Abstract
With about 200 000 phytochemicals in existence, identifying those of biomedical significance is a mammoth task. In the postgenomic era, relating metabolite fingerprints, abundances, and profiles to genotype is also a large task. Ion analysis using Fourier transformed ion cyclotron resonance mass spectrometry (FT-ICR-MS) may provide a high-throughput approach to measure genotype dependency of the inferred metabolome if reproducible techniques can be established. Ion profile inferred metabolite fingerprints are coproducts. We used FT-ICR-MS-derived ion analysis to examine gdhA (glutamate dehydrogenase (GDH; EC 1.4.1.1)) transgenic Nicotiana tabacum (tobacco) carrying out altered glutamate, amino acid, and carbon metabolisms, that fundamentally alter plant productivity. Cause and effect between gdhA expression, glutamate metabolism, and plant phenotypes was analyzed by labeling of amino acid fractions, and by FT-ICR-MS analysis of metabolites. The gdhA transgenic plants increased 13N labeling of glutamate and glutamine significantly. FT-ICR-MS detected 2 012 ions reproducible in 2 to 4 ionization protocols. There were 283 ions in roots and 98 ions in leaves that appeared to significantly change abundance due to the measured GDH activity. About 58% percent of ions could not be used to infer a corresponding metabolite. From the 42% of ions that inferred known metabolites we found that certain amino acids, organic acids, and sugars increased and some fatty acids decreased. The transgene caused increased ammonium assimilation and detectable ion variation. Thirty-two compounds with biomedical significance were altered in abundance by GDH including 9 known carcinogens and 14 potential drugs. Therefore, the GDH transgene may lead to new uses for crops like tobacco.
INTRODUCTION
Due to improvements in mass spectrometry (MS), the methods of metabolite analysis are becoming fast, reliable, sensitive, and automated [1] with broad applications to biological phenomena [2, 3, 4]. A range of analytical techniques can be used with complex biological samples. However, the development of ionization techniques such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) have provided robust techniques that can be widely applied [1]. Electron impact quadrupole MS is also evolving toward a robust technology for metabolite analysis [2, 3, 4]. Libraries of compound identities have been developed at a mass accuracy of 10 ppm (about 0.01 d), often by MS-MS fragmentation. In contrast, the mass accuracy of full-scan MS in a Fourier transformed ion cyclotron resonance mass spectrometer (FT-ICR-MS) format provides for mass accuracy to 1 ppm (about 0.001–0.0001 d) if the ion cyclotron is not filled [5, 6, 7]. The greater potential for mass accuracy is derived from the longer path length that allows for separation of a larger number of compounds, protein fragments, or DNA molecules per analysis.
However, with FT-ICR-MS the techniques for robust identification of ions, the methods for inference of the underlying metabolites, the supporting databases, and the methods for quantification are at an earlier stage of development and are less well known than for other MS formats [8]. The abundance of specific ions in total infusion mass spectra is the result of the combined ion suppression effects of all other components, pH and salinity of the solution, flow rate, tip opening, and electrospray current [9]. Small effects that alter the overall matrix composition may have large effects on total mass spectra. Therefore, although ion fingerprinting by FT-ICR-MS is a valuable tool for detecting subtle effects for mutant classification [10], exact masses alone may not be sufficient to identify specific compounds in more complex comparisons.
Post-genomic research that aims to determine gene function(s) and relationships among pathways and products will require more tools for metabolite analyses [1]. While multiparallel analyses of mRNA and protein abundance provide indirect information on the biochemical function of genes, metabolic analysis can provide direct information on instantiations [4]. Biological function is the sum of gene interactions and metabolic network interactions; both are affected by environment and genetics [11]. Many changes in mutants and transgenic organisms are cryptic, silent, or unpredictable [12, 13, 14, 15, 16]. Metabolite analysis, particularly metabolite fingerprinting and metabolomics, can detect cryptic changes and link unpredictable phenotypes to their biochemistry [4, 16]. Both metabolomics and metabolite profiling can provide information on how the central metabolites regulate cellular metabolism [11].
Glutamate dehydrogenases (GDH; EC 1.4.1.1 and EC 1.4.1.2) catalyze the reversible amination of alpha-ketoglutarate to form glutamate. In plants, they are not expected to assimilate ammonium because the enzyme is located in the mitochondria, is homo-octameric in structure, and has a high Km for substrates compared to glutamine synthetase (EC 6.4.2.1). The effects of genetic modification of nitrogen metabolism via the bacterial glutamate dehydrogenase (homo-hexameric GDH; EC 1.4.1.1) on plant growth and metabolism were not as expected [12, 13, 14, 15]. In the greenhouse and growth chamber herbicide tolerance is provided, biomass increase is increased, and water deficit resistance is increased [12, 13, 14, 15]. In the field, over three consecutive years, relative yield increase was caused by GDH [12]. An overall increase in the concentration of sugars, amino acids, and ammonium ions occurs within the cell [12, 13, 14]. A biochemical alteration may cause this effect, related to increased production of glutamate in one intracellular compartment, the cytoplasm. Increased total carbohydrate and amino acid compositions show that both carbon and nitrogen metabolism are altered in gdhA plants [13].
Reported here are the detected ion inferred metabolic fingerprint and changes in ion peak size inferred metabolite abundance among tobacco roots and leaves in plants transgenic for GDH compared to nontransgenic plants. The extent of glutamate synthesis was measured by 13N labeling. These data illustrate the use of FT-ICR-MS as a tool to analyze transgenic plants and to identify chemicals with biomedical significance.
RESULTS
Production of homozygous lines for biochemical evaluations
We had previously generated r2 seed from a series of independently regenerated plants that showed a range of GDH activity of 2–25 μmol min−1mg−1 protein [12]. Each line was an independent transformant, with genetic architecture consistent with one or two copies of the gdhA transgene [15]. The mRNA abundance and GDH activity were correlated via Northern hybridization. The mRNA was of high abundance for the GDH10 that produced between 20 and 23 μmol min−1mg−1 protein GDH activity. GDH10 line was selected for further analyses compared to vector and nontransgenic controls.
Analysis of glutamate fraction labeling
For comparison of glutamate fraction labeling we selected GDH10, GUS, and BAR transgenic tobacco lines because only GDH is expected to be resistant to methionine sulfoximine (MSX), an inhibitor of photorespiratory ammonium assimilation. Comparisons (Tables 1, 2, 3, and 4) did show organ specific differences.
Table 1.
Labeling of the glutamate pool via absorption of in intact roots of transgenic plants not treated with 1 mM MSX. Tracer exposure was for 15 minutes. Incorporation is expressed as a percentage of the label input plus or minus the range detected among 3 individual plant replicates and three measurement replicates.
| BAR | GDH10 | GUS | ||
| Glu | 9.5 ± 1.5 | 21.3 ± 4.9 | 9.4 ± 1.4 | |
| Gln | 32.9 ± 4.3 | 41.7 ± 3.8 | 32.7 ± 4.2 | |
| 57.5 ± 5.7 | 36.3 ± 3.5 | 57.3 ± 6.0 | ||
Table 2.
