Abstract
N-Myc is a transcription factor that forms heterodimers with the protein Max and binds gene promoters by recognizing a DNA sequence, CACGTG, called E-box. The identification of N-myc target genes is an important step for understanding N-Myc biological functions in both physiological and pathological contexts. In this study, we describe the identification of N-Myc-responsive genes through chromatin immunoprecipitation and methylation-sensitive restriction analysis. Results show that N-Myc is a direct regulator of several identified genes, and that methylation of the CpG dinucleotide within the E-box prevents the access of N-Myc to gene promoters in vivo. Furthermore, methylation profile of the E-box within the promoters of EGFR and CASP8, two genes directly controlled by Myc, is cell type-specific, suggesting that differential E-box methylation may contribute to generating unique patterns of Myc-dependent transcription. This study illuminates a central role of DNA methylation in controlling N-Myc occupancy at gene promoters and modulating its transcriptional activity in cancer cells.
Keywords: chromatin immunoprecipitation, neuroblastoma, C-Myc, EGFR, Caspase 8
The Myc oncoproteins, c-Myc, N-Myc, and L-Myc, share a conserved basic helix-loop-helix zip DNA binding motif that can dimerize with proteins of the Max subfamily (1-3). Besides their involvement in many normal biological processes (4), Myc proteins are implicated in the etiology of several types of human cancer (5). Although c-Myc activity has been well characterized, much less is known about the specific role of N-Myc, mainly because of the assumption that c-Myc and N-Myc are redundant (6). However, gene inactivation studies have shown that functional loss of either c-myc or N-myc leads to embryonic lethality, suggesting that both genes are essential and play distinct roles in mammalian development (7-9). This idea has been further reinforced by recent studies showing that conditional inactivation of N-myc specifically impairs the regulation of neuronal progenitor cell proliferation, differentiation, and nuclear size (10).
The identification of genes directly controlled by N-Myc represents an important step toward understanding the role of N-Myc in cancer biology, and particularly in tumors, in which the N-MYC gene is commonly amplified and/or overexpressed (11, 12). Despite the many strategies adopted to identify genes regulated by Myc proteins (13-19), a full comprehension of which genes are genuinely regulated by Myc, and how, is far from being achieved. This aspect has been also complicated by the finding that Myc appears to be associated with several DNA sites interspersed within the genomic DNA (17, 19, 20).
Myc preferentially associates with sites in genomic DNA with a high CpG dinucleotide content, called CpG islands (17). Dense methylation of the cytosine residue within the island causes gene inactivation (21). Interestingly, the E-box DNA sequence (CACGTG) recognized by Myc proteins contains a central CpG dinucleotide in which the cytosine may become methylated. This modification could affect the binding of Myc/Max to DNA and contribute to promoter specificity (17, 22, 23). It has also been shown that Myc associates with acetylated chromatin (17, 24-26), typical of unmethylated promoter regions, suggesting that the access of Myc to promoters may require specific DNA and chromatin modifications.
Based on these premises, we analyzed N-Myc binding and transcriptional activity in human neuroblastoma cells to identify direct N-Myc target genes and evaluate the role of E-box methylation as a determinant of N-Myc-mediated gene regulation. Our results provide compelling evidence that E-box methylation is critical in controlling Myc occupancy at gene promoters and in modulating its transcriptional activity in cancer cells.
Materials and Methods
Cell Cultures. Human neuroblastoma SK-N-BE, SH-SY-5Y, and LAN-1 cells were grown in DMEM containing 10% FBS and 50 μg/ml gentamycin. HL-60 and HEK-293 cells were grown in RPMI medium 1640 containing 10% heat-inactivated FBS and 50 μg/ml gentamycin. SK-N-BE9N and Tet-21/N cells have been described (27, 28).
Construction and Analysis of Genomic Libraries. Immunoprecipitated genomic DNA was double-digested with PmlI and EcoRI, HindIII, or BamHI. DNA fragments were cloned into pBlueScript-SKII (Stratagene) and sequenced. Sequences were analyzed by blast and map viewer software tools from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov). E-box sites were identified by using the match 1.0 software from the BIOBASE Database (www.generegulation.com).
Chromatin Immunoprecipitation (ChIP) Assay. ChIP was performed as described (29). The following antibodies were obtained: IgG (Santa Cruz Biotechnology sc-2027); N-Myc (BD Pharmingen B8.4.B); c-Myc (Santa Cruz Biotechnology N-262); and Max (Santa Cruz Biotechnology C-17). Promoter regions were PCR-amplified with specific pairs of primers listed in Table 1, which is published as supporting information on the PNAS web site.
EMSA. Nuclear extracts were prepared according to Dignam et al. (30). DNA probes were as follows: WT E-box, 5′-CTTCAGCGAGCCACGTGGACCAACTC-3′; mutated E-box, 5′-CTTCAGCGAGCCTCGAGGACCAACT-3′; and a WT E-box containing the central cytosine methylated on both strands.
