatinib | 14 (46.7) | 154 (10.7) | ||
| Other | 16 (53.3) | 1282 (89.3) | ||
| Lines of dasatinib | ||||
| None | 16 (53.3) | 1282 (89.3) | ||
| 1st-line | 2 (6.7) | 25 (1.7) | 0.011 | |
| ≥ 2nd-line | 12 (40.0) | 129 (9.0) | < 0.0001 | |
| Duration of dasatinib (months) | 0.0 (0.0, 15.3) | 0.0 (0.0, 0.0) | 0.54 | |
| EMR at 3 months | < 0.0001 | 5.692 (1.146,28.262) | ||
| Yes | 2 (6.7) | 1105 (76.9) | ||
| No | 28 (93.3) | 331 (23.1) | ||
| BCR::ABLIS ≤ 1% at 6 months | < 0.0001 | 22.794 (2.529,205.450) | ||
| Yes | 1 (3.3) | 1066 (74.2) | ||
| No | 29 (96.7) | 370 (25.8) | ||
| MMR at 12 months | 0.995 | |||
| Yes | 0 (0.0) | 1044 (72.7) | ||
| No | 30 (100.0) | 392 (27.3) | ||
ACA additional chromosomal abnormalities, EMR early molecular response, MMR major molecular response.
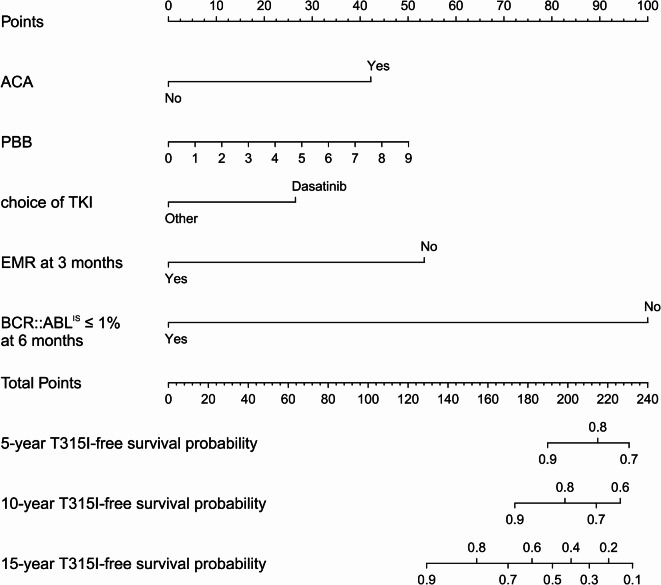
By integrating the above five factors, a nomogram model was constructed to predict the T315I-free survival probability at different time points for CP-CML patients (Fig. 2). Points of each variable in the nomogram were shown in the Table S2 in the supplement. As time progresses, higher scores indicate a lower T315I-free survival probability. For example, if a CP-CML patient presents with ACA, has a PBB of 2%, has a history of using dasatinib, does not achieve EMR at 3 months, and has a BCR::ABLIS > 1% at 6 months, the corresponding scores would be approximately 42 points, 11 points, 26 points, 53 points, and 100 points, respectively. The total score would be 232 points, corresponding to a 5-year T315I-free survival probability of 68.7%, a 10-year survival rate of 55.0%, and a 15-year survival rate of 10.0%.
Fig. 2.
Nomogram for the prediction of the T315I-free survival probability. Based on the individual patient’s data, a vertical line was drawn on each variable axis to obtain a specific score corresponding to the scale at the top. The obtained scores were summed to obtain the total score, and a vertical line was drawn from the total score on the scale at the bottom to determine the 5-year, 10-year, and 15-year T315I-free survival probabilities.
