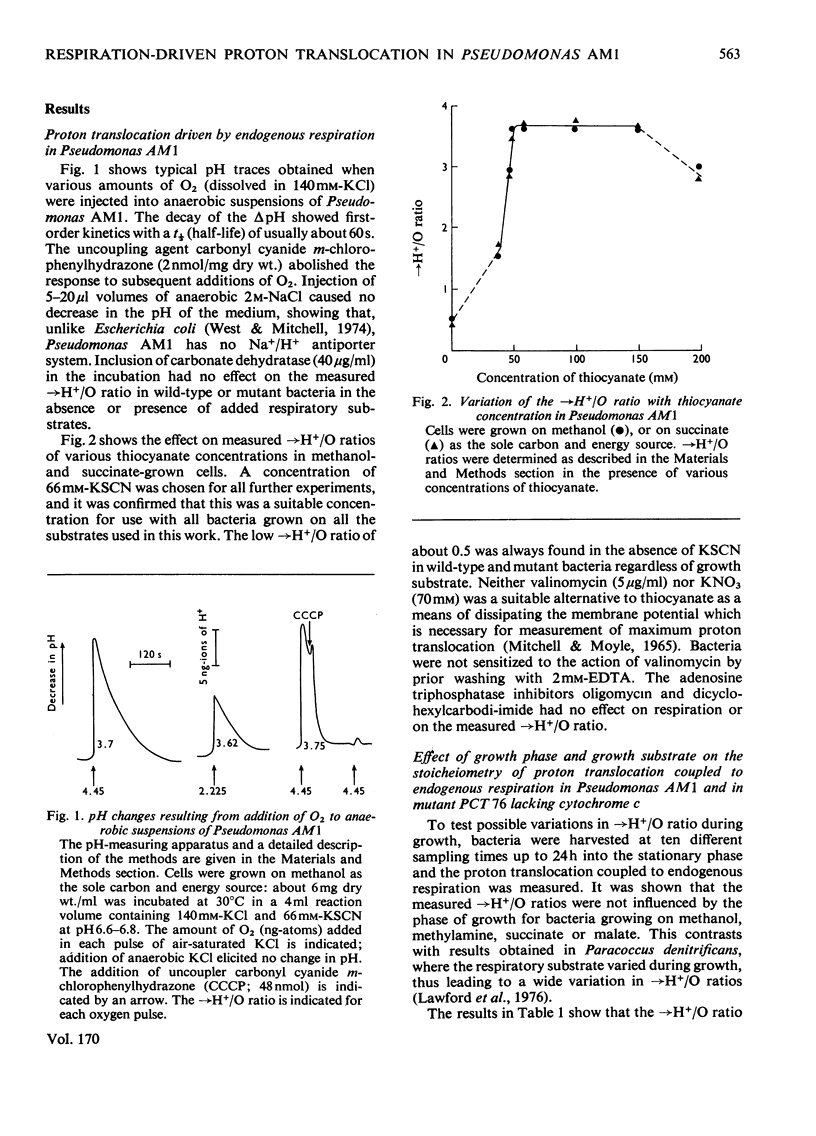
Abstract
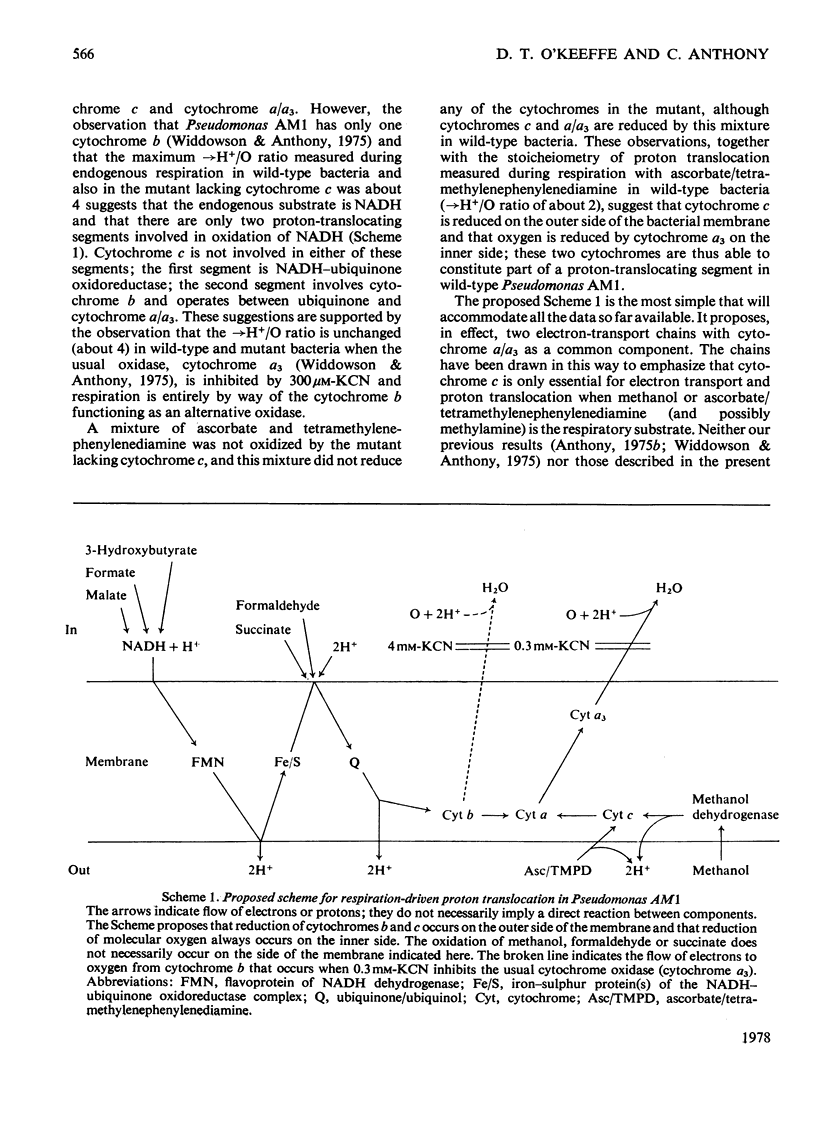
This paper clarifies the role of cytochrome c in Pseudomonas AM1 by measuring the stoicheiometry of proton translocation driven by respiration of endogenous or added substrates in wild-type bacteria and in a mutant lacking cytochrome c (mutant PCT76). The maximum →H+/O ratio (protons translocated out of the bacteria per atom of oxygen consumed during respiration) was about 4 and, except when respiration was markedly affected, this ratio was similar in mutant and wild-type bacteria. The →H+/O ratios were unaltered when the usual oxidase (cytochrome a3) was inhibited by 300μm-KCN and respiration involved the single cytochrome b functioning as an alternative oxidase. Ratios measured in cells respiring endogenous substrate and in cells loaded with malate or 3-hydroxybutyrate suggest that there are two proton-translocating segments operating during the oxidation of NADH. By contrast, during oxidation of formaldehyde or methylamine only one pair of protons is translocated. Proton translocation could not be measured with methanol as substrate, because its oxidation was inhibited (90–95%) by 5mm-KSCN. It is tentatively proposed that the electron-transport chain for NADH oxidation in Pseudomonas AM1 is arranged such that the NADH–ubiquinone oxidoreductase forms one proton-translocating segment and the second segment consists of ubiquinone and cytochromes b and a/a3. The cytochrome c appears to be essential only for respiration and proton translocation from methanol (and possibly from methylamine); there is no conclusive evidence that cytochrome c ever mediates between cytochromes b and a/a3 in Pseudomonas AM1.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C. The biochemistry of methylotrophic micro-organisms. Sci Prog. 1975 Summer;62(246):167–206. [PubMed] [Google Scholar]
- Anthony C. The microbial metabolism of C1 compounds. The cytochromes of Pseudomaonas AM1. Biochem J. 1975 Feb;146(2):289–298. doi: 10.1042/bj1460289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 1. Isolation and properties of Pseudomonas sp. M27. Biochem J. 1964 Sep;92(3):609–614. doi: 10.1042/bj0920609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg I., Rock J. S., Ben-Bassat A., Mateles R. I. Bacterial yields on methanol, methylamine, formaldehyde, and formate. Biotechnol Bioeng. 1976 Dec;18(12):1657–1668. doi: 10.1002/bit.260181202. [DOI] [PubMed] [Google Scholar]