Immunoblotting Analysis. Western blots were performed according to standard procedures with 50 μg of whole-cell extracts. The antibodies used were as follows: anti-N-Myc mAb (OP13, Oncogene); anti-β-actin antibody (A2066, Sigma-Aldrich); and anti-c-Myc antibody (9E10, Santa Cruz Biotechnology).
Northern Blot Analysis, RT-PCR, and Real-Time RT-PCR Analyses. Northern blot hybridizations were carried out as described (31). Probes were obtained by PCR (Table 1). RNAs were prepared by using Tri-Reagent (Sigma) and treated with DNase (DNA-free, Ambion). Reverse transcription and PCR were performed with the Thermoscript RT-PCR kit (Invitrogen). Real-time PCR was performed by using iQ SYBR green Supermix and the iQCycler thermocycler (Bio-Rad). Primers for real-time PCR are listed in Table 1.
Methylation-Sensitive PCR (MS-PCR). This protocol is an adapted version of arbitrary primer PCR (32). Genomic DNA, digested with the appropriate restriction enzyme (RE), was used for the PCR. Primers are described in Table 1.
Luciferase Assay. The pGL2-basic, pGL2-SV40, and Renilla-TK vectors were obtained from Promega. The pGL2-6XEBOX vector was generated by inserting a 70-bp fragment containing six E-box sites upstream of the TK-TATA box of pGL2-basic. CMV-N-MYC and CMV-C-MYC expression vectors were obtained by cloning the N-MYC or C-MYC coding regions into the 3XFLAG-CMV14 expression vector (Sigma). Activity of firefly or Renilla luciferase was measured with the dual luciferase assay kit (Promega) according to the manufacturer's instructions.
Results
Identification of N-Myc Target Genes. To identify N-Myc-responsive genes, we adopted a strategy based on cross-linking ChIP (29). Genomic libraries were generated by direct cloning of the immunoprecipitated DNA into appropriate vectors. However, our preliminary analysis revealed that these libraries contained a high level of nonspecific DNA, which compromised the search for genuine N-Myc DNA targets. Based on previous findings showing that the methylation of the E-box on its central cytosine can inhibit c-Myc/Max binding to DNA in vitro (17, 29), we devised a strategy to enrich for N-Myc-responsive genes by digesting the immunoprecipitated DNA with REs that cleave the unmethylated E-box sequence. PmlI is a commercial RE that meets this requirement because it specifically recognizes and cleaves the unmethylated CACGTG sequence only.
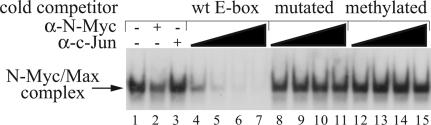
Although a few studies have demonstrated that E-box methylation can inhibit in vitro c-Myc/Max binding (29), there are no published data, to our knowledge, showing that this inhibition also applies to N-Myc. To address this issue, nuclear protein extracts from SK-N-BE, a human neuroblastoma cell line expressing N-Myc but not c-Myc, were tested in an EMSA to look for the formation of an N-Myc/Max-E-box complex. The presence of N-Myc in the complex was determined by using either α-N-Myc antibodies or α-c-Jun antibodies (negative control). The binding specificity of N-Myc was verified by competition analysis using a battery of cold DNAs. Results show that the methylated competitor failed to compete effectively (Fig. 1). Hence, the N-Myc/Max heterodimer binds to unmethylated E-box sequences only. Based on this result, we reasoned that selecting for genomic DNA fragments by PmlI digestion will increase the stringency of our selection of N-Myc-associated DNA. Cross-linked chromatin from SK-N-BE cells was immunoprecipitated with α-N-Myc antibodies, subsequently digested with PmlI together with EcoRI, BamHI, or HindIII and cloned into pBlueScript-SKII. Recombinant colonies were selected for further analyses.
Fig. 1.
N-Myc binds unmethylated E-boxes in vitro. Nuclear extracts from SK-N-BE cells were incubated in the presence of a radiolabeled canonical E-box probe. Protein composition of the DNA/protein complex was determined by using a specific anti-N-Myc antibody (lane 2) or an anti-c-Jun antibody used as a negative control (lane 3). Competition analysis was performed by incubating the binding reaction with 10-, 25-, 50-, and 100-fold molar excess of unlabeled double-stranded oligonucleotides: wt E-box (lanes 4-7); mutated E-box (lanes 8-11); and wt-E-box methylated on the central cytosine of both strands (lanes 12-15).