Labeling of the glutamate pool via absorption of in intact roots of transgenic plants treated with 1 mM MSX for 2 hours before feeding. Tracer exposure was for 15 minutes. Incorporation is expressed as a percentage of the label input plus or minus the range detected among 3 individual plant replicates and three measurement replicates.
| BAR | GDH10 | GUS | ||
| Glu | 1.3 ± 0.5 | 3.2 ± 0.6 | 1.3 + 0.5 | |
| Gln | 1.5 ± 0.6 | 1.9 ± 0.4 | 1.5 + 0.6 | |
| 97.2 ± 0.4 | 94.9 ± 1.0 | 97.1 + 0.3 | ||
Table 3.
Labeling of amino acid fractions in leaves fed through the petiole after 15 minutes and held in nutrient solution. Entire leaves were cut from 3 replicates of tobacco plants that were 6 weeks old grown in soil in a 16/8 walk in growth room at 26°C with light at about 500 microEinsteins. Incorporation is expressed as a percentage of the label input plus or minus the range detected among 3 individual plant replicates and three measurement replicates.
| GUS | GDH10 | BAR | ||
| Glu | 29.7 + 4.0 | 18.6 + 2.7 | 23.4 + 3.1 | |
| Gln | 18.8 + 6.0 | 12.0 + 1.1 | 16.3 + 3.3 | |
| 51.3 + 4.3 | 69.5 + 1.8 | 51.3 + 2.5 | ||
Table 4.
Labeling of the glutamate pool via absorption of in entire leaves of transgenic plants treated with 1 mM MSX for 1.5 hours before feeding. Leaf petioles were recut under water. Tracer exposure was for 15 minutes. Incorporation is expressed as a percentage of the label input plus or minus the range detected among 3 individual plant replicates and three measurement replicates.
| GUS | GDH10 | BAR | ||
| Glu | 2.9 ± 1.3 | 7.0 ± 0.6 | 2.8 ± 0.7 | |
| Gln | 1.7 ± 0.1 | 8.5 ± 1.9 | 1.5 ± 0.9 | |
| 95.3 ± 2.3 | 84.0 ± 2.4 | 95.0 ± 4.1 | ||
In the roots, labeling of the fraction containing 13N-glutamate (from administered during a 15-minute period) was increased 2.2 fold in GDH10 compared to BAR and GUS plants as a result of the introduced GDH activity, representing 21% (dpm/dpm) of the 13NH3 applied (Table 1). Treatment with MSX, an inhibitor of glutamine synthetase, reduced glutamate fraction labeling 7 fold among the GDH, GUS, and BAR transgenics suggesting the GS/GOGAT cycle accounts for 86% of the labeling in the absence of MSX. However, in MSX-inhibited GDH10 roots, glutamate labeling remained 2.2 fold higher than GUS and BAR roots (Table 2). Therefore, GDH was not inhibited by MSX. As expected BAR did not inactivate MSX.
In leaves, both glutamate fraction labeling and total labeling were decreased by 1.2 to 1.5 fold in GDH10 compared to GUS and BAR control plants (Table 3). The decrease was not significant in this experiment or experiments with leaf discs (data not shown). However, glutamate fraction labeling in presence of MSX was decreased 10 fold in GUS and BAR plants but only 2.6 fold in the GDH10 (Table 4). In addition, glutamine fraction labeling in presence of MSX was decreased 10 fold in GUS and BAR plants but only 0.6 fold in the GDH10. Therefore, in MSX-inhibited leaves; GDH10 assimilated 5 fold more 13N than GUS control and BAR plants. The GDH10 line, in the presence of MSX, also left less unincorporated (84% compared to 95%, Table 4) reflecting the contribution of the gdhA gene in assimilation in MSX-inhibited leaves. The glutamine labeling in MSX-treated GDH10 leaves was not related to incomplete inhibition of GS since the same degree of labeling was observed in 1 cm3 leaf discs floating in labeling solution (data not shown).
In GDH10 there was 2–3 fold more label in the glutamate fraction of both MSX-treated leaves (7.0% of the absorbed , Table 4) and roots (3.2%, Table 2) than BAR leaves (2.9%, Table 4) and roots (1.3%, Table 2). However, in non-MSX-treated GDH10 transgenic leaves, compared to the roots, the very high activity of GS, the larger pool sizes of glutamate and the greater flux through pathways involving glutamate may have resulted in less labeling by 13N (Tables 1 and 3). The amount of label in the glutamate and glutamine fractions that could be attributed to GDH activity was modest in roots, about 2.3%. ((3.2–1.3) + (1.9–1.5)). However, in leaves, labeling was significantly greater, about 11.9% ((7.0–2.9) + (8.5–1.7)).
Analysis of ion fingerprints and profiles
Experiments with tobacco [13, 14, 15] and corn [17, 18] had indicated that the total soluble amino acid, ammonium, and carbohydrate contents of GDH transgenic plants were each increased. The transgenic seedlings were shown to reproduce this phenotype (Table 5(a)). Ions were separated and characterized to infer the detectable metabolite complement using four ionization protocols for FT-ICR-MS. There were 2 012 ions detected within 2–4 ionization protocols (unique ions and isotope ions were removed). Regardless of genotype, ion fingerprints of leaves and roots differed significantly judged by FT-ICR-MS. Qualitative differences (compounds only detected in one organ) approached 23% (462/2012). Ion masses were validated by internal calibration with compounds of known mass and concentrations. Among the ions common in roots and leaves, apparent quantitative differences were in the majority 60% (929/1550). Quantitative differences were inferred from peak areas and validated by internal calibration. However, many factors can interfere with peak detection so that the estimates of differences in quantity are not unequivocal and some may be erroneous. Within that context some of the data observed were consistent with known organ-specific metabolisms in plants and some were not.
| (a) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percentage changeb | |
| (1) N-alpha-phenylacetyl-glutamine | C13H16N2O4 | 264.1110 | 227 |
| (2) 3-aryl-5-oxoproline ethyl ester | C13H15NO3 | 233.1052 | 303 |
| (3) 5-methyl-DL-tryptophan | C12H14N2O2 | 218.1055 | 40 |
| (4) N-alpha-BOC-L-tryptophan | C16H20N2O4 | 304.1423 | 333 |
The metabolites we putatively inferred from ion masses that were altered in abundance by GDH activity are depicted in Figures 1, 2, 3, 4, and listed in Table 5. The majority of the metabolites increased or decreased in leaves and/or roots by less than 10 fold. Between 5% and 14% of detectable metabolites were altered in abundance. This portion of the database can be examined at http://www.siu.edu/~pbgc/metabolite-profiles/GDH/.
Figure 1.
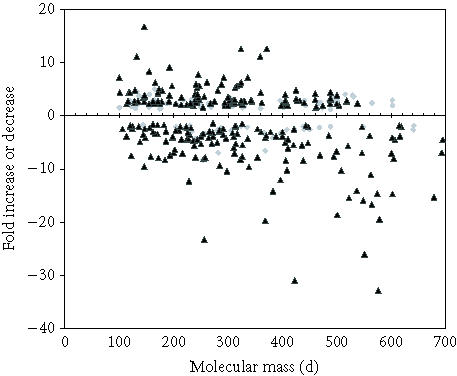
Distribution of metabolites (judged by mass) altered in relative abundance in leaves and roots. Grey diamonds are leaf metabolites and black triangles represent metabolites altered in roots.
Figure 2.
