Abstract
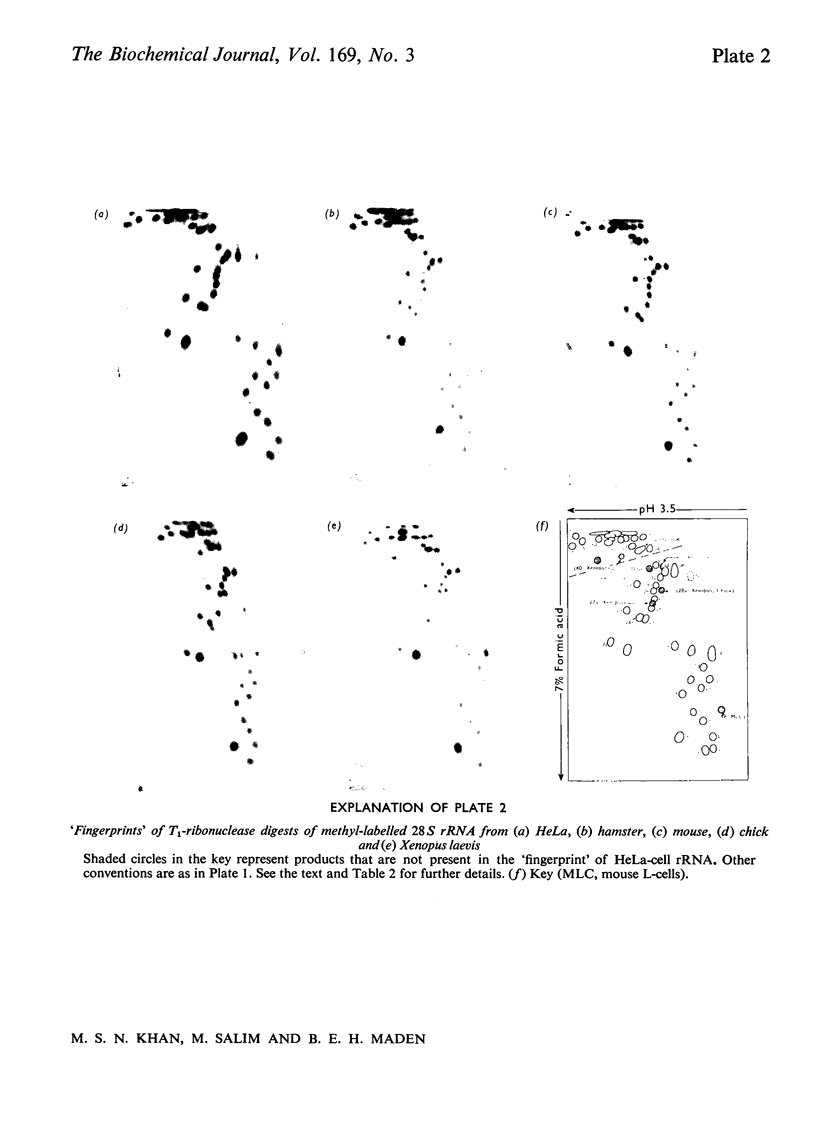
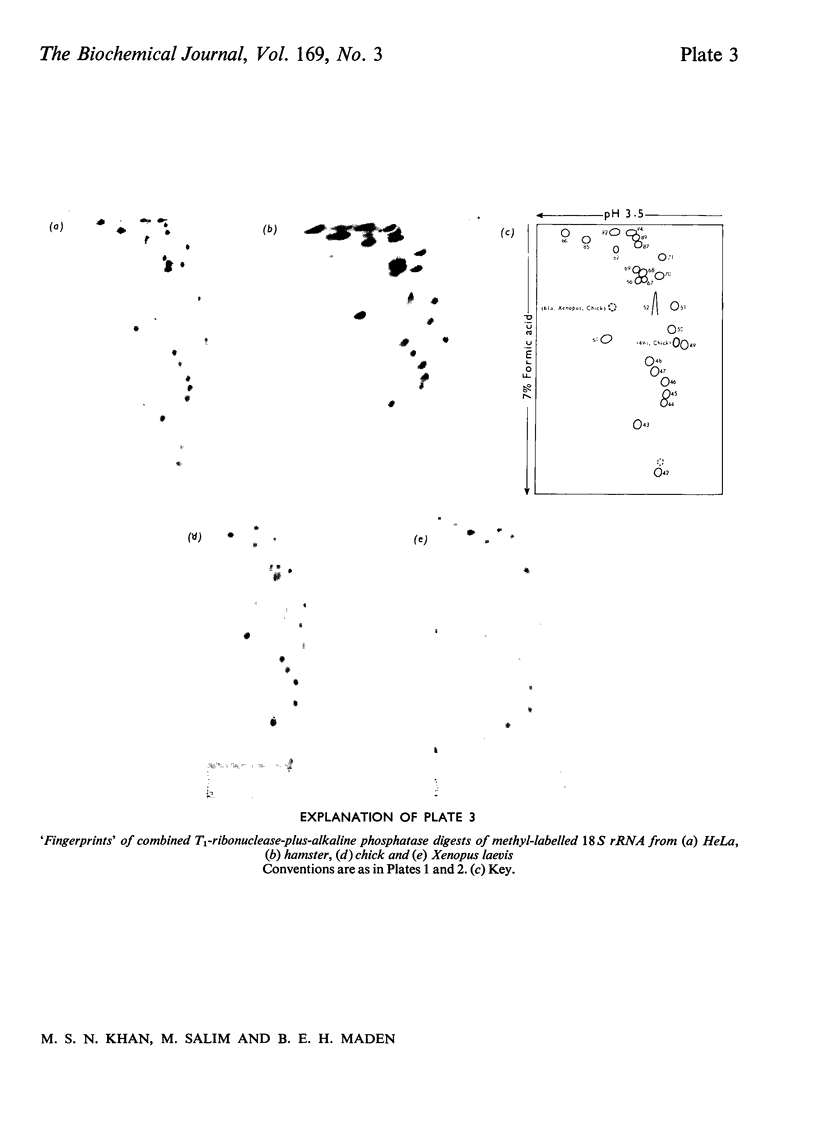
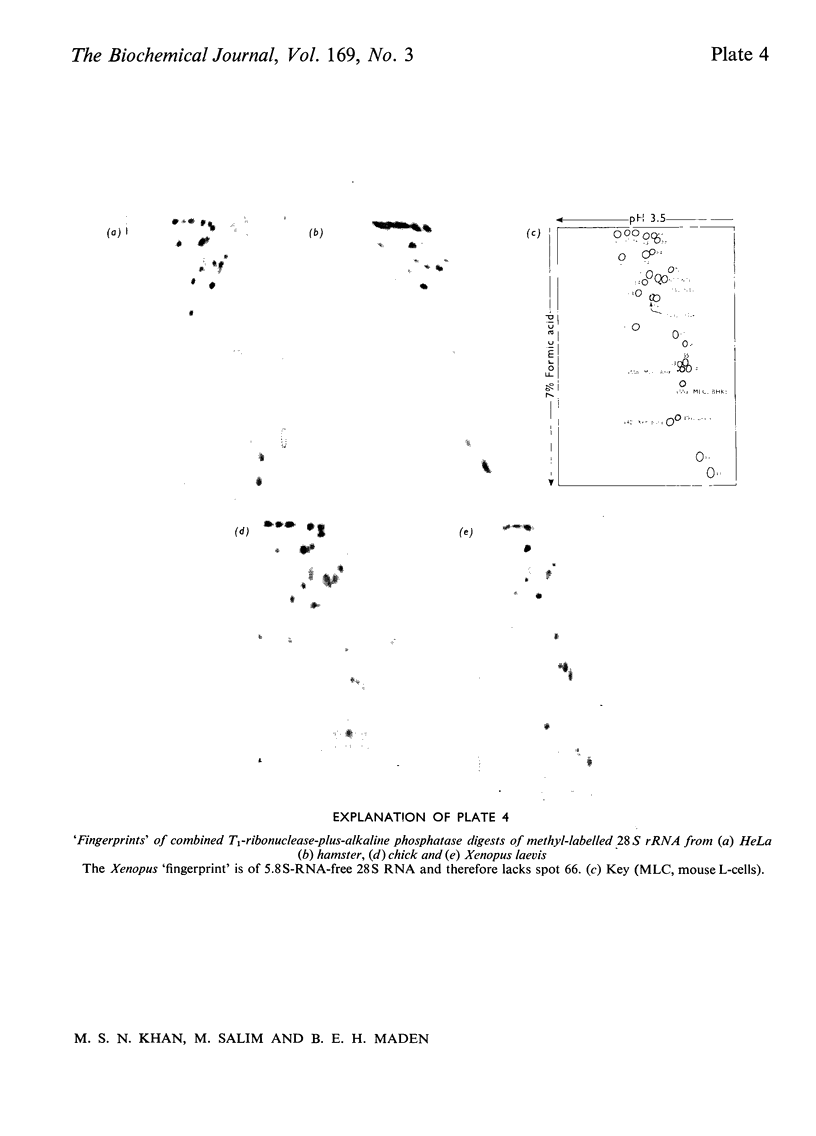
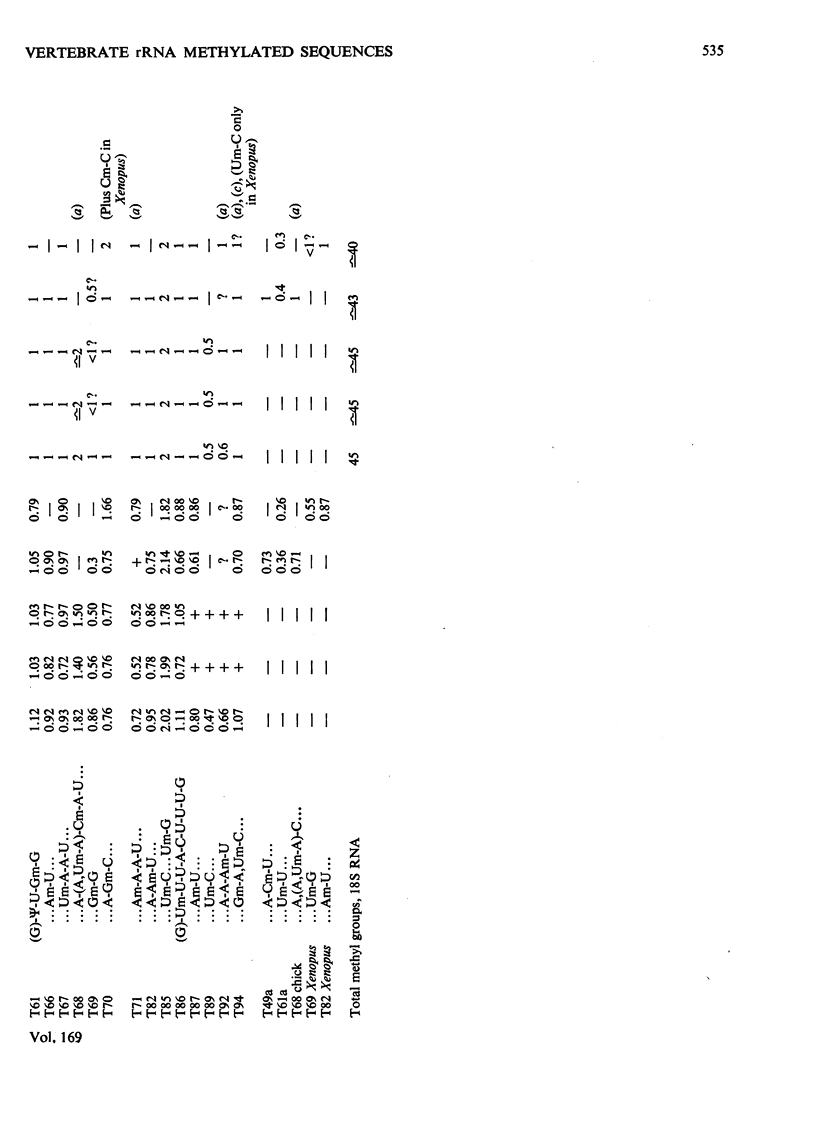
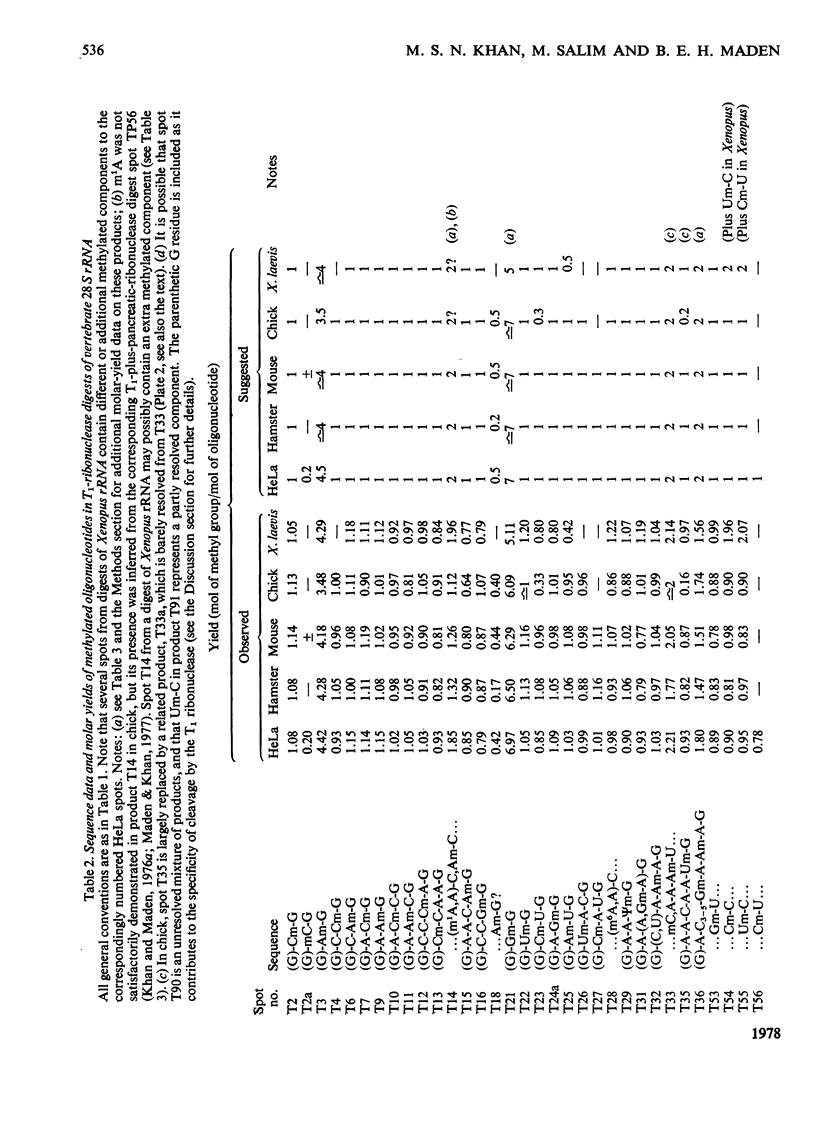
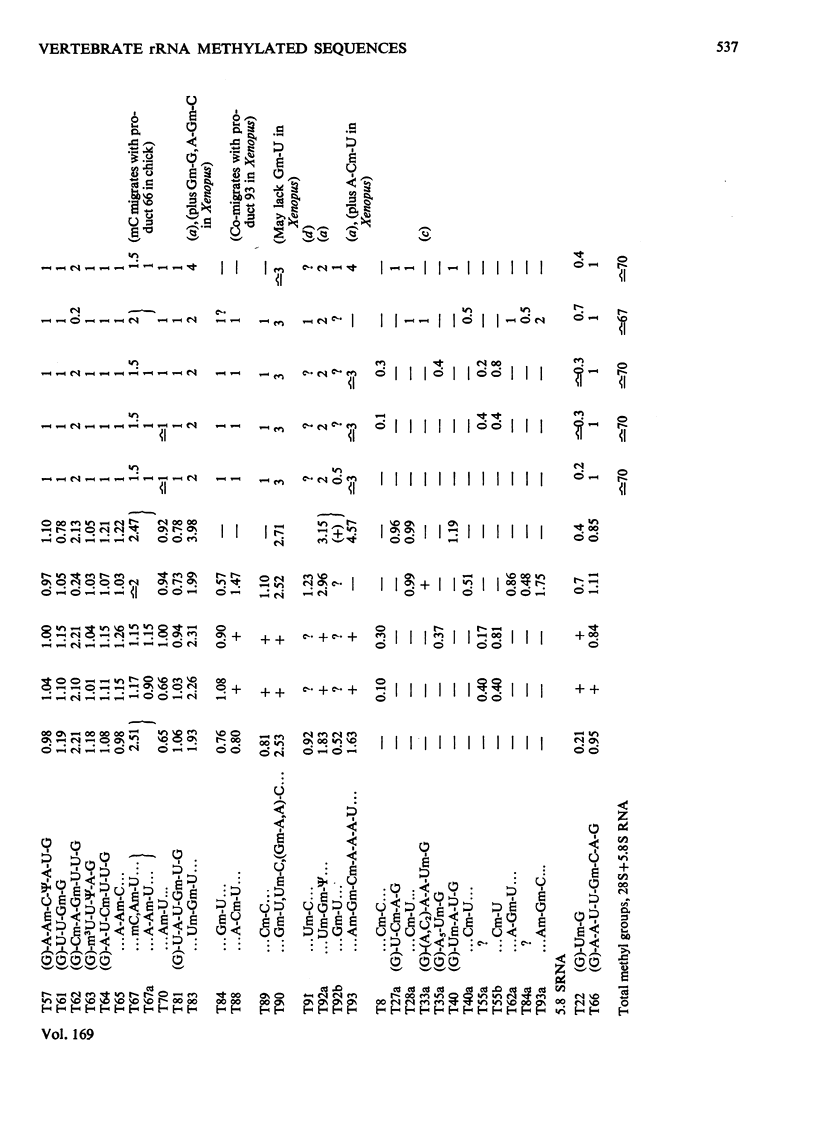
The methylated nucleotide sequences in the rRNA molecules of the following vertebrate cultured cells were compared: human (HeLa); hamster (BHK/C13); mouse (L); chick-embryo fibroblast; Xenopus laevis kidney. In each species the combined 18S, 28S and 5.8S molecules possess approx. 110-115 methyl groups, and the methylated oligonucleotides released after complete digestion of the rRNA by T1 ribonuclease encompass several hundred nucleotides. "Fingerprints" of the three mammalian methyl-labelled 18S rRNA species were qualitatively indistinguishable. "Fingerprints" of digests of 28S rRNA of hamster and mouse L-cells were extremely similar to