Identification and Analysis of N-Myc Targets. A total of 163 unique inserts were sequenced and analyzed by blast and map viewer software. Most DNA fragments were found to map at the 5′ end of genes, close to the predicted transcription start site. However, a significant portion of DNA inserts corresponds to repetitive DNA sequences (16.3%) or with regions located far from transcribed genes (30.4%). Clones corresponding to genes with known or inferred function represented 31.2% of the total, whereas 22.1% of the clones were derived from genes of unknown function. Known genes were grouped into functional classes based on their biological activity (Table 2, which is published as supporting information on the PNAS web site). The fact that several DNA inserts matched genes previously identified as c-Myc targets suggests that the library is enriched for genuine N-Myc-responsive promoters. Importantly, most promoters mapped within CpG islands. The entire promoter regions of the genes isolated in our library were analyzed by using match software. Interestingly, many contain additional E-boxes in addition to those present in cloned fragments, suggesting that we have identified bona fide MYC-regulated genes. Overall, our results show that the library was significantly enriched for DNA fragments corresponding to gene promoters carrying multiple canonical Myc binding sites.
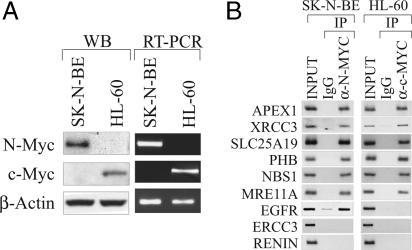
N-Myc Regulates Transcription of Identified Genes by Binding Directly to Their Promoters. To confirm direct association of N-Myc with the promoters of genes identified in our library, ChIP experiments were conducted in SK-N-BE cells. Immunoprecipitated DNA was analyzed by PCR using pairs of primers directed to a selected set of N-Myc-responsive promoters (APEX1, XRCC3, SLC25A19, PHB, NBS1, MRE11A, EGFR, and ERCC3). The analysis was extended to the HL-60 acute promyelocytic leukemia cell line, which in contrast to SK-N-BE cells, overexpresses c-Myc but lacks expression of N-Myc (Fig. 2A). ChIP results indicated that both N-Myc and c-Myc recognize the same array of gene promoters, but the EGFR promoter was an exception (Fig. 2B). N-Myc bound the EGFR promoter in neuroblastoma cells, whereas c-Myc did not bind to it in HL-60 cells. One gene, ERCC3, was not bound either by N-Myc or c-Myc, suggesting that a minor fraction of genes of the library might not be Myc-responsive targets. Thus, ERCC3 was used as a negative control in subsequent experiments.
Fig. 2.
N-Myc binds to the promoter of candidate genes in vivo. (A) Western blot and RT-PCR analysis of N-Myc and c-Myc expression in SK-N-BE and HL-60 cells. (B) Immunoprecipitation of cross-linked chromatin with control (IgG lane) or α-N-Myc antibodies. ChIP was performed in HL-60 cells with respect to c-Myc. Enrichment of individual promoters in both the N-Myc and c-Myc immunoprecipitation samples was determined by PCR. PCR detection of the RENIN promoter, which lacks canonical E-box sites, was used as a negative control. PCR products were visualized by ethidium bromide staining.
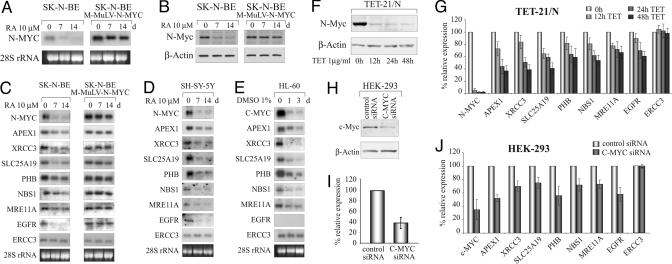
Next, the transcription profile of the N-Myc-responsive genes as a function of N-Myc expression was examined. SK-N-BE cells were treated with retinoic acid (RA), which turns off N-Myc expression (Fig. 3 A and B), and the levels of mRNA of selected target genes isolated after 0, 7, and 14 days of RA treatment were determined by Northern blotting. Results show that the mRNA level of most genes decreased upon silencing N-Myc transcription (Fig. 3C). The results were also validated in SH-SY-5Y cells, another human neuroblastoma cell line expressing N-Myc, although in the absence of gene amplification (Fig. 3D). Moreover, the analysis repeated in HL-60 cells differentiated with 1% DMSO showed that c-Myc affects the expression of the same set of genes (Fig. 3E). Interestingly the EGFR gene was an exception again in that it was expressed and regulated by N-Myc in SK-N-BE cells, but was silent in HL-60 cells lacking N-myc. The expression data mirrored the MYC occupancy data generated by ChIP analysis (Fig. 2B).
Fig. 3.
N-Myc directly regulates transcription of the identified genes. (A) mRNA expression of N-Myc in SK-N-BE and SK-N-BE9N cells as a function of RA treatment. (B) Expression of the N-Myc protein in SK-N-BE and SK-N-BE9N cells by Western blotting. The levels of Myc protein were normalized to β-actin. (C) Northern blotting analysis of genes in SK-N-BE versus SK-N-BE9N cells treated with RA for 0, 7, and 14 days. (D and E) mRNA expression analysis performed in SH-SY-5Y and HL-60 cells. The latter were differentiated with DMSO for 0, 1, and 3 days. (F) Expression level of N-Myc protein in Tet-21/N cells after treatment with tetracycline (TET). (G) Relative expression of N-Myc and target genes at different time points of tetracycline treatment by real-time PCR. (H and I) Repression of c-Myc by RNA interference in HEK-293 cells using specific C-MYC small interfering RNAs (siRNAs) (Ambion). Negative control was performed with a silencer duplex RNA (control siRNA lane). c-Myc protein was detected by Western blot and quantified by densitometric analysis. (J) Measurement of mRNA expression of c-MYC and tested genes by real-time PCR in HEK-293 cells.