Nomogram model validation
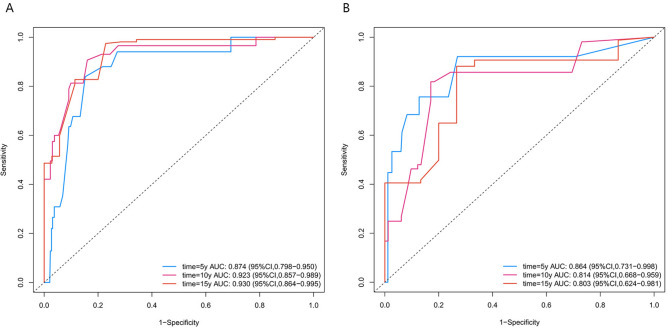
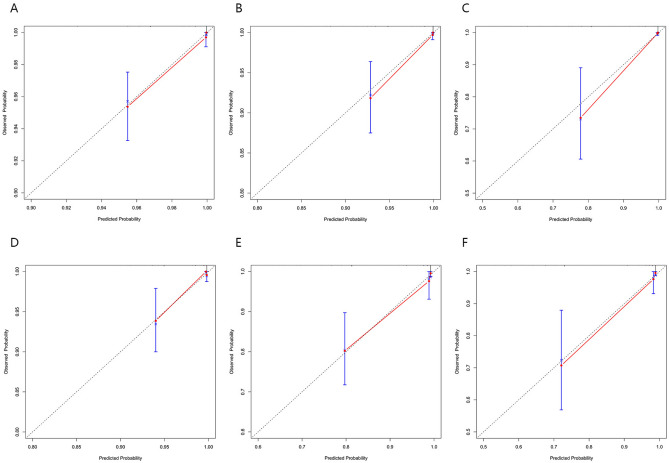
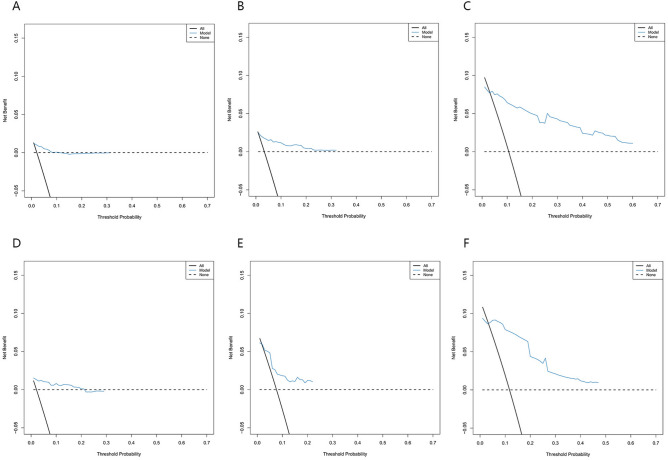
In the training cohort, the C-index was 0.894, and the AUC values for 5, 10, and 15 years were 0.874 (95% CI 0.798–0.950), 0.925 (95% CI 0.857–0.989), and 0.930 (95% CI 0.864–0.995), respectively, indicating the model’s long-term discriminative ability (Fig. 3A). The calibration curve was close to the ideal diagonal line, reflecting good accuracy (Fig. 4A–C). The DCA curve demonstrated a clinical benefit for intervention in patients whose predicted risk was within the threshold range (Fig. 5A–C). The nomogram model was validated in the validation cohort via the same methods. Analysis revealed a C-index of 0.857, and the AUC values at 5, 10, and 15 years were 0.864 (95% CI 0.731–0.998), 0.814 (95% CI 0.668–0.959), and 0.803 (95% CI 0.624–0.981), respectively (Fig. 3B). The calibration curve in the validation cohort was also close to the diagonal (Fig. 4D–F), and the DCA curve demonstrated clinical net benefit within a certain range (Fig. 5D–F).
Fig. 3.
ROC curves of the nomogram. (A) Training cohort. (B) Validation cohort. ROC receiver operating characteristic; AUC area under the ROC curve.
Fig. 4.
Calibration curves of the nomogram. (A–C) The 5-, 10-, and 15-year T315I-free survival probabilities in the training cohort. (D–F) The 5-, 10-, and 15-year T315I-free survival probabilities in the validation cohort. The x- and y-axes represent the nomogram-predicted and actual generalization probabilities, respectively. The 45° dashed line represents the ideal reference line. The closer the solid red line is to the reference line, the higher the accuracy of the model.
Fig. 5.
DCA curves of the nomogram. (A–C) The 5-, 10-, and 15-year T315I-free survival probabilities in the training cohort. (D–F) The 5-, 10-, and 15-year T315I-free survival probabilities in the validation cohort. DCA = decision curve analysis.
