Abstract
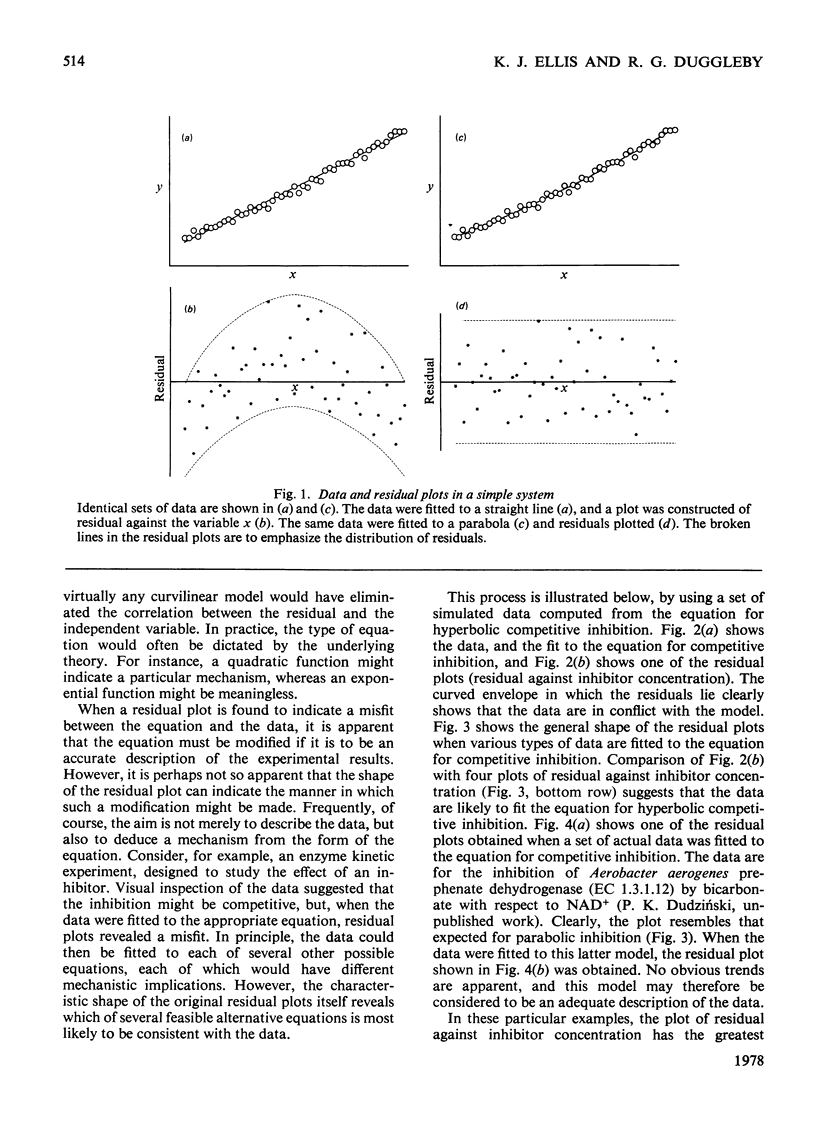
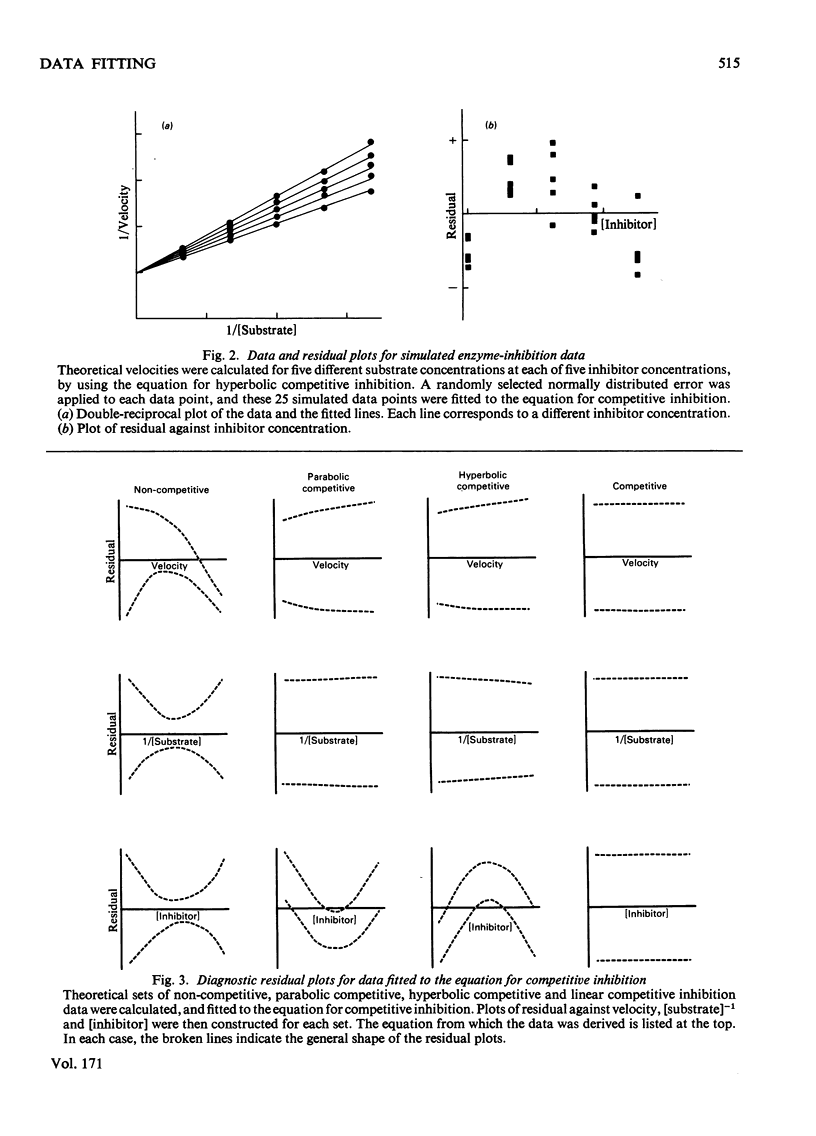
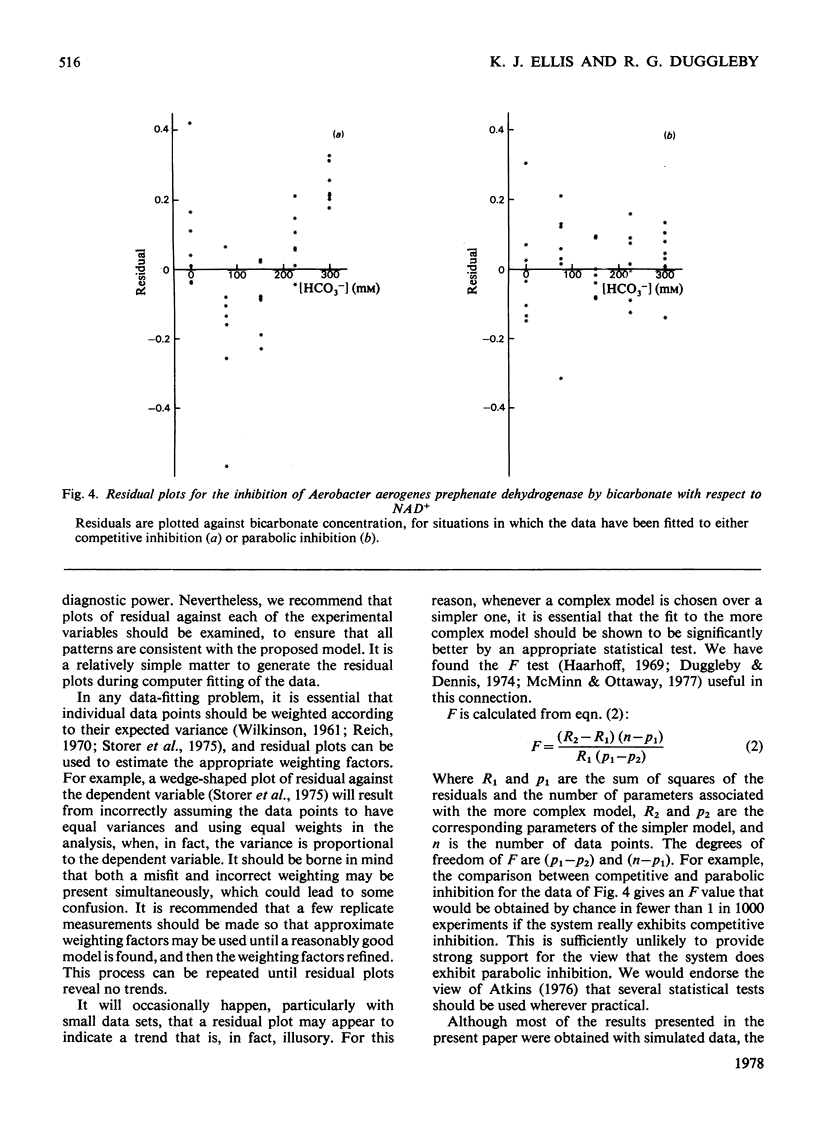
In many problems of data analysis it is necessary to fit the data to a mathematical equation. Random errors of measurement will be responsible for deviations between the data and the equation, but superimposed on this there may be deviations that result from the equation being an inadequate description of the system from which the data were obtained. Plots of the residual (i.e. the difference between the experimental and calculated values of the dependent variable) against each of the experimental variables have been previously used to detect a misfit between the data and the equation. In the present paper, we show that the shape of the residual plots may be used as a guide in choosing a more appropriate equation. In addition, residual plots give useful information on the error structure of the data, and hence the weighting factors that should be used in the analysis.