To exclude that secondary effects of RA treatment account for our observations, SK-N-BE cell clones were constructed that express N-Myc under the control of the strong LTR promoter of the Molony murine leukemia virus. Expression analysis of one clone (SK-N-BE9N) showed that, in contrast to parental cells, RA does not affect N-Myc mRNA and protein levels (Fig. 3 A and B). Consistent with this finding, expression of genes tested did not change as a function of RA treatment (Fig. 3C). Our findings were also validated in Tet-21/N, a human neuroblastoma cell line in which N-Myc expression is controlled through a Tet-off inducible promoter (28). N-Myc transcript and protein levels were determined by real-time PCR and Western blotting, respectively, as a function of tetracycline treatment (Fig. 3 F and G). Fig. 3G shows that all gene transcripts, with the exception of ERCC3, decrease as a function of the reduction in N-Myc levels. Similar results were also obtained in HEK-293 cells in which c-Myc was silenced through RNA interference by means of a c-myc-specific duplex small interfering RNA (Ambion) (Fig. 3 H-J). Taken together, these data indicate that N-Myc and c-Myc are direct and positive transcriptional regulators of most genes identified by means of the screening performed in this study.
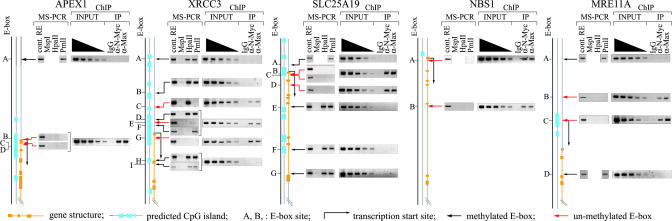
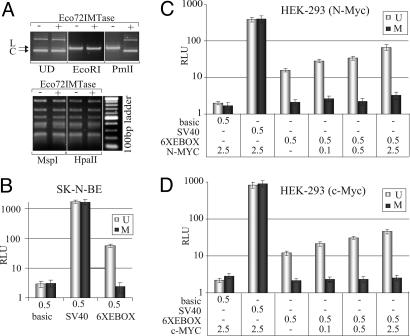
N-Myc Occupies Promoters at Unmethylated E-Box Sites in Vivo. To check whether N-Myc associates with unmethylated E-box sites in vivo, and if E-box methylation may contribute to controlling N-Myc function, SK-N-BE genomic DNA was digested with PmlI and tested in a MS-PCR assay. Primers, flanking each E-box, were designed for PCR. Briefly, if the E-box is unmethylated, then PmlI cleaves the E-box and prevents synthesis of the PCR product, whereas if the E-box is methylated, cleavage by PmlI is prevented, thus allowing synthesis of a specific PCR product. To verify that the technique was working properly, primers were designed to flank another methylation-sensitive restriction site (MspI/HpaII) in addition to PmlI. This control, however, could not be used in the analysis of every gene (e.g., E-box A of the APEX1 gene). Results of five gene promoters show that only one subset of E-boxes is unmethylated (Fig. 4, MS-PCR lanes). To determine which E-box N-Myc/Max binds in vivo, promoters were scanned by ChIP with an anti-N-Myc or anti-Max antibody. As shown in Fig. 4 (ChIP lanes), only those DNA regions containing unmethylated E-boxes could be immunoprecipitated. In a few cases, the resolution of the ChIP assay could not discriminate among clustered E-box sites (e.g., D, E, and F E-box sites of the XRCC3 promoter), because of the average size of the DNA fragments (≈500 bp). To establish whether methylation of the E-box can affect Myc association to DNA in vivo and transcription, we tested the effect of methylation on Myc activity on a luciferase reporter (pGL2-6XEBOX) amplified in either the DH10B or DHB10-MT strain of Escherichia coli; the latter expresses the Eco72I methyltransferase shown to specifically methylate the central cytosine of the CACGTG sequence (33). The methylation status of the E-boxes in the pGL2-6XEBOX vector was verified by restriction digestion of the plasmid with PmlI. Results show that the plasmid amplified in DH10B-MT was resistant to PmlI cleavage, although it could be digested by HpaII, another methylation-sensitive RE (Fig. 5A). The methylated and unmethylated forms of pGL2-6XEBOX were separately transfected into SK-N-BE cells and tested for their ability to drive luciferase expression. Results show that in contrast to the unmethylated vector, which displayed a high level of activity, the methylated version could not drive luciferase expression (Fig. 5B). To exclude the possibility that methylation might generically affect the stability of the reporter vector, we performed two controls. First, the differentially methylated pGL2-6XEBOX plasmids were separately transfected into HEK-293 cells and purified 48 h later from cell nuclei. The integrity of the plasmid DNA was analyzed by Southern blotting. Results show that both plasmids were intact and recovered in comparable amounts from cell nuclei (see Fig. 7, which is published as supporting information on the PNAS web site). Second, a pGL2-SV40 reporter vector that did not contain any E-box sequence was prepared from both E. coli strains and subsequently transfected into SK-N-BE and HEK-293 cells. As shown in Fig. 4 B-D, the pGL2-SV40 vector was equally efficient in expressing luciferase, regardless of the E. coli strain in which it was amplified. Based on these results we exclude possible generic effects of methylation on reporter transcription.