Clinical application of the nomogram
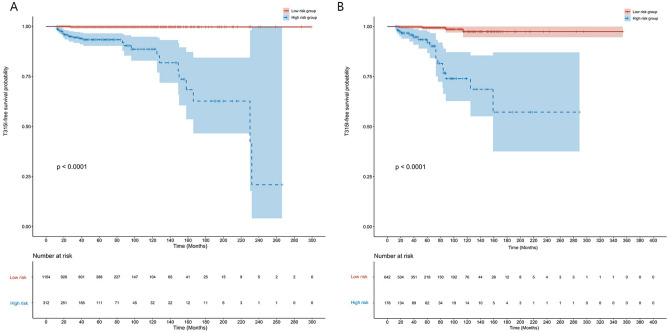
According to the ROC curve, the cutoff value for the total score was approximately 128 points. Patients with scores > 128 were defined as the high-risk group, whereas those with scores ≤ 128 were defined as the low-risk group. In both the training and validation cohorts, the Kaplan‒Meier curves revealed that patients in the high-risk group had a lower T315I-free survival probability than patients in the low-risk group did (Fig. 6A,B). In the overall population, the 5-year and 10-year T315I-free survival probabilities for the low-risk group were 99.6% (95% CI 99.3–100.0%) and 98.8% (95% CI 97.6–100.0%), respectively, whereas they were 92.9% (95% CI 90.1–95.7%), 83.3% (95% CI 77.6–89.5%), and 60.6% (95% CI 47.4%–77.4%), respectively, for the high-risk group.
Fig. 6.
Kaplan–Meier curves in the low- and high-risk groups. Kaplan–Meier curves of T315I-free survival probability in the low- and high-risk groups in the (A) training cohort and (B) validation cohort.
Discussion
The survival rate of CP-CML patients is close to that of the general population; however, CP-CML patients with concurrent T315I mutations have a poorer prognosis. In the imatinib era, the median OS of CP-CML patients with the T315I mutation was found to be 22.4 months19. In the ponatinib era, the 5-year survival rate for CP-CML patients with the T315I mutation was 70%9. Currently, treatment options for patients with the T315I mutation primarily include third-generation TKIs (such as ponatinib, asciminib, and olverembatinib) and allogeneic hematopoietic stem cell transplantation (HSCT)5. Research indicates that in CP-CML patients with the T315I mutation, ponatinib is superior to allogeneic HSCT20. Typically, the timing for detecting BCR::ABL1 KD mutations is based on increasing levels of BCR::ABL1 transcripts7, but low-level T315I mutations often occur before clinical detection21, Therefore, timely identification of patients with the T315I mutation can help physicians implement appropriate interventions to improve disease prognosis.
Several studies have explored the relevant factors and predictive models for molecular response, TKI resistance, and treatment failure in CML patients22–24. However, a model for predicting the T315I mutation in CP-CML patients has yet to be developed. Here, we developed an innovative and meaningful nomogram model for evaluating the risk of T315I mutation in CP-CML patients. We included 1,466 eligible CP-CML patients from 24 hematology centers. The incidence of the T315I mutation was 2.0%, similar to the 4.2% incidence reported in a recent study9. From the training cohort of 1,466 patients, we identified five independent risk factors influencing the occurrence of the T315I mutation in CP-CML patients: PBB, ACA, choice of dasatinib as a TKI, non-EMR at 3 months, and BCR::ABLIS > 1% at 6 months. We developed a nomogram model based on these factors to predict the 5-year, 10-year, and 15-year T315I-free survival probabilities. The model was validated with 820 patients from an additional 20 hospitals and demonstrated good discrimination, accuracy, and clinical net benefit. In addition, the nomogram effectively stratified patients into low-risk and high-risk categories, showing valuable clinical applicability.