those of HeLa cells, differing in one and three methylated oligonucleotides respectively. "Fingerprints" of methyl-labelled rRNA from chick and Xenopus strongly resembled those of mammals in most respects, but differed in several oligonucleotides in both 18S and 28S rRNA. At least some of the differences between "fingerprints" appear to be due to single base changes or to the presence or absence of methyl groups at particular points in the primary sequence. The findings strongly suggest that the methylated-nucleotide sequences are at least 95% homologous between the rRNA molecules of the two most distantly related vertebrates compared, man and Xenopus laevis.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand R. C., Klootwijk J., Planta R. J., Maden B. E. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J. 1978 Jan 1;169(1):71–77. doi: 10.1042/bj1690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Nucleotide sequences from the low molecular weight ribosomal RNA of Escherichia coli. J Mol Biol. 1967 Feb 14;23(3):337–353. doi: 10.1016/s0022-2836(67)80109-8. [DOI] [PubMed] [Google Scholar]
- Fuke M., Busch H. Partial methylation of 18 S ribosomal RNA detected by T1 ribonuclease digestion and homochromatography fingerprinting. FEBS Lett. 1977 May 15;77(2):287–290. doi: 10.1016/0014-5793(77)80253-6. [DOI] [PubMed] [Google Scholar]
- Fuke M., Busch H., Rao P. N. Evolutionary trends in 18S ribosomal RNA nucleotide sequences of rat, mouse, hamster and man. Nucleic Acids Res. 1976 Nov;3(11):2939–2957. doi: 10.1093/nar/3.11.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold M. D., Brown R. D., Tocchini-Valentini G. P. An analysis of the degree of homology between 28S rRNA from Xenopus laevis and Xenopus mulleri. Biochem Biophys Res Commun. 