Fig. 4.
Unmethylated E-boxes are preferentially bound by N-Myc/Max in vivo. To determine the methylation status of the E-boxes within selected promoters, genomic DNA was digested with PmlI or appropriate REs (EcoRI, MspI, or HpaII) as controls and used as a template for PCR (MS-PCR). DNA, immunoprecipitated with N-Myc or Max antiserum, was PCR-amplified by using the same set of primers used for MS-PCR. Enrichment of E-box-containing DNA regions was determined by comparing the intensity of the PCR product with a scale of the input (from 250 to 10 ng). Preserum was used as a negative control (IgG lanes). Clustered E-boxes are indicated by the square brackets. The position of E-boxes relative to the transcription start site is listed in Table 3.
Fig. 5.
E-box methylation can affect Myc transcription activity in vivo. (A) The pGL2-6XEBOX vector, prepared from either DH10B(-) or DH10B-MT (+) E. coli strains, was separated on a 1% agarose gel and stained with ethidium bromide. Migration profiles of both plasmid forms were compared. UD, undigested; EcoRI, digestion with EcoRI used as a control; PmlI, digestion with PmlI to discriminate between methylated and unmethylated E-box sequences; MspI, digestion with MspI that cleaves the CCGG sequence regardless of cytosine methylation; HpaII, digestion with HpaII that cleaves the unmethylated CCGG sequence only. L, linear form of the plasmid; C, supercoiled form of the plasmid. (B) Luciferase activity of reporters prepared from DH10B (U, unmethylated) or DH10B-MT (M, methylated) after transfection into SK-N-BE cells. (C and D) Luciferase activity of reporters measured as a function of increased N-Myc or c-Myc expression in HEK-293 cells. The activity of the reporter firefly luciferase was normalized to that of the Renilla, used as an internal control. Relative luciferase units (RLU) were plotted on a semilogarithmic scale. Micrograms of transfected plasmids are indicated on the x axes of respective graphs.
Finally, to confirm that transcription was genuinely Myc-dependent, luciferase expression from both pGL2-6XEBOX plasmid forms was tested as a function of increasing concentration of c-Myc or N-Myc. Results show that the unmethylated reporter responds to both N-Myc and c-Myc in a dose-dependent fashion, whereas the methylated one is refractory to increased Myc expression (Fig. 5 C and D). We conclude that E-box methylation inhibits Myc-mediated transcription, possibly by preventing in vivo occupancy of the Myc/Max complex at promoters.
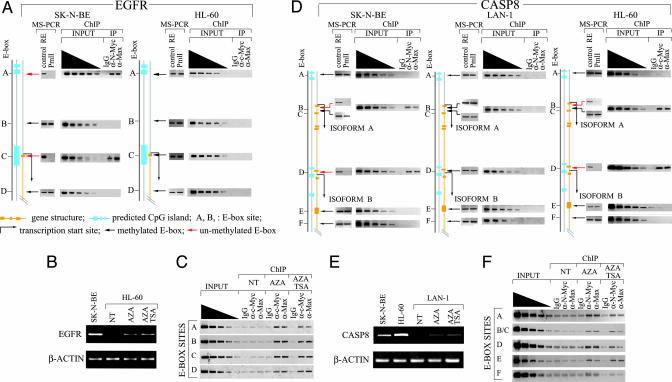
E-Box Methylation Controls the Binding of Myc Proteins to Their Cognate Sites in a Cell Type-Specific Fashion. EGFR was expressed in SK-N-BE but not in HL-60 cells. The EGFR promoter contains three E-boxes near the transcription start site, and another ≈13 kb away from it (Fig. 6A and Table 3, which is published as supporting information on the PNAS web site). ChIP data show that N-Myc, but not c-Myc, is associated with the EGFR promoter. A possible explanation for this finding is that E-box methylation contributes to dictating such a transcription pattern. To address this point, the EGFR expression profile was correlated with its E-box methylation status and Myc/Max occupancy in both SK-N-BE and HL-60 cells. Results show that two of four E-box sites are unmethylated in SK-N-BE cells and are also bound by N-Myc/Max in vivo. On the other hand, we did not observe association of the c-Myc/Max complex to any of the E-box sites in HL-60 cells and found that these sites were methylated in vivo (Fig. 6A). This finding suggests that methylation controls Myc/Max occupancy and accounts for the cell type-specific expression. To demonstrate a causal link between E-box methylation and Myc binding activity in vivo, HL-60 cells were treated with 5-aza-2′-deoxycytidine (5-aza-C), a demethylating agent that blocks the cellular DNA methyltransferase activity. If our hypothesis is correct, causing demethylation of the E-boxes by 5-aza-C treatment should allow for the binding of the Myc/Max complex to the EGFR promoter and the enhancement of EGFR promoter activity in HL-60 cells. As predicted, both the ChIP and transcription analyses showed that 5-aza-C could restore the binding of c-Myc to EGFR and increase gene transcription (Fig. 6 B and C). To demonstrate that this was also true for N-Myc, the same analysis