This study revealed that the percentage of PBB was a predictor of the occurrence of the T315I mutation in CP-CML patients. An increased percentage of PBB indicates disease progression and is associated with higher grades of bone marrow fibrosis and difficulty in achieving treatment-free remission25. The incidence of ACA in CP-CML patients has been found to be approximately 5%26, similar to the findings of our study. ACAs are categorized into major and minor routes and have been shown to negatively affect survival and prognosis14. A German study revealed that ACA is common in the T315I mutation subtype, with the 3q26.2 rearrangement highly correlated with BCR::ABL1 mutations27. Patients with genetically unstable CML are more likely to develop KD mutations, possibly related to changes in the DNA damage response, cell cycle checkpoints, and DNA repair28,29. In CP-CML patients with the T315I mutation treated with ≥ 1 TKI, baseline data revealed that 73.3% of patients used dasatinib, whereas 4.4–57.8% of patients used other TKIs30. The 2-year follow-up of the DASISION study revealed that among patients with BCR::ABL mutations in the dasatinib group, 70% had the T315I mutation, whereas no T315I mutation was found in the imatinib group31. Our results are consistent with these findings, suggesting that a history of dasatinib use predicts a greater likelihood of the T315I mutation. Therefore, monitoring for the occurrence of the T315I mutation in patients currently using or who have previously used dasatinib is crucial. EMR has become an effective predictor of long-term progression-free survival (PFS) and OS4. Studies have shown that adjusting early intervention strategies on the basis of EMR at 3 months can be beneficial32. A BCR::ABL1IS ≤ 1% at 6 months is also a recognized predictor of long-term survival33. In the IRIS study, the estimated 6-year PFS rate for patients with a BCR::ABL1IS ≤ 1% at 6 months was 97%, whereas it was 80% for those with a BCR::ABL1IS > 1% at 6 months. This finding supports the rationale that failure to achieve a molecular response is associated with the T315I mutation.
Additional mutations other than BCR::ABL1 KD mutations are gaining increasing attention. A position paper recommended the use of next-generation sequencing (NGS) for mutation detection in CP-CML patients experiencing “failure” in first- or second-line TKI treatment34. Compared with Sanger sequencing, NGS can detect the T315I mutation three months earlier and identify mutations undetectable by Sanger sequencing35. In CML patients, the presence of epigenetic gene mutations at diagnosis, such as ASXL1, IKZF1, BCOR, TET1/2, IDH1/2, DNMT3A/3B, and EZH2, is associated with lower molecular response rates and reduced PFS7,36,37. A novel HDAC I/IIb inhibitor has been shown to prevent T315I-induced CML progression and target leukemia stem cells (LSCs) responsible for CML relapse by inhibiting EZH238. ROS-induced oxidative DNA damage in LSCs can lead to the T315I mutation and IKZF1 deletion. Studies on the correlation between non-BCR::ABL1 KD mutations and T315I mutations are limited. We anticipate incorporating parameters related to gene mutations into our model in future research.
Our study has certain limitations. First, as a retrospective study, it inevitably has some degree of internal bias. Second, given the complexity of clinical situations, we did not strictly screen patients on the basis of the different first-line TKIs used or the various TKI switch scenarios, which may have reduced the model’s accuracy but also increased its applicability. And it does not impede the model’s ability to identify patients who require close monitoring. Third, existing warning criteria and BCR::ABL1 elevation thresholds typically occur after mutations have already developed. In contrast, our nomogram integrates more clinical information to enable earlier risk prediction, providing alerts before the mutation becomes apparent and facilitating earlier intervention. However, it should still be used in conjunction with existing standards to enhance the overall management of patients with the T315I mutation.
Conclusion
In this study, we identified PBB, ACA, the choice of dasatinib as a TKI, a non-EMR at 3 months, and a BCR::ABLIS > 1% at 6 months as predictors of the T315I mutation in CP-CML patients. Both the training set and the validation set demonstrated good performance of this nomogram. This visualized nomogram model provides clinicians with an intuitive tool to help identify patients requiring close monitoring. In the future, we will conduct prospective studies to further validate the accuracy of this model, and we will aim to incorporate clinical parameters during follow-up and gene data determined by NGS to optimize this nomogram model.
Supplementary Information
Acknowledgements
This study was performed by the CML Cooperation Group of Zhejiang Hematology and Zhejiang Clinical Medical Research Center of Hematology in China.
Author contributions
H.-J., J.-J., and T.-H.Y. designed the study; H.-S.E., and Y.,-X.D. performed the analysis and wrote the draft; H.-S.E., W.-D.J., W.-Y., Z.-X.Q., F.-W.Y., Q.-H.L., L.-Y., C.-L.L., C.-L.H., L.-J., Z.-L., S.-Y.P., L.-L.L., T.-G.Y., Z.-H., C.-Y., Y.-X.F., F.-X.N., H.-L., Z.-K.F., X.-Y.M., X.-L.J., Z.-H.Q., Z.-G., J,-Y.Q., Z.-X.J., W.-G.Q., T.-J.M., Z.-F., Z.-Y.F., and Y.-G.L. collected the data; Y.,-X.D., W.-D.J., W.-Y. and Z.-J.J. aided in interpreting the results and verified the underlying study data; H.-J., J.-J., and T.-H.Y. supervised the work; and all the authors discussed the results and approved the final manuscript.