- Haddock B. A., Jones C. W. Bacterial respiration. Bacteriol Rev. 1977 Mar;41(1):47–99. doi: 10.1128/br.41.1.47-99.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawford H. G., Cox J. C., Garland P. B., Haddock B. A. Electron transport in aerobically grown Paracoccus denitrificans: kinetic characterization of the membrane-bound cytochromes and the stoichiometry of respiration-driven proton translocation. FEBS Lett. 1976 May 1;64(2):369–374. doi: 10.1016/0014-5793(76)80330-4. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Moyle J. Respiration-driven proton translocation in rat liver mitochondria. Biochem J. 1967 Dec;105(3):1147–1162. doi: 10.1042/bj1051147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Moyle J. Stoichiometry of proton translocation through the respiratory chain and adenosine triphosphatase systems of rat liver mitochondria. Nature. 1965 Oct 9;208(5006):147–151. doi: 10.1038/208147a0. [DOI] [PubMed] [Google Scholar]
- Netrusov A. I., Rodionov Y. V., Kondratieva E. N. ATP-generation coupled with C1-compound oxidation by methylotrophic bacterium Pseudomonas sp.2. FEBS Lett. 1977 Apr 1;76(1):56–58. doi: 10.1016/0014-5793(77)80119-1. [DOI] [PubMed] [Google Scholar]
- Scholes P., Mitchell P. Respiration-driven proton translocation in Micrococcus denitrificans. J Bioenerg. 1971 Sep;1(3):309–323. doi: 10.1007/BF01516290. [DOI] [PubMed] [Google Scholar]
- West I. C., Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974 Oct;144(1):87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West I., Mitchell P. Proton-coupled beta-galactoside translocation in non-metabolizing Escherichia coli. J Bioenerg. 1972 Aug;3(5):445–462. doi: 10.1007/BF01516082. [DOI] [PubMed] [Google Scholar]
- Widdowson D., Anthony C. The microbial metabolism of C1 compounds. The electron-transport chain of Pseudomonas am1. Biochem J. 1975 Nov;152(2):349–356. doi: 10.1042/bj1520349. [DOI] [PMC free article] [PubMed] [Google Scholar]