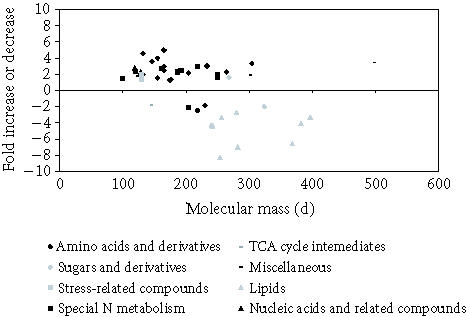
Scatter plot distribution of all classes of metabolites identified in leaf extracts.
Figure 3.
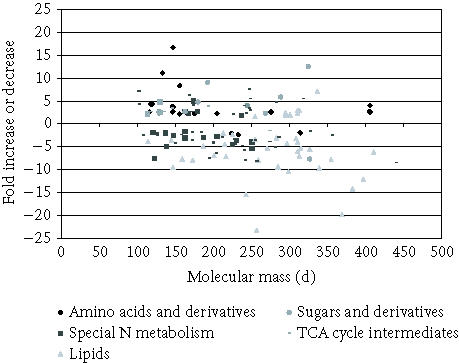
Scatter plot distribution of all classes of metabolites identified in root extracts.
Figure 4.
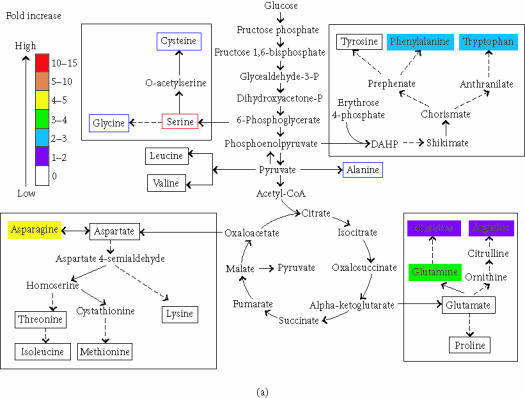
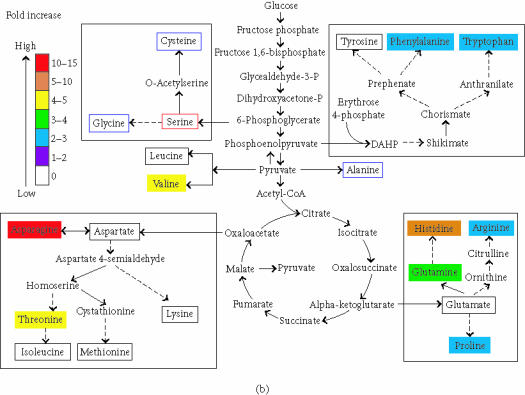
Metabolites in blue boxes were not detected. Metabolites in red boxes were used as internal standards and therefore detected. Metabolites in black boxes were detected and not changed. (a) Amino acids in leaves increased by gdhA. (b) Amino acids in roots increased by gdhA. Metabolites that are not protein amino acids are not annotated for changes here.
In leaves, 98 (5%) of the ions detected were changed in abundance between GDH and non-GDH plants. Only 91 empirical formulas could be inferred because seven were equivocal. Forty-one matched the formulas and predicted masses of the ions of compounds found in the databases we searched. The masses of the remaining fifty unidentified metabolite ions are available at http://www.siu.edu/~pbgc/metabolite-profiles/GDH but not discussed further here for brevity. The 41 putatively identified compounds were categorized as follows: 11 amino acids, 2 sugars, 8 fatty acids, 6 compounds of special nitrogen metabolism, 2 nucleic acid derivatives, 1 TCA cycle intermediate, 1 stress-related compound, and 10 miscellaneous metabolites not of those classes (Figure 4, Table 5). Not all of these compounds are common metabolites. Some are compounds not previously detected in plant cells, possibly reflecting the animal and microbial fauna present on tobacco samples. Some identified compounds were not previously detected in vivo possibly reflecting ionization artifacts. However, for brevity hereafter the metabolite putatively inferred from a detected ion will be referred to as just the metabolite.
In roots, there were 283 ions (14%) that changed in abundance among the 2 012 ion species repeatedly detected. Only 268 empirical formulas could be inferred. Database searches putatively identified only 117 of the 283 changed metabolites (Figure 3). Masses of the unidentified ions are available at http://www.siu.edu/∼pbgc/metabolite-profiles/GDH but not reported further here for brevity. Among the 117 altered metabolites, there were 14 amino acids, 6 sugars, 34 fatty acids, 15 compounds of special nitrogen metabolism, 2 nucleic acid derivatives, 4 TCA cycle intermediates, 2 stress-related compounds, and 40 metabolites not of those classes (Figure 4, Table 5). Judged by the correspondence of ion mass estimates, 90% of the compounds that changed in abundance in leaves also increased or decreased in the same way in roots. Only three metabolites were altered so that the increase in one organ was accompanied by decrease in the other organ (63 and 75, 86 and 87, and 60 and 67, Table 5).
Amino acids, precursors, and derivatives
In leaf extracts, consistent with previous reports of increased free amino acids [12], we found 6 amino acids that increased in abundance (1.3 to 4.5 fold, Figure 4a) in GDH plants. Arginine, phenylalanine, tryptophan, asparagine, glutamine, and histidine were inferred to be altered in abundance. Most of the known pathway intermediates involved in the biosynthesis of protein amino acids were detected, but were not altered in abundance. Four amino acid derivatives changed in abundance (Table 5(a)), one decreased, and three increased. The nonprotein amino acid ornithine increased 2.3 fold.
In roots, 9 amino acids appeared to be increased in abundance in GDH plants by 2 to 11 fold (Table 5(b)). Arginine, phenylalanine, tryptophan, asparagine, glutamine, histidine, proline, threonine, and valine were inferred to be altered in abundance. The root increases in proline, threonine, and valine were not detected in leaves. Many of the known pathway intermediates involved in the biosynthesis of protein amino acids were detected but not altered in abundance, except for the proline precursor delta-pyrroline-5-carboxylate (91, Table 5). No amino acid was decreased in abundance in GDH plants but three of the five amino acid derivatives that were changed decreased (Table 5(b)).
Sugars and derivatives
In leaves, consistent with a previous report of a modest increase in free carbohydrates [12], two sugar derivatives appeared to be increased 1.5–2 fold (10,11, Table 5) in GDH plants. In roots (Table 5(d)) six sugars appeared to be increased by 2.3–12.5 fold between GDH and non-GDH plants and included key intermediates involved in the regeneration of ribulose-5-phosphate in the Benson-Calvin cycle. One sugar derivative increased in both leaves and roots (10,15, Table 5).
Fatty acids
In leaves the six fatty acids and two derivatives that appeared to be changed in abundance all decreased in the GDH plants (18–25, Table 5). Two 16-carbon fatty acids (19,20, 16:0 and 16:1, Table 5) and two 18-carbon plant membrane fatty acids were reduced (21,22, 18:1, 18:2, Table 5). These changed fatty acids are minor components of both plant cell membranes and chloroplast membranes. However, α-linolenic acid (18:3), the main constituent of both membranes was not altered in abundance. Two rare unsaturated fatty acids (18 and 23; 15:0 and 24:0) were significantly reduced (Table 5(e)). Two fatty acid derivatives also decreased (Table 5(f)).