Funding
This research was funded by the Key R&D Program of Zhejiang (No. 2022C03137) and the Zhejiang Medical Association Clinical Medical Research Special Fund Project (No. 2022ZYC-D09).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study protocol was approved by the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University Institutional Review Board, with a waiver for informed consent (2022, No469). All methods were carried out in accordance with relevant guidelines and regulations.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shi-wei Hu, Xiu-di Yang, Di-jiong Wu and Yi Wang contributed equally to this work.
Contributor Information
Hong-yan Tong, Email: tonghongyan@zju.edu.cn.
Jie Jin, Email: jiej0503@zju.edu.cn.
Jian Huang, Email: househuang@zju.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-89851-y.
References
- 1.Faderl, S. et al. The biology of chronic myeloid leukemia. N. Engl. J. Med.341, 164–172. 10.1056/NEJM199907153410306 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Jabbour, E. & Kantarjian, H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am. J. Hematol.97, 1236–1256. 10.1002/ajh.26642 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Brummendorf, T. H. et al. Bosutinib versus imatinib for newly diagnosed chronic phase chronic myeloid leukemia: Final results from the BFORE trial. Leukemia36, 1825–1833. 10.1038/s41375-022-01589-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kantarjian, H. M. et al. Long-term outcomes with frontline nilotinib versus imatinib in newly diagnosed chronic myeloid leukemia in chronic phase: ENESTnd 10-year analysis. Leukemia35, 440–453. 10.1038/s41375-020-01111-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senapati, J. et al. Management of chronic myeloid leukemia in 2023 - common ground and common sense. Blood Cancer J.13, 58. 10.1038/s41408-023-00823-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, J. et al. Recent advances in Bcr-Abl tyrosine kinase inhibitors for overriding T315I mutation. Chem. Biol. Drug Des.97, 649–664. 10.1111/cbdd.13801 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Shah, N. P. et al. Chronic Myeloid Leukemia Version, 2.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw.22, 43–69. 10.6004/jnccn.2024.0007 (2024). [DOI] [PubMed] [Google Scholar]
- 8.Al Hamad, M. Contribution of BCR-ABL molecular variants and leukemic stem cells in response and resistance to tyrosine kinase inhibitors: A review. F1000Res10, 1288. 10.12688/f1000research.74570.2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddad, F. G. et al. Characteristics and outcomes of patients with chronic myeloid leukemia and T315I mutation treated in the pre- and post-ponatinib era. Am. J. Hematol.98, 1619–1626. 10.1002/ajh.27037 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balachandran, V. P., Gonen, M., Smith, J. J. & DeMatteo, R. P. Nomograms in oncology: More than meets the eye. Lancet Oncol.16, e173-180. 10.1016/S1470-2045(14)71116-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochhaus, A. et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia34, 966–984. 10.1038/s41375-020-0776-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laboratory Diagnosis Group of Hematology Branch of Chinese Medical Association. Chinese expert consensus on the laboratory practice of BCR- ABL tyrosine kinase domain mutation detection. Zhonghua Xue Ye Xue Za Zhi36, 899–901. 10.3760/cma.j.issn.0253-2727.2015.11.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabarius, A. et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: Long-term observation of 1151 patients from the randomized CML Study IV. Blood118, 6760–6768. 10.1182/blood-2011-08-373902 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hehlmann, R. et al. High-risk additional chromosomal abnormalities at low blast counts herald death by CML. Leukemia34, 2074–2086. 10.1038/s41375-020-0826-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrell, F. E. Jr., Califf, R. M., Pryor, D. B., Lee, K. L. & Rosati, R. A. Evaluating the yield of medical tests. JAMA247, 2543–2546 (1982). [PubMed] [Google Scholar]
- 16.Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology143, 29–36. 10.1148/radiology.143.1.7063747 (1982). [DOI] [PubMed] [Google Scholar]