was applied to the Caspase 8 (CASP8) gene, which, although it was not isolated through our screening, has been shown to be a direct target of c-Myc in lymphoblastoid cells (17). Moreover CASP8 promoter has been found hypermethylated in a significant percentage of human neuroblastomas (34). By screening a few neuroblastoma cell lines we found that LAN-1 cells carried a silenced CASP8 gene in which all E-box sites of the promoter were methylated and were not occupied by the N-Myc/Max complex (Fig. 6D). These cells, like SK-N-BE cells, express high levels of N-Myc but lack expression of c-Myc (data not shown). Moreover, unlike SK-N-BE cells, LAN-1 cells are sensitive to 5-aza-C, and thus it is possible to test whether N-Myc can bind the CASP8 promoter after demethylation treatment. As expected, treatment of LAN-1 cells with 5-aza-C reactivates the expression of CASP8 mRNA (Fig. 6E) and allows the binding of N-Myc to E-box sites (Fig. 6F).
Fig. 6.
The cell type-specific binding of Myc to promoters correlate with the E-box methylation status. (A) The methylation status of the EGFR E-boxes (MS-PCR) correlated with c-Myc/Max occupancy (ChIP) in SK-N-BE and HL-60 cells. PCR products were visualized by ethidium bromide staining. (B) 5-Aza-C treatment reactivates EGFR transcription in HL-60 cells. RNA levels were measured by RT-PCR. (C) Occupancy of c-Myc/Max complex at EGFR E-boxes in HL-60 cells was determined by ChIP. NT, not treated; AZA, cells treated with 3 μM 5-aza-C for 48 h; AZA+TSA, cells treated with 3 μM 5-aza-C and 500 nM TSA for 48 h. (D) The methylation status of the CASP8 E-box sites (MS-PCR) is correlated with N-Myc/Max occupancy (ChIP) in SK-N-BE, LAN1, and HL-60 cells. (E) Reactivation of CASP8 transcription in LAN-1 cells treated with 5-aza-C (3 μM) or a combination of 5-aza-C (3 μM) and TSA (500 nM). RNA levels were measured by RT-PCR. (F) Occupancy of the N-Myc/Max complex at CASP8 E-box sites in LAN-1 cells treated with 5-aza-C or 5-aza-C plus TSA.
It is interesting to note that after 5-aza-C treatment all E-box sites within the EGFR promoter of HL-60 cells and within the CASP8 promoter of LAN-1 cells become available for Myc/Max occupancy. This finding is consistent with 5-aza-C acting unselectively on methylated cytosines. Finally, treatment of HL-60 cells with both 5-aza-C and trichostatin A (TSA), a histone deacetylase inhibitor, was slightly more efficient in promoting EGFR transactivation than treatment with 5-aza-C alone (Fig. 6C), thus supporting previous studies showing that, for certain genes, Myc may preferentially function within the context of acetylated chromatin (17, 24-26).
Taken together, these findings provide strong evidence that E-box methylation prevents Myc binding to canonical E-box sites in vivo and support the notion that differential methylation of the E-box sites may contribute to the determination of unique patterns of Myc transcription.
Discussion
In this work, we describe the identification of genes whose transcriptional regulation depends on the direct association of the N-Myc/Max complex to their promoters through the recognition of canonical E-box sites. The identification of such genes is based on a strategy that integrates ChIP and DNA methylation analysis. Although our library was significantly enriched for N-Myc-responsive promoters the few hundred colonies generated by the library suggest that our approach, as compared with other screenings (17, 19, 20), underestimated the number of Myc sites within the genomic DNA of neuroblastoma cells. This result may be in part caused by the cloning strategy that did not take into account noncanonical Myc DNA binding sites and perhaps to the complex procedure used to clone N-Myc-bound promoters. Nonetheless, extensive bioinformatic analysis of screened clones shows that 80 of 163 inserts were derived from the promoters of genes, most of which carry multiple canonical E-box sites. Transcription analysis performed in SK-N-BE, SK-N-BE9N, SH-SY-5Y, and Tet-21/N cells reveals that N-Myc is a direct transactivator of many of the genes identified by our strategy. In agreement with previous reports (17), we observed that most Myc-bound sites localize to predicted CpG islands and are proximal to the gene transcription start site, suggesting that Myc may preferentially function close to the transcription initiation site.