In roots, seventeen of the eighteen fatty acids and twelve of the sixteen fatty acid derivatives that appeared to be changed abundance decreased in GDH plants (26–40, 42,43, Table 5). The decreased fatty acids included 5 of those that decreased in leaves (33--35,38,43, Table 5) but the 18:2 was not decreased. The decreased fatty acid derivatives included both those that decreased in leaves. The four fatty acid derivatives that increased included 3 di-enoates or tri-enoates of 18-C fatty acids that may be biosynthetically related (45,48,51,55, Table 5). None of the common diacylglycerol lipids were detected by FT-ICR-MS so whether decreases in fatty acids were reflected by decreases in lipids is not known.
Special nitrogen metabolism
Six metabolites that appeared to be increased in abundance between GDH and non-GDH plants in leaf extracts were amines (1), alkaloids (2), and phenolics (3), three classes of products derived from special nitrogen metabolism (Table 5(i)).
From four classes of special nitrogen metabolites sixteen appeared to be altered in abundance between GDH and non-GDH plants in roots. There were amines (2), alkaloids (7), phenolics (5), and isoprenoids (1) (Table 5(j)). Five increased and nine decreased. Only N-caffeoylputrescine was altered in both organs. However, it increased in leaves but decreased in roots.
Nucleic acids and derivatives
Only two derivatives of nucleic acids appeared to be increased 2–3 fold in leaf extracts between GDH and non-GDH plants (Table 5(k)). Two compounds were increased in roots more than 2 fold, including the common ribonucleotide uridine (Table 5(l)).
TCA cycle intermediates and derivatives
The monoethyl ester of fumaric acid was the sole metabolite identified that appeared to be changed by GDH in leaves (Table 5(m)). In roots, all four metabolites that changed increased in abundance (2.7–4 fold) including three TCA cycle intermediates, fumaric, malic, and citric acids (Table 5(n)).
Stress-related compounds
Only one member of this group appeared to be altered in leaf extracts (Table 5(o)); two increased more than 2 fold in roots (Table 5(p)).
Miscellaneous
Ten metabolites appeared to be altered in leaves and eight contained nitrogen. Five of the compounds identified in leaf extracts represent known drugs and cigarette toxins (Table 5(q)).
Forty metabolites appeared to be altered in roots and twenty-two contained nitrogen. Among these are five drugs, five flavoring agents, four pesticides, three carcinogens, and five toxins. There were two compounds that were also coordinately altered in leaves: N-nitrosopyrollidone and menthyl acetoacetate.
DISCUSSION
Metabolite analysis
This study used metabolite analysis with FT-ICR-MS [5, 8] to associate phenotype with biochemical changes resulting from endogenous effects of ectopic glutamate synthesis in transgenic plants. The GDH plants were a suitable test for FT-ICR-MS because they have cell composition alterations that result from a specific biochemical alteration in a well-characterized pathway targeting the cellular glutamate pools [12].
The identification of ions and the inference of a metabolite were relatively inefficient, with less than half the ions detected having known metabolites of corresponding masses. The rest of the ions may represent reactions occurring before sample quenching, multiple ionization effects, ion suppression effects, or ion fragmentation [9]. Some of these ions may represent new metabolites not previously reported in plants. An estimate of the extent of artifact ions compared to new products will be a future goal.
Although not reliable or fully quantitative, the changed abundances inferred from ions detected by FT-ICR-MS that appear to correspond to metabolites such as amino acids, sugars, and fatty acids largely agreed with quantitative spectrophotometer assays [12, 17, 18, 19] HPLC separation of sixteen individual amino acids from the methanol soluble, low molecular weight fraction of cell extracts showed eight were significantly changed. Four of the eight amino acids had been inferred to increase in abundance by FT-ICR-MS; the remainder had not been detected as quality peaks. Three amino acids, histidine, valine, and threonine, were not increased as expected from FT-ICR-MS. The difference appears to be due to interference by other ions [9].
Given the only partial agreement among three different measurements of the amino acids, we conclude that the abundance of specific ions in total infusion mass spectra were significantly affected by combined ion suppression effects of all other components, pH and salinity of the solution, flow rate, tip opening, and electrospray current [9]. Further the samples may have differed in the rapidity of turnover of intracellular metabolites, the rate at which metabolism was quenched and the time for which metabolites were separated from the cell debris [10]. The evidence of reproducibility for some amino acid measurements may be related to handling samples simultaneously [9, 10]. Samples analyzed separately either temporally or spatially will be more difficult to compare.
However, the exact masses alone are not sufficient to identify specific compounds unequivocally. Several compounds were identified that are not metabolites in plants (eg, alpha-tert.butoxycarbonyl-L-tryptophan compound no 4, Table 5(a), a synthetic intermediate in peptide synthesis); some artificial pesticide-like metabolites (119,126,127,133,138, Table 5); and metabolites found in insects not plants (eg, no 114, a component of bee royal jelly). Therefore, data from FT-ICR-MS analysis should be used as preliminary evidence to suggest further experiments [8]. In this publication we focused on amino acid metabolism and the effect on central metabolism.
Among the effects detected, those altering amino acid metabolism and fatty acid metabolism were most profound and appear to underlie a doubling of free amino acids and halving of free fatty acid content [12, 13, 18]. In comparison the effects of GDH on carbohydrate metabolism were comparatively trivial and may not solely underlie the increased content reported [12]. The increases in three abundant organic acids may have contributed to the carbohydrate content reported by spectrophotometer assays of reducing sugar content. In addition some of the unidentified ions may have been sugars or carbohydrates.
The majority of ions detected by FT-ICR-MS could not be identified from their predicted formulas or mass. In comparison about 30% of ions identified in plants by GC-MS could not be identified [4, 7, 11]. The unidentified ions detected may represent novel constituents of tobacco leaves or roots [38] or ionization artifacts of MS [5, 6]. Different abundances could also be experimental artifacts. To reduce artifacts we used pooled samples for each genotype from plants grown in an RCB in a growth chamber; accepted only those ions derived from two ionization methods of the four applied; and by repeating the entire experiment.
Masses of the unidentified metabolite ions are available online at http://www.siu.edu/∼pbgc/metabolite-profiles/GDH/Ntabacum/IONS1-4. html but not discussed further here for brevity.
The concurrence between FT-ICR-MS and spectrophotometer data [12] appears to validate the use of the method for metabolite analyses. However, the informational content of FT-ICR-MS is orders of magnitude greater than other high-throughput methods [1]. FT-ICR-MS detected 2 012 ions that were consistent across ionization methods from each replicated extract. In comparison, tandem MS required several independent extractions to identify 326 metabolites [4] or 88 metabolites [11]. However, there is no doubt that GC-MS in a tandem format is a superior technique for unequivocal identification of ions and therefore metabolites [1]. In addition GC-MS is superior in that ion concentrations can be derived. Both methods suffer from tuning artifacts among spatially separated runs that can only be partly compensated for by internal standards. We conclude that FT-ICR-MS will have a role in functional genomics where sample throughput is more important than chemical identification and relative quantifications, a situation analogous to the decision to employ microarray or macroarray for analysis of the transcriptome.
