Abstract
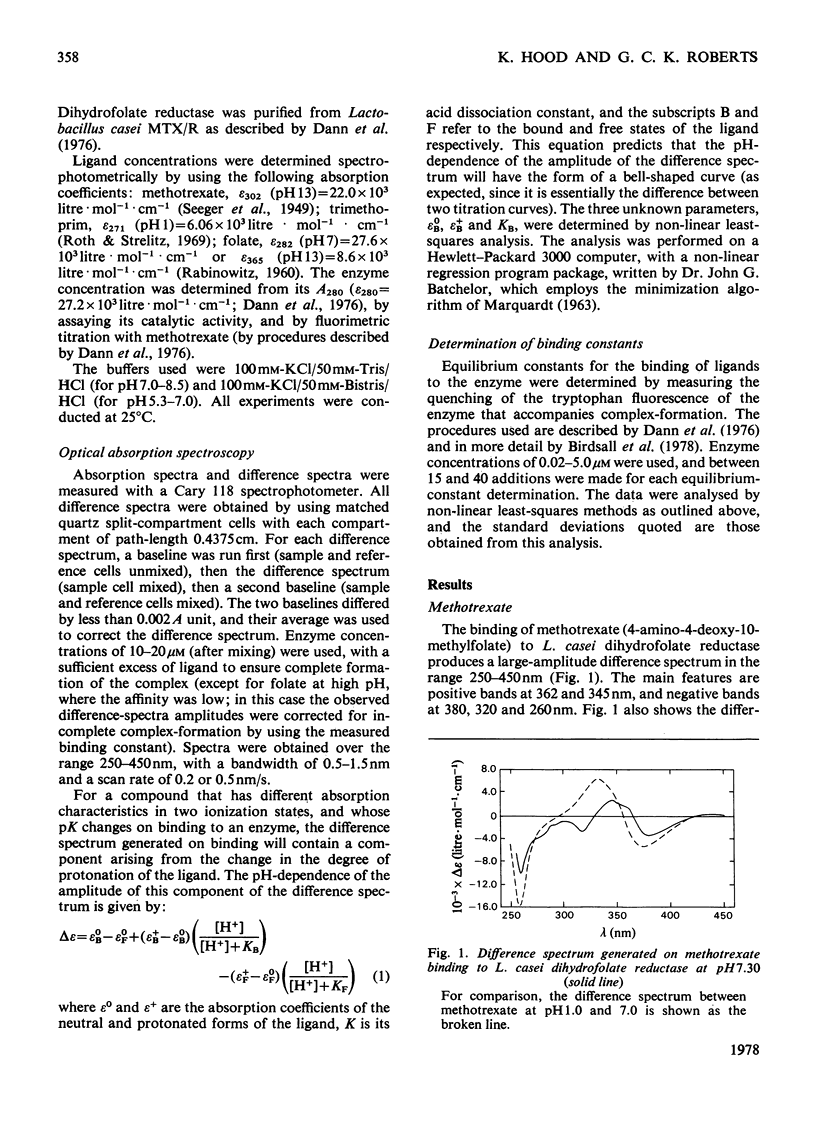
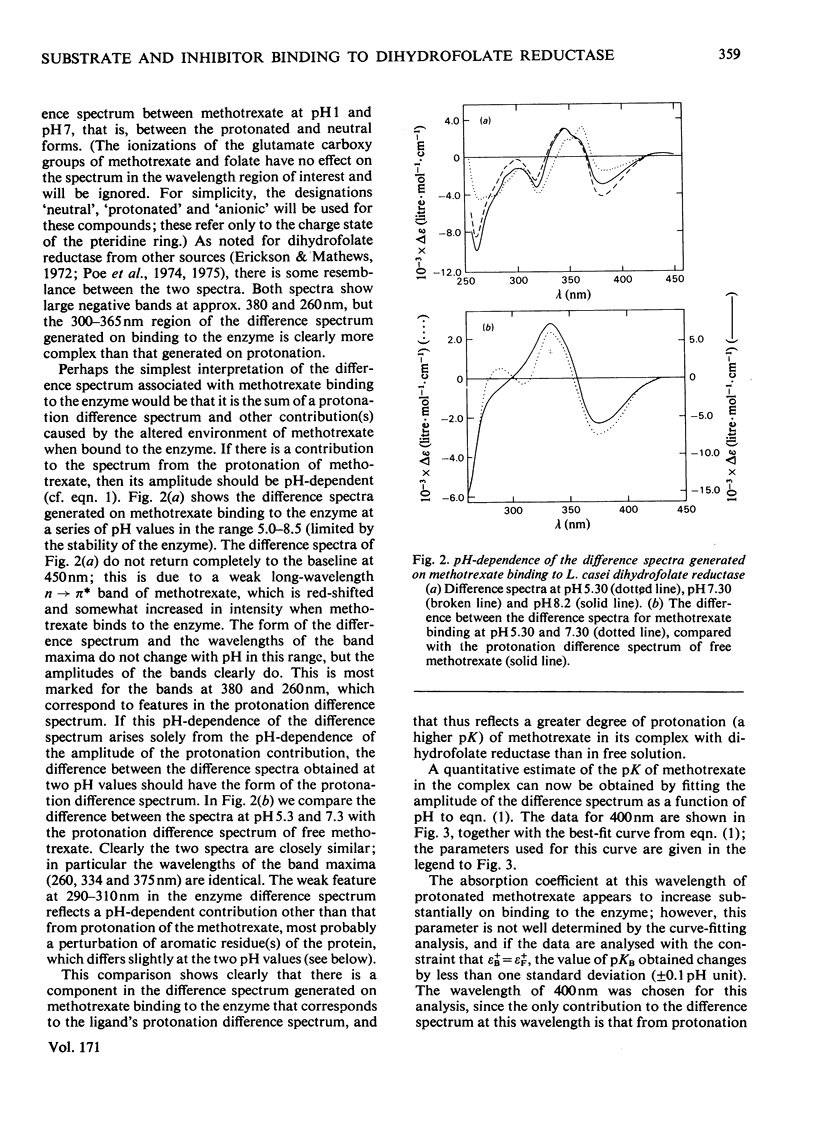
The u.v. difference spectra generated when methotrexate, trimethoprim or folate bind to Lactobacillus casei dihydrofolate reductase were analysed. The difference spectrum producted by methotrexate binding is shown to consist of three components: (a) one closely resembling that observed on protonation of methotrexate, reflecting an increased degree of protonation on binding; (b) a pH-independent contribution corresponding to a 40 nm shift to longer wavelengths of a single absorption band of methotrexate: (c) a component arising from perturbation of tryptophan residue(s) of the enzyme. Quantitative analysis of the pH-dependence of component (a) shows that pK of methotrexate is increased from 5.35 to 8.55 (+/-0.10) on binding. In contrast, folate is not protonated when bound to the enzyme at neutral pH. At pH7.5, where methotrexate is bound 2000 times more tightly than folate, one-third of the difference in binding energy between the two compounds arises from the difference in chaarge stage. A similar analysis of the difference spectra generated on trimethoprim binding demonstrates that this compound, too, shows an increase in pK on binding but only from 7.22 to 7.90 (+/-0.10), suggesting that its 2,4-diaminopyrimidine ring does not bind to the enzyme in precisely the same way as the corresponding moiety of methotrexate.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birdsall B., Griffiths D. V., Roberts G. C., Feeney J., Burgen A. 1H nuclear magnetic resonance studies of Lactobacillus casei dihydrofolate reductase: effects of substrate and inhibitor binding on the histidine residues. Proc R Soc Lond B Biol Sci. 1977 Mar 18;196(1124):251–265. doi: 10.1098/rspb.1977.0040. [DOI] [PubMed] [Google Scholar]