- 17.Alba, A. C. et al. Discrimination and calibration of clinical prediction models: Users’ guides to the medical literature. JAMA318, 1377–1384. 10.1001/jama.2017.12126 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Vickers, A. J., van Calster, B. & Steyerberg, E. W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res.3, 18. 10.1186/s41512-019-0064-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolini, F. E. et al. Epidemiologic study on survival of chronic myeloid leukemia and Ph(+) acute lymphoblastic leukemia patients with BCR-ABL T315I mutation. Blood114, 5271–5278. 10.1182/blood-2009-04-219410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolini, F. E. et al. Overall survival with ponatinib versus allogeneic stem cell transplantation in Philadelphia chromosome-positive leukemias with the T315I mutation. Cancer123, 2875–2880. 10.1002/cncr.30558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soverini, S. et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: The NEXT-in-CML study. Blood135, 534–541. 10.1182/blood.2019002969 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Zhang, X. S. et al. Predictive scoring systems for molecular responses in persons with chronic phase chronic myeloid leukemia receiving initial imatinib therapy. Leukemia36, 2042–2049. 10.1038/s41375-022-01616-y (2022). [DOI] [PubMed] [Google Scholar]
- 23.Krishnan, V. et al. A single-cell atlas identifies pretreatment features of primary imatinib resistance in chronic myeloid leukemia. Blood141, 2738–2755. 10.1182/blood.2022017295 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Zhang, X. S., Gale, R. P., Zhang, M. J., Huang, X. J. & Jiang, Q. A predictive scoring system for therapy-failure in persons with chronic myeloid leukemia receiving initial imatinib therapy. Leukemia36, 1336–1342. 10.1038/s41375-022-01527-y (2022). [DOI] [PubMed] [Google Scholar]
- 25.Jacobi, H. et al. Myelofibrosis at diagnosis is associated with the failure of treatment-free remission in CML patients. Front. Pharmacol.14, 1212392. 10.3389/fphar.2023.1212392 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molica, M., Massaro, F. & Breccia, M. Diagnostic and prognostic cytogenetics of chronic myeloid leukaemia: An update. Expert Rev. Mol. Diagn.17, 1001–1008. 10.1080/14737159.2017.1383156 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Chen, Z. et al. Cytogenetic landscape and impact in blast phase of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Leukemia31, 585–592. 10.1038/leu.2016.231 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Muvarak, N., Nagaria, P. & Rassool, F. V. Genomic instability in chronic myeloid leukemia: Targets for therapy?. Curr. Hematol. Malig. Rep.7, 94–102. 10.1007/s11899-012-0119-0 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Popp, H. D. et al. DNA damage and DNA damage response in chronic myeloid leukemia. Int. J. Mol. Sci.21, 1177. 10.3390/ijms21041177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes, J. E. et al. Asciminib monotherapy in patients with chronic-phase chronic myeloid leukemia with the T315I mutation after >/=1 prior tyrosine kinase inhibitor: 2-year follow-up results. Leukemia10.1038/s41375-024-02278-8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kantarjian, H. M. et al. Dasatinib or imatinib in newly diagnosed chronic-phase chronic myeloid leukemia: 2-year follow-up from a randomized phase 3 trial (DASISION). Blood119, 1123–1129. 10.1182/blood-2011-08-376087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neelakantan, P. et al. Combining BCR-ABL1 transcript levels at 3 and 6 months in chronic myeloid leukemia: Implications for early intervention strategies. Blood121, 2739–2742. 10.1182/blood-2012-11-466037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jabbour, E. et al. The achievement of an early complete cytogenetic response is a major determinant for outcome in patients with early chronic phase chronic myeloid leukemia treated with tyrosine kinase inhibitors. Blood118, 4541–4546. 10.1182/blood-2011-04-348110 (2011) (quiz 4759). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soverini, S. et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: A position paper. J. Hematol. Oncol.12, 131. 10.1186/s13045-019-0815-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baer, C. et al. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica101, 830–838. 10.3324/haematol.2016.145888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schonfeld, L. et al. ASXL1 mutations predict inferior molecular response to nilotinib treatment in chronic myeloid leukemia. Leukemia36, 2242–2249. 10.1038/s41375-022-01648-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochi, Y. et al. Clonal evolution and clinical implications of genetic abnormalities in blastic transformation of chronic myeloid leukaemia. Nat. Commun.12, 2833. 10.1038/s41467-021-23097-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu, Q. et al. HDAC I/IIb selective inhibitor Purinostat Mesylate combined with GLS1 inhibition effectively eliminates CML stem cells. Bioact. Mater.21, 483–498. 10.1016/j.bioactmat.2022.08.006 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.