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins G. L. Tests for the goodness of fit of models. Biochem Soc Trans. 1976;4(2):357–361. doi: 10.1042/bst0040357. [DOI] [PubMed] [Google Scholar]
- Bártfai T., Mannervik B. A procedure based on statistical criteria for discrimination between steady state kinetic models. FEBS Lett. 1972 Oct 1;26(1):252–256. doi: 10.1016/0014-5793(72)80585-4. [DOI] [PubMed] [Google Scholar]
- Duggleby R. G., Dennis D. T. Nicotinamide adenine dinucleotide-specific glyceraldehyde 3-phosphate dehydrogenase from Pisum sativum. Assay and steady state kinetics. J Biol Chem. 1974 Jan 10;249(1):167–174. [PubMed] [Google Scholar]
- Haarhoff K. N. Use of multivariate non-linear regression analysis in fitting enzyme kinetic models. An empirical study of the inhibition of aspartate aminotransferase by dicarboxylic acid substrate analogues. J Theor Biol. 1969 Jan;22(1):117–150. doi: 10.1016/0022-5193(69)90083-6. [DOI] [PubMed] [Google Scholar]
- McMinn C. L., Ottaway J. H. Studies on the mechanism and kinetics of the 2-oxoglutarate dehydrogenase system from pig heart. Biochem J. 1977 Mar 1;161(3):569–581. doi: 10.1042/bj1610569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich J. G. Parameter estimation and enzyme kinetic models. FEBS Lett. 1970 Aug 31;9(5):245–251. doi: 10.1016/0014-5793(70)80367-2. [DOI] [PubMed] [Google Scholar]
- Reich J. G., Wangermann G., Falck M., Rohde K. A general strategy for parameter estimation from isosteric and allosteric-kinetic data and binding measurements. Eur J Biochem. 1972 Apr 11;26(3):368–379. doi: 10.1111/j.1432-1033.1972.tb01776.x. [DOI] [PubMed] [Google Scholar]
- Storer A. C., Darlison M. G., Cornish-Bowden A. The nature of experimental error in enzyme kinetic measurments. Biochem J. 1975 Nov;151(2):361–367. doi: 10.1042/bj1510361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]