- COLLIN R., PULLMAN B. ON THE MECHANISM OF FOLIC ACID REDUCTASE INHIBITION. Biochim Biophys Acta. 1964 Aug 26;89:232–241. doi: 10.1016/0926-6569(64)90212-3. [DOI] [PubMed] [Google Scholar]
- DIXON M. The effect of pH on the affinities of enzymes for substrates and inhibitors. Biochem J. 1953 Aug;55(1):161–170. doi: 10.1042/bj0550161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann J. G., Ostler G., Bjur R. A., King R. W., Scudder P., Turner P. C., Roberts G. C., Burgen A. S. Large-scale purification and characterization of dihydrofolate reductase from a methotrexate-resistant strain of Lactobacillus casei. Biochem J. 1976 Sep 1;157(3):559–571. doi: 10.1042/bj1570559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. S., Mathews C. K. Spectral changes associated with binding of folate compounds to bacteriophage T4 dihydrofolate reductase. J Biol Chem. 1972 Sep 25;247(18):5661–5667. [PubMed] [Google Scholar]
- Feeney J., Roberts G. C., Birdsall B., Griffiths D. V., King R. W., Scudder P., Burgen A. 1H nuclear magnetic resonance studies of the tyrosine residues of selectively deuterated Lactobacillus casei dihydrofolate reductase. Proc R Soc Lond B Biol Sci. 1977 Mar 18;196(1124):267–290. doi: 10.1098/rspb.1977.0041. [DOI] [PubMed] [Google Scholar]
- Freisheim J. H., Ericsson L. H., Bitar K. G., Dunlap R. B., Reddy A. V. An active center tryptophan residue in dihydrofolate reductase: chemical modification, sequence surrounding the critical residue, and structural homology considerations. Arch Biochem Biophys. 1977 Apr 30;180(2):310–317. doi: 10.1016/0003-9861(77)90043-1. [DOI] [PubMed] [Google Scholar]
- Hitchings G. H., Burchall J. J. Inhibition of folate biosynthesis and function as a basis for chemotherapy. Adv Enzymol Relat Areas Mol Biol. 1965;27:417–468. doi: 10.1002/9780470122723.ch9. [DOI] [PubMed] [Google Scholar]