Importantly, a comparative analysis of Myc occupancy at gene promoters and examination of the methylation status of specific E-boxes indicate that unmethylated E-boxes are the preferred binding sites of the Myc/Max complex in vivo. In most cases, at least two E-box sites were unmethylated and bound by Myc/Max in the promoters tested; this fact appears to be consistent with the recently proposed structural model in which Myc/Max heterotetramers may be needed to activate transcription (35). Finally, a correlative analysis carried out in SK-N-BE and HL-60 cells suggests that the E-box methylation profile and Myc occupancy of gene promoters are essentially conserved (data not shown). However, this status does not apply to the EGFR and CASP8 promoters whose E-box sites are all methylated in HL-60 and LAN-1 cells, respectively. Interestingly 5-aza-C can restore binding, respectively, of c-Myc to the EGFR promoter and N-Myc to the CASP8 promoter and reactivate their transcription, thus supporting a causal link between methylation of the E-box and Myc binding and transcription.
The present findings have two main implications. First, they support the notion that a specific E-box methylation pattern may determine which sites can be bound by Myc with consequential effects on gene transcription. Second, altered E-box methylation may play a critical role in misregulating Myc transcription functions during tumor development. It has been proposed that either DNA hypermethylation or hypomethylation may be responsible for tumor progression, possibly by repressing tumor suppressor genes and/or by inducing genomic instability (36, 37). Interestingly, hypomethylation correlates with overexpression of c-myc and an increased frequency of hematological tumors in mice (37). Our results extend this view and suggest that genome-wide hypomethylation in tumors, along with Myc overexpression, may redirect Myc to unmethylated E-box sites not bound in normal cells (17). This hypothesis may explain the relatively high number of Myc-bound sites within intergenic regions of genes in tumor cells (refs. 17 and 20 and this study). Overall, our study points to E-box methylation as another relevant regulatory level through which cells can control Myc access to DNA and modulate transcription of Myc-responsive genes.
Supplementary Material
Acknowledgments
We thank M. Schwab (German Cancer Research Center, Heidelberg) for providing the Tet-21/N cells, A. Lubis (Institute of Biotechnology, Vilnius, Lithuania) for providing the Eco72IMTase plasmid, and J. Reese and S. Biondi for critical reading of the manuscript. D.D. and A.P. were supported by the Ph.D. Program in Cell Biology and Physiology, University of Bologna. This work was supported by Italian Association for Research on Cancer Grants AIRC-2002 and AIRC-2003 and the European Union-Programma Regionale per la Ricerca Industriale, L'Innovazione et Il Trasferimento Tecnologico of the Emilia Romagna Region (to G.D.V.).
Author contributions: G.P. and G.D.V. designed research; D.D. and A.P. performed research; G.D.V. contributed new reagents/analytical tools; G.P., D.D., A.P., and G.D.V. analyzed data; and G.P. and D.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; RE, restriction enzyme; MS-PCR, methylation-sensitive PCR; RA, retinoic acid; 5-aza-C, 5-aza-2′-deoxycytidine; CASP8, Caspase 8; TSA, Trichostatin A.
References
- 1.Blackwood, E. M. & Eisenman, R. N. (1991) Science 251, 1211-1217. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel, A., Cziepluch, C., Hamann, U., Schurmann, J. & Schwab, M. (1991) EMBO J. 10, 3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amati, B., Dalton, S., Brooks, M. W., Littlewood, T. D., Evan, G. I. & Land, H. (1992) Nature 359, 423-426. [DOI] [PubMed] [Google Scholar]
- 4.Amati., B. & Land, H. (1994) Curr. Opin. Genet. Dev. 4, 102-108. [DOI] [PubMed] [Google Scholar]
- 5.Eisenman, R. N. (1989) in Oncogenes and the Molecular Origins of Cancer, ed. Weinberg, R. A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 175-221.