Amino acid metabolism
Ammonium assimilation fluxes in roots showed that the introduced GDH contributed to total labeling of glutamate regardless of GS inhibition, suggesting that the enzymes compete for . In leaves, the GDH reduced net 13NH3 assimilation, possibly by suppression of GS activity [20, 21]. This is consistent with the increase in leaf reported [12]. However, in the presence of MSX, GDH partially substituted GS by increasing glutamate labeling (7% label incorporated compared to 2.9% for controls). It is possible that the higher Km of GDH for [22] and the 7–10 fold greater fluxes in nitrogen (resulting from photorespiration) [23] drive the NAD(P)H-dependent GDH reaction forward to produce glutamate in large quantities during GS inhibition [20]. From labeling we conclude that the modifications in transgenic plants are not the product of greatly increased efficiency of nitrogen assimilation by GDH plants. Instead, the glutamate generated in the cytoplasm may result in altered metabolic fluxes and profiles.
Metabolite analysis apparently contradicts 13N flux labeling because the steady-state of extractable glutamate is not altered between GDH and control transgenic roots or leaves. Short-term flux does not always predict steady-state concentration because plants have mechanisms for sensing nitrogen fluxes and maintaining homeostasis [24, 25]. Flux away from glutamate appears to equal the extra flux into glutamate as many major nitrogen sinks were increased and few decreased (in leaves 19 increased and 4 decreased, in roots 29 increased and 17 decreased). Since plant mRNA abundances did not change (data not shown), allosteric effectors of many enzymes may be involved [26]. The effects of GDH expression on phenotype may result from the signaling effects of increased cytosolic glutamate seen in plants grown at low light intensities, nia and rbc mutants [27] and the status of certain inorganic N compounds [28, 29, 30]. Metabolites derived from nitrate, such as ammonium, glutamine, and glutamate all may act as signals to report on organic N status [25]. Cytosolic glutamate may act directly as a ligand to activate ion channels.
Metabolites that shared C skeletons were coordinately altered in abundance in both roots and leaves in response to GDH activity. Among the amino acids, 4 that derived from alpha-ketoglutarate were coordinately changed in roots and 3 in leaves. Ornithine, the nonprotein amino acid derived from alpha-ketoglutarate, was also increased in roots. Also changed to the same extent in both organs were phosphoenol pyruvate derivatives, phenylalanine, and tryptophan. However, tyrosine, the only other amino acid originating from phosphoenolpyruvate C skeletons, was not altered in abundance. Asparagine was the only amino acid, derived from oxaloacetate, changed in both leaves and roots but threonine increased 4–5 fold in roots. Among amino acids derived from pyruvate C skeletons, valine was increased 4–5 fold in roots but no changes in leucine were seen and alanine was used as an internal standard and therefore changes could not be detected. Similarly, the amino acids derived from 6-phosphoglycerate could not be detected. The pattern of amino acid changes is similar to that in maize endosperm with the opaque mutation [29] where endogenous GDH activity is increased, but different from that caused by photosynthesis [31, 32]. Therefore, the metabolic alterations are GDH specific, not systemic, implying that the effect of GDH on metabolism depends on the metabolism occurring in the cell.
Changes related to water deficit tolerance
Increases in sugar concentrations could also significantly increase the water deficit tolerance [33]. However, the sugars increased by GDH were complex sugars, not the monosaccharides or disaccharides normally associated with tolerance. The notion of sugar sensing is also gaining momentum [25]. The FT-ICR-MS assays would not detect polysaccharides over 700 d, so again flux and steady state may differ.
None of the following compatible solutes were changed in abundance in either leaves or roots [33]: trigonelline, trehalose, dimethylsulfoniopropionate, glycerol, sorbitol, mannitol, choline-O-sulphate, beta alanine betaine, glycinebetaine, prolinebetaine, N-methyl-proline, hydroxyproline, hydroxyprolinebetaine, and pipecolic acid. However, since the association of water deficit tolerance with any single solute is imperfect, we expect the phenotype was derived from a combination of increased compatible solutes. One or a few of the unidentified metabolites may also participate. Stomatal behavior, GS activity and resistance to photooxidation may contribute to the tolerant phenotype [34]. Plant morphology does not appear to contribute as the root to shoot ratios were not changed [13, 14]. The mechanism by which water deficit tolerance is afforded remains to be unraveled.
Fatty acids
Oil and protein contents are inversely related to each another, to carbohydrate content and to yield in many crop plants. The synthetic pathways for fatty acid, protein, and carbohydrate compete for carbon skeletons [35]. Therefore, the increase in protein and sugar caused by GDH was expected to cause the reduction in fatty acid content observed in leaves and more pronouncedly in roots. The 16-carbon and 18-carbon fatty acids changed were common constituents of the diacylglycerols in plant cell membranes and chloroplast membranes. However, the most abundant fatty acid in cell membranes, α-linolenic acid (18:3), was not altered in abundance in either leaves or roots. The other fatty acids unaltered in abundance are of the 16:3 class. These are mainly found in chloroplast membranes, albeit in quantities far lower than α-linolenic acid (18:3). Interestingly, only the fatty acids whose contribution to plant membrane composition is minor were reduced. The cells of GDH transgenics appear to be regulating closely the abundance of the most common fatty acids necessary for normal cellular function.
The TCA cycle intermediates that are increased by GDH, fumarate, malate, and citrate (Figure 1) may be associated with the redirection of C away from fatty acid synthesis and toward amino acid synthesis. Fumarate and malate are immediate precursors to pyruvate. Citrate is produced from the catabolism of acetyl-CoA.
Special nitrogen metabolism
Special nitrogen metabolites (amines, alkaloids, phenolics, and isoprenoids) may represent more than 50% of the compounds in plants in the 100–700 d range [36]. They provide defense against herbivores, microorganisms, or competing plants and color or scent to attract pollinating insects and seed-dispersing or fruit-dispersing animals. Their nitrogen is derived from ammonium assimilation via the amino acids (the carbon skeletons may derive from many diverse pathways) so it was surprising that none were decreased in leaves and only nine were decreased in roots in GDH plants.
The abundance of just 2 amines (of the 48 detected) altered in response to GDH (Figure 4, Table 5, 60 and 67, 66). Amines are products of arginine or ornithine metabolism (that were affected by GDH) so the unchanged amine contents were surprising. The amine N-caffeoylputrescine that was increased in shoots and decreased in roots by GDH (by transport) accumulates during abiotic or biotic stress [37, 38] and will stabilize histones, stabilize biomembranes, inhibit viral replication, and regulate cellular growth [36]. Such changes directed by GDH may be useful for the economic production of plant secondary metabolites.
The alkaloids that were altered by GDH (9 of 34 detected) were mainly coumarin (68, 72, 73, Table 5) and quinone (61, 69, 71, 74, Table 5) derivatives. Alkaloids occur in about 15% of plant taxa, including N tabacum [36]. Most derive from amines that are synthesized from amino acids. They accumulate in tissues that are important for survival and reproduction providing chemical defense. Targets include heart, liver, lung, kidney, CNS, and reproductive organs. Toxic alkaloids may have pharmacological uses at nontoxic doses (eg, 62, 68, Table 5) [36]. Scopoletin (62, Table 5) inhibits Escherichia coli O157, is antiviral, is anti-inflammatory (5 fold more than aspirin), and is an asthma treatment [36, 39]. Increasing leaf concentrations 2–3 fold with GDH may be a useful approach to finding new uses for the tobacco crop. Coumarin (68, Table 5), a perfumed liver and lung toxicant [40] was decreased 10 fold by GDH in roots, potentially useful for the manipulation of diets based on root crops.