- Hood K., Roberts G. C. The charge state of substrates and inhibitors when bound to Lactobacillus casei dihydrofolate reductase [proceedings]. Biochem Soc Trans. 1977;5(3):771–772. doi: 10.1042/bst0050771. [DOI] [PubMed] [Google Scholar]
- Kimber B. J., Griffiths D. V., Birdsall B., King R. W., Scudder P., Feeney J., Roberts G. C., Burgen A. S. 19 Fnuclear magnetic resonance studies of ligand binding to 3-fluorotyrosine-and 6-fluorotryptophan-containing dihydrofolate reductase from Lactobacillus casei. Biochemistry. 1977 Jul 26;16(15):3492–3500. doi: 10.1021/bi00634a032. [DOI] [PubMed] [Google Scholar]
- Liu J. K., Dunlap R. B. Implication of a tryptophyl residue in the active site of dihydrofolate reductase. Biochemistry. 1974 Apr 23;13(9):1807–1814. doi: 10.1021/bi00706a005. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Alden R. A., Bolin J. T., Freer S. T., Hamlin R., Xuong N., Kraut J., Poe M., Williams M., Hoogsteen K. Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate. Science. 1977 Jul 29;197(4302):452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- PERAULT A. M., PULLMAN B. [Electronic structure and mode of action of folic acid antimetabolites]. Biochim Biophys Acta. 1961 Sep 16;52:266–280. doi: 10.1016/0006-3002(61)90676-x. [DOI] [PubMed] [Google Scholar]
- Warwick P. E., D'Souza L., Freisheim J. H. Role of tryptophan in dihydrofolate reductase. Biochemistry. 1972 Sep 26;11(20):3775–3779. doi: 10.1021/bi00770a016. [DOI] [PubMed] [Google Scholar]
- Williams M. N. Effect of N-bromosuccinimide modification on dihydrofolate reductase from a methotrexate-resistant strain of Escherichia coli. Activity, spectrophotometric, fluorescence and circular dichroism studies. J Biol Chem. 1975 Jan 10;250(1):322–330. [PubMed] [Google Scholar]
- ZAKRZEWSKI S. F. The mechanism of binding of folate analogues by folate reductase. J Biol Chem. 1963 Apr;238:1485–1490. [PubMed] [Google Scholar]