- 6.Malynn, B. A., de Alboran, I. M., O'Hagan, R. C., Bronson, R., Davidson, L., DePinho, R. A. & Alt, F. W. (2000) Genes Dev. 14, 1390-1399. [PMC free article] [PubMed] [Google Scholar]
- 7.Charron, J., Malynn, B. A., Fisher, P., Stewart, V., Jeannotte, L., Goff, S. P., Robertson, E. J. & Alt, F. W. (1992) Genes Dev. 6, 2248-2257. [DOI] [PubMed] [Google Scholar]
- 8.Sawai, S., Shimono, A., Wakamatsu, Y., Palmes, C., Hanaoka, K. & Kondoh, H. (1993) Development (Cambridge, U.K.) 117, 1445-1455. [DOI] [PubMed] [Google Scholar]
- 9.Nagy, A., Moens, C., Ivanyi, E., Pawling, J., Gertsenstein, M., Hadjantonakis, A. K., Pirity, M. & Rossant, J. (1998) Curr. Biol. 8, 661-664. [DOI] [PubMed] [Google Scholar]
- 10.Knoepfler, P. S., Cheng, P. F. & Eisenman, R. N. (2002) Genes Dev. 16, 2699-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwab, M., Alitalo, K., Klempnauer, K. H., Varmus, H. E., Bishop. J. M., Gilbert, F., Brodeur, G., Goldstein, M. & Trent, J. (1983) Nature 305, 245-248. [DOI] [PubMed] [Google Scholar]
- 12.Seeger, R. C., Brodeur, G. M., Sather, H., Dalton, A., Siegel, S. E., Wong, K. Y. & Hammond, D. (1985) N. Engl. J. Med. 313, 1111-1116. [DOI] [PubMed] [Google Scholar]
- 13.Boyd, K. E., Wells, J., Gutman, J., Bartley, S. M. & Farnham, P. J. (1998) Proc. Natl. Acad. Sci. USA 95, 13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coller, H. A., Grandori, C., Tamayo, P., Colbert, T., Lander, E. S., Eisenman, R. N. & Golub, T. R. (2000) Proc. Natl. Acad. Sci. USA 97, 3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuldiner, O. & Benvenisty, N. (2001) Oncogene 20, 4984-4994. [DOI] [PubMed] [Google Scholar]
- 16.Watson, J. D., Oster, S. K., Shago, M., Khosravi, F. & Penn, L. Z. (2002) J. Biol. Chem. 277, 36921-36930. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez, P. C., Frank, S. R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A. & Amati, B. (2003) Genes Dev. 17, 1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell, B. C., Cheung, A. F., Simkevich, C. P., Tam, W., Ren, X., Mateyak, M. K. & Sedivy, J. M. (2003) J. Biol. Chem. 278, 12563-12573. [DOI] [PubMed] [Google Scholar]
- 19.Orian, A., van Steensel, B., Delrow, J., Bussemaker, H. J., Li, L., Sawado, T., Williams, E., Loo, L. W., Cowley, S. M., Yost, C., et al. (2003) Genes Dev. 17, 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A. J., et al. (2004) Cell 116, 499-509. [DOI] [PubMed] [Google Scholar]
- 21.Bird, A. P. (1987) Trends Genet. 3, 342-347. [Google Scholar]
- 22.Patel, J. H., Loboda, A. P., Showe, M. K., Showe, L. C. & McMahon, S. B. (2004) Nat. Rev. Cancer 4, 562-568. [DOI] [PubMed] [Google Scholar]
- 23.Prendergast, G. C. & Ziff, E. B. (1991) Science 251, 186-189. [DOI] [PubMed] [Google Scholar]
- 24.Eberhardy, S. R., D'Cunha, C. A. & Farnham, P. J. (2000) J. Biol. Chem. 275, 33798-33805. [DOI] [PubMed] [Google Scholar]
- 25.McMahon, S. B., Wood, M. A. & Cole, M. D. (2000) Mol. Cell. Biol. 20, 556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. (2001) Genes Dev. 15, 2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peverali, F. A., Orioli, D., Tonon, L., Ciana, P., Bunone, G., Negri, M. & Della-Valle, G. (1996) Oncogene 12, 457-462. [PubMed] [Google Scholar]
- 28.Lutz, W., Stohr, M., Schurmann, J., Wenzel, A., Lohr, A. & Schwab, M. (1996) Oncogene 13, 803-812. [PubMed] [Google Scholar]
- 29.Weinmann, A. S. & Farnham, P. J. (2002) Methods 26, 37-47. [DOI] [PubMed] [Google Scholar]
- 30.Dignam, J. D., Lebowitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Church, G. M. & Gilbert, W. (1991) Proc. Natl. Acad. Sci. USA 81, 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, G., Gonzalgo, M. L., Salem, C. & Jones, P. A. (2002) Methods 27, 150-155. [DOI] [PubMed] [Google Scholar]
- 33.Rimseliene, R., Vaisvila, R. & Janulaitis, A. (1995) Gene 157, 217-219. [DOI] [PubMed] [Google Scholar]
- 34.Teitz, T., Wei, T., Valentine, M. B., Vanin, E.F., Grenet, J., Valentine, V. A., Behm, F. G., Look, A. T., Lahti, G. M. & Kidd, V. J. (2000) Nat. Med. 6, 529-535. [DOI] [PubMed] [Google Scholar]
- 35.Nair, S. K. & Burley, S. K. (2003) Cell 112, 193-205. [DOI] [PubMed] [Google Scholar]
- 36.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415-428. [DOI] [PubMed] [Google Scholar]
- 37.Gaudet, F., Hodgson, J. G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J. W., Leonhardt, H. & Jaenisch, R. (2003) Science 300, 489-492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.