Some (8 of the 186 detected) phenolic compounds were altered by GDH. The production of phenylpropanoids occurs predominantly from the amino acid phenylalanine [41]. Quinones, monoterpenes, and modified side chains derive from other pathways. The 2-fold increase in phenylalanine caused by GDH (Figure 3) may explain why phenolics are the predominant class of special nitrogen metabolites increased in leaves (3) and roots (3) of GDH transgenics. Phenolics provide mechanical support and barriers; insect attractant or repellents; antioxidants used in leather making; and flavor components in wines and herbal teas. Swainsonine (78, Table 5) is an inhibitor of mannosidase II, used as a cancer therapy [42]. The 5-fold increase in abundance could be useful. Nicotine (synthesized from ornithine), an animal stimulant and insect repellent, was increased in roots (77, Table 5) but not in leaves [41]. Nicotine is synthesized in the roots and transported to the leaves so increased synthesis may not produce a desirable outcome. Nopinone (80, Table 5) was the only isoprenoid affected by GDH but it has no important pharmacological properties [41].
Nucleic acids
The synthesis of nucleic acids from glutamine is a major nitrogen sink in plant cells [36]. GDH did not alter the abundance of the common phosphonucleotides. Uridine was increased 4 fold. Uridine is a precursor of important biosynthetic compounds UMP, UDP, UTP and their glycosyl derivatives. However, these compounds were not altered in abundance. Clearly the altered amino acid fluxes caused by GDH are being directed toward specific pathways and intermediates, leaving others unaffected.
Miscellaneous compounds
The 46 compounds we termed miscellaneous that were altered by GDH in tobacco included 29 that contain nitrogen but structurally cannot be classified with the special nitrogen metabolites [36]. The predominance of N-containing compounds suggests these alterations are directed by GDH-induced metabolism (Table 5(q), Table 5(r), and Table 5(s)). This group of compounds includes some of medicinal relevance (http://www.cieer.org/geirs/); an antihelmitic (98, Table 5); a tumorstatic that binds to nucleic acids (104, Table 5); a vitamin C metabolite that causes increased absorption, cellular uptake, accumulation and reduced excretion (96, Table 5); a nootropic (a drug that enhances mental function; 99, Table 5); treatments for diabetes, high blood pressure and arteriosclerosis (also found in bee royal jelly; 114, Table 5). An inhibitor of neutral sphingomyelinase (117, Table 5); a mycotoxin and an antitumor agent (134, Table 5); and a GABA uptake inhibitor (116, Table 5). Some constituents of cosmetics (135, 113, Table 5), flavoring agents (131, 124, Table 5), and a solvent (110, Table 5) were altered in GDH plants. Altering abundance of these compounds may provide alternate uses for the tobacco crop.
Some pesticide-like metabolites were altered by GDH although the plants were not exposed to pesticides (119,126,127,133,138, Table 5). These compounds may be enzyme substrates occurring naturally in plants that are structurally similar to pesticides. Cataloging metabolites may lead to new leads for pesticidal chemical discovery [1].
Carcinogens and poisons were primarily altered in abundance in the roots (100 and 131, 105, 107, 108, 111, 115, 128, 132, Table 5) consistent with the root synthesis of these compounds early in development and later translocation to the leaf [41]. The carcinogens and cigarette components detected are specific to tobacco and most probably part of its inherent secondary metabolism. Detection of carcinogens and poisons may serve to validate the use of FT-ICR-MS and suggests applications in the association of smoking with cancer incidence.
Plant pigments, haem, and other porphyrins are major sinks for glutamate source pools in plants [27]. However, the glutamate flux perturbation caused by GDH does not alter the regulation or intermediates of pigment biosynthesis.
We conclude that GDH can be useful for plant metabolic engineering to increase or decrease the yield of a large number of chemical compounds. GDH may be a useful tool as the pharmaceutical industry discovers new plant-derived compounds of therapeutic value.
The work presented here demonstrated that metabolite analyses by FT-ICR-MS provide a useful tool for the analysis of cryptic phenotypes in transgenic plants. The analysis of data from extracts without derivatization allows analysis of the relationships between various metabolites and the equivalence of samples. If there are 40 000 different molecules among all extant plant species in the range of 100–700 d [36], cataloging them by means other than FT-ICR-MS would be a mammoth task [3, 4]. Assuming there are 3–4 thousand different molecules in individual plant species in the range of 100–700 d [4, 11] we will have sampled about 50%–60% (2 012) in two analyses (replicated). However, it is clear from our data that about 50% of the molecules detected are not in the databases we interrogated. Therefore, estimates of the chemical diversity of plant may be grossly underestimated. The development of a cell map and exploration of metabolic instantiations with that map will be impossible without cataloging the consequences of metabolism accurately.
The sensitivity and resolution of FT-ICR-MS provides a useful method for cataloging chemical diversity. Within the existing limits, differences may be measured between samples comprising more than ten thousand cells. Therefore, the occurrence of novel compounds of biomedical significance in individuals, populations, species, and genera may be cataloged.
MATERIALS AND METHODS
Gene manipulations and construction of plasmids
To examine the effects of NADPH-GDH in plants we used three lines, GDH10, GUS, and BAR, described previously [12, 13, 14, 15, 17]. GDH10 is a well-characterized independent line of Nicotiana tabacum var “Petite Havana SR1” that expresses the E coli gdhA gene. The line represents an early regenerant and lacks noticeable variation from the wild type under normal growth conditions. The gdhA gene inserted in GDH10 plants has an architecture and segregation pattern consistent with a single site of insertion. The gene is under the control of the CaMV 35S promoter. Transcript abundances are equal when comparing roots and leaves. Enzyme activity is found in the cytoplasm but not plastids and is equal in roots and leaves. GUS is a well-characterized independent line of N tabacum var Petite Havana SR1 that expresses the modified gusA gene. The line represents an early regenerant and lacks noticeable variation from the wild type under normal growth conditions. The gusA gene inserted in GUS plants has an architecture and segregation pattern consistent with a single site of insertion. The gene is under the control of the CaMV 35S promoter. Enzyme activity is found in the cytoplasm but not plastids and is equal in roots and leaves. BAR is a well-characterized independent line of N tabacum var Petite Havana SR1 that expresses the S hygroscopicus bar gene. The line represents an early regenerant and lacks noticeable variation from the wild type under normal growth conditions. The bar gene inserted in BAR plants has an architecture and segregation pattern consistent with a single site of insertion. The gene is under the control of the CaMV 35S promoter. Enzyme activity provides tolerance to phosphinothricin herbicide to roots, leaves, and cell culture derived from them. BAR and GUS were chosen as adequate controls because they were not significantly different from wild-type SR1 across a wide range of growth conditions, locations, and years [12, 13, 14, 15, 17].
Seeds of the lines described and clones used for transformation are freely available on request and are being widely used for transformation of other plant species.
Plant material and growth conditions
Tobacco seeds were obtained from the seed stocks at the Agriculture Research Center, Southern Illinois University at Carbondale (Carbondale, Ill). Seeds were sown in 4-inch pots [14] containing a mixture of sand and soil (1:1). Seedlings were thinned to one plant per pot, watered daily, and grown on unshaded benches and in the Horticulture Research Center, Southern Illinois University, from 9/99 to 9/03. The conditions for the growth of plants for 13N labeling, and metabolomics are described in the coming sections. Seeds of each line used are available on request.
Preparation of cell free extracts and GDH assays
GDH assays were performed exactly as described [12]. All preparative steps were carried out at 4°C. The specific activity of aminating NADPH-GDH was quantified by measuring the rate of oxidation of NADPH dependent on reductive amination of alpha-ketoglutarate. Assays were performed at 25°C. The amount of protein in the extracts was determined by Bradford assay.
Labeling of the glutamate pool by 13N
Three individual plants were fed for 15 minutes via hydroponic solutions for root feeding and via excised stems for leaf feeding, then treated with liquid N, ground up, extracted with distiled water, filtered through glass wool, and separated on an anion-exchange column (Dowex 2X8-100) which retained glutamate. The eluate was washed through the column with another 10 mL of distiled water and passed through a cation-exchange resin (Dowex 50WX8-100) which bound . Glutamine came through in the eluate. The columns were washed with 10 mL of 2M KCl to elute glutamate (anion-exchange column) and (cation-exchange column). Eluates were collected in 20 mL scintillation vials and counted in a Canberra Packard gamma counter that was automatically corrected for decay time (13N has a half-life of 10 minutes). The percent label incorporated was calculated using the following formula: [{(percent 13N as glutamate in the presence of MSX by GDH10 line)-(percent 13N as glutamate in the presence of MSX by non-GDH line)}/(percent 13N as glutamate in the absence of MSX by the GDH10 line)].
Preparation of metabolite extracts for FT-ICR-MS assays
Three pooled leaf and root samples from each control and transgenic genotype were used to remove spatial and genetic variation not associated with GDH activity. About 100 mg of tissue was ground to which 1.0 mL of 50/50 (v/v) methanol/0.1% (w/v) formic acid was added [8]. The samples were homogenized and centrifuged. The supernatant was used for the analyses. Each sample was mixed with a known and equal amount of a standard mix of serine, tetra-alanine, reserpine, Hewlett-Packard tuning mix, and the adrenocorticotrophic hormone fragment 4–10. These internal calibration compounds produced 4–5 ions of mass encompassing the range reported allowing for control of spectra used for mass reports. The internal calibration compound peak area was used to detect nonbiological variations in abundance reported allowing for control of spectra used for the quantities reported. All analytes we purchased from Sigma-Aldrich (St Louis, Mo) and used without further purification.
FT-ICR-MS assays
Briefly, we used the Bruker Daltonics APEX III FT-ICR-MS equipped with a 7.0 Tesla magnet, electrospray, and APCI ionization sources [8]. Both positive and negative ionizations were carried out. Tips were prepared as previously described [8]. For negative ionization, samples were introduced by capillary, diluted 1:19 in 50% (v/v) methanol, 0.2% (v/v) formic acid, 49.8% (v/v) water. For positive ionization, samples were introduced by capillary, diluted 1:19 in 50% (v/v) methanol, 0.2% (v/v) ammonium hydroxide, 49.8% (v/v) water. Flow rates were 5 μL/min for electrospray and 100 μL/min for APCI ionization sources. ESI, APCI, and ion transfer conditions were optimized using a standard mix of serine, tetra-alanine, reserpine, Hewlett-Packard tuning mix, and the adrenocorticotrophic hormone fragment 4–10. Instrument conditions were optimized for ion intensity and broadband accumulation over the mass range of 100–1000 d. One-megaword data files were acquired and a sinm data transformation was performed prior to Fourier transform and magnitude calculations.
(a) Calibration
All samples were internally calibrated for mass accuracy over the approximate mass range of 100–1000 d using a mixture of the above-mentioned standards. The results for each ionization method can be viewed at http://www.siu.edu/∼pbgc/metabolite-profiles/GDH/Ntabacum/ions1-4.html.
All mass deviances from the standard curves were less than 1.0 ppm over the mass range studied, although most of them were typically in the 0.1 to 0.2 ppm range. The value for each peak reported by each ionization method can be viewed at http://www.siu.edu/∼pbgc/metabolite-profiles/GDH/Ntabacum/ions1-4.html.
(b) Matrix effects and reproducibility
Mass spectra were recorded by averaging 10 single spectra of 3-seconds acquisition time each. Some suppression was observed but based on several random samples, spectrum to spectrum fluctuations of signal intensity ratios varied from one another by less than 30%. This value was used to indicate the confidence of each data point. Absolute interference is reported for each ion. It ranged from 1.01E+60 to 5.82E+80. The average noise peak was 1.01E+60. Data is an open source, each value and each spectra can be downloaded from http://www.siu.edu/∼pbgc/metabolite-profiles/GDH/.
Preparation of database searching for FT-MS assays
(a) Empirical formula inference
Empirical formulas were inferred for those ions for which the area under the peak changed between treatments. Excluded were peaks with inaccurate mass estimates and peaks with multiple likely empirical formulas. Final empirical formulas and masses for the metabolite inferred from each ion followed the addition or subtraction of a single hydrogen ion or electron depending on the mode of ionization used. An assumption made was that all ions represented single ionization events. When there were specific metabolites that we were interested in evaluating, we used the spreadsheet calculator to identify corresponding ion mass to charge ratios. We then manually examined the raw peak list for the corresponding ion mass to charge ratio.
(b) Database searching
We identified compounds by manually interrogating two publicly available databases, one at Chemfinder (http://chemfinder.cambridgesoft.com) and the second at NIST (http://webbook.nist.gov/chemistry/mwser.html). As the mass of the metabolites increases, the number of possible isomer combinations increases. Determination of the isotope expected in tobacco extracts was made manually with reference to plant biochemistry texts and databases. We did not use Phenomenome PLC. proprietary software (Saskatchewan, Canada), only publicly available databases were used so that data and databases of ions would remain open source.
Table 5.
a: mass is ± 1 ppm, or 0.0002–0.00001 d. b: % changes are ±2%. N/A denotes not applicable.