1974 Jun 18;58(4):1093–1103. doi: 10.1016/s0006-291x(74)80256-1. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Sakai M., Muramatsu M. 2'-O-methylated oligonucleotides in ribosomal 18S and 28S RNA of a mouse hepatoma, MH 134. Biochemistry. 1975 May 6;14(9):1956–1964. doi: 10.1021/bi00680a024. [DOI] [PubMed] [Google Scholar]
- Judes C., Jacob M. Compartmentation of the S-adenosylmethionne pool in developing chick embryo cerebral hemispheres, as demonstrated by a fingerprint study of 18 S ribosomal RNA. FEBS Lett. 1972 Nov 1;27(2):289–292. doi: 10.1016/0014-5793(72)80643-4. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Conformation of mammalian 5.8 S ribosomal RNA: S1 nuclease as a probe. FEBS Lett. 1976 Dec 15;72(1):105–110. doi: 10.1016/0014-5793(76)80823-x. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequences within the ribosomal ribonucleic acids of HeLa cells, Xenopus laevis and chick embryo fibroblasts. J Mol Biol. 1976 Feb 25;101(2):235–254. doi: 10.1016/0022-2836(76)90375-2. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Forbes J., de Jonge P., Klootwijk J. Presence of a hypermodified nucleotide in HeLa cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. FEBS Lett. 1975 Nov 1;59(1):60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Khan M. S. Methylated nucleotide sequences in HeLa-cell ribosomal ribonucleic acid. Correlation between the results from 'fingerprinting' hydrolysates obtained by digestion with T1 ribonuclease and with T1 plus pancreatic ribonuclease. Biochem J. 1977 Oct 1;167(1):211–221. doi: 10.1042/bj1670211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Robertson J. S. Demonstration of the "5-8 S" ribosomal sequence in HeLa cell ribosomal precursor RNA. J Mol Biol. 1974 Aug 5;87(2):227–235. doi: 10.1016/0022-2836(74)90145-4. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol. 1974 Sep 5;88(1):133–152. doi: 10.1016/0022-2836(74)90299-x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Tissue specific differences in the 2'-O-methylation of eukaryotic 5.8S ribosomal RNA. FEBS Lett. 1975 Nov 1;59(1):83–87. doi: 10.1016/0014-5793(75)80346-2. [DOI] [PubMed] [Google Scholar]
- Saponara A. G., Enger M. D. The isolation from ribonucleic acid of substituted uridines containing alpha-aminobutyrate moieties derived from methionine. Biochim Biophys Acta. 1974 Apr 27;349(1):61–77. doi: 10.1016/0005-2787(74)90009-4. [DOI] [PubMed] [Google Scholar]
- Vaughan M. H., Jr, Soeiro R., Warner J. R., Darnell J. E., Jr The effects of methionine deprivation on ribosome synthesis in HeLa cells. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1527–1534. doi: 10.1073/pnas.58.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]