| (b) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (5) N-acetyl-L-tyrosine | C11H13NO4 | 223.0845 | 49 |
| (6) PTH-proline | C12H12N2O3 | 232.0670 | 43 |
| (7) (gamma-L-glutamyl)-L-glutamine | C10H17N3O6 | 275.1117 | 263 |
| (8) N-Benzoyl-L-tyrosine ethylester | C18H19NO4 | 314.1201 | 50 |
| (9) 1-[N-(1-carboxy-3-phenylpropyl)-L-lysyl]-L-proline | C21H31N3O5 | 405.2264 | 278 |
| (c) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (10) 3-deoxy-D-glycero-D-galacto-2-nonulosonic acid | C9H16O9 | 268.0794 | 159 |
| (11) Bis-D-fructose 2′,1:2,1′-dianhydride | C12H20O10 | 324.1056 | 208 |
| (d) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (12) 1,6-anhydro-beta-D-glucopyranose | C6H10O5 | 162.0528 | 263 |
| (13) 2-amino-2-deoxy-D-glucose | C6H13NO5 | 179.0794 | 276 |
| (14) Sedoheptulose anhydride | C7H12O6 | 192.0634 | 909 |
| (15) 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid | C9H16O9 | 268.0794 | 233 |
| (16) 1,6-anhydro-beta-D-glucopyranose 2,3,4-triacetate | C12H16O8 | 288.0845 | 588 |
| (17) Bis-D-fructose 2′,1:2,1′-dianhydride | C12H20O10 | 324.1056 | 1250 |
| (e) | |||||
|---|---|---|---|---|---|
| Common name | Systematic name | Empirical | Molecular | Degree of | Percent |
| formula | massa | saturation | changeb | ||
| (18) Pentadecanoic acid | n-pentadecanoic acid | C15H30O2 | 242.2246 | 15:0 | 23 |
| (19) Palmitoleic acid | Hexadecenoic acid | C16H30O2 | 254.2246 | 16:1 | 12 |
| (20) Palmitic acid | Hexadecanoic acid | C16H32O2 | 256.2402 | 16:0 | 30 |
| (21) Linoleic acid | 9,12-octadecanedioic acid | C18H32O2 | 280.2402 | 18:2 | 36 |
| (22) Oleic acid | 9-octadecenoic acid | C18H34O2 | 282.2559 | 18:1 | 14 |
| (23) Lignoceric acid | Tetracosanoic acid | C24H48O2 | 368.3654 | 24:0 | 15 |
| (f) | |||
|---|---|---|---|
| Systematic name | Empirical formula | Molecular massa | changeb |
| (24) Ethyl tricosanoate | C25H50O2 | 382.3811 | 24 |
| (25) Ethyl tetracosanoate | C26H52O2 | 396.3967 | 30 |
| (g) | |||||
|---|---|---|---|---|---|
| Common name | Systematic name | Empirical | Degree of | Molecular | Percent |
| formula | saturation | massa | changeb | ||
| (26) Pelargonic acid | n-nonanoic acid | C9H18O2 | 9:0 | 158.1380 | 13 |
| (27) Capric acid | n-decanoic acid | C10H20O2 | 10:0 | 172.1463 | 13 |
| (28) Undecanoic acid | n-hendecanoic acid | C11H22O2 | 11:0 | 186.1620 | 21 |
| (29) Lauric acid | Dodecanoic acid | C12H24O2 | 12:0 | 200.1776 | 14 |
| (30) N/A | Trans-2-tridecenoic acid | C13H24O2 | 13:1 | 212.1776 | 50 |
| (31) N/A | Tridecanoic acid | C13H26O2 | 13:0 | 214.1933 | 22 |
| (32) Undecanedioic acid | N/A | C11H20O4 | 11:2 | 216.1362 | 14 |
| (33) Pentadecanoic acid | n-pentadecanoic acid | C15H30O2 | 15:0 | 242.2246 | 6 |
| (34) Palmitoleic acid | Hexadecenoic acid | C16H30O2 | 16:1 | 254.2246 | 29 |
| (35) Palmitic acid | Hexadecanoic acid | C16H32O2 | 16:0 | 256.2402 | 4 |
| (36) Myristic acid | Tetradecanoic acid | C14H26O4 | 14:2 | 258.1831 | 13 |
| (37) Margaric acid | n-heptanoic acid | C17H34O2 | 17:0 | 270.2559 | 19 |
| (38) Oleic acid | 9,12-octadecanedioic acid | C18H32O2 | 18:1 | 282.2559 | 32 |
| (39) Stearic acid | Octadecenoic acid | C18H34O2 | 18:0 | 284.2715 | 11 |
| (40) N/A | n-nonanoic acid | C19H38O2 | 19:0 | 298.2872 | 10 |
| (41) DL-12-hydroxystearic acid | N/A | C18H36O3 | 18:0 | 300.2664 | 196 |
| (42) Tricosanois acid | n-tricosanoic acid | C23H46O2 | 23:0 | 354.3498 | 13 |
| (43) Lignoceric acid | Tetracosanoic acid | C24H48O2 | 24:0 | 368.3654 | 5 |
| (h) | |||
|---|---|---|---|
| Systematic name | Empirical | Molecular | Percent |
| formula | massa | changeb | |
| (44) Tetradecanoic acid, 7-oxo-, methyl ester | C15H28O | 224.2140 | 43 |
| (45) (9Z)-(13S)-12,13-epoxyoctadeca-9,11-dienoate | C18H30O3 | 294.2195 | 192 |
| (46) 9-Octadecenoic acid, methyl ester | C19H36O2 | 296.2715 | 23 |
| (47) Ethyl linoleate | C20H36O2 | 308.2715 | 31 |
| (48) (9Z,11E,14Z)-(13S)-hydroperoxyoctadeca-(9,11,14)-trienoate | C18H30O4 | 310.2144 | 238 |
| (49) Methyl 12-oxo-trans-10-octadecenoate | C19H34O3 | 310.2508 | 25 |
| (50) Octadecanoic acid, ethenyl ester | C20H38O2 | 310.2872 | 17 |
| (51) (9Z,11E)-(13S)-13-hydroperoxyoctadeca-9,11-dienoate | C18H32O4 | 312.2301 | 194 |
| (52) Octadecanoic acid, 12-oxo-, methyl ester | C19H36O3 | 312.2664 | 14 |
| (53) Diethyl tetradecanedioate | C18H34O4 | 314.2457 | 19 |
| (54) propyl stearate | C21H32O2 | 326.3185 | 18 |
| (55) 5(S)-hydroperoxy-arachidonate | C20H32O4 | 336.2301 | 714 |
| (56) Octadecanoic acid, 9,10-epoxy-, allyl ester | C21H38O3 | 338.2821 | 10 |
| (57) Ethyl tricosanoate | C25H50O2 | 382.3811 | 7 |
| (58) Ethyl tetracosanoate | C26H52O2 | 396.3967 | 8 |
| (59) 4,4′-Dimethylcholestatrienol | C29H46O | 410.3549 | 16 |
| (i) | |||
|---|---|---|---|
| Class amines | Empirical formula | Molecular massa | Percent changeb |
| (60) N-caffeoylputrescine | C13H18N2O3 | 250.1317 | 196 |
| Alkaloids | |||
| (61) 8-acetyl quinoline | C11H0NO2 | 187.0633 | 227 |
| (62) Scopoletin | C10H8O4 | 192.0423 | 244 |
| Phenolics | |||
| (63) Acetophenone | C8H8O | 120.0575 | 238 |
| (64) 4-hydroxycoumarin | C9H6O3 | 162.0317 | 270 |
| (65) N,N-dimethyl-5-methoxytryptamine | C13H18N2O | 218.1419 | 294 |
| (j) | |||
|---|---|---|---|
| Class amines | Empirical formula | Molecular massa | Percent changeb |
| (66) Epinine | C9H13NO2 | 167.0946 | 222 |
| (67) N-caffeoylputrescine | C13H18N2O3 | 250.1317 | 19 |
| Alkaloids | |||
| (68) Coumarin | C9H6O2 | 146.0368 | 10 |
| (69) Indole-5,6-quinone | C8H5NO2 | 147.0393 | 40 |
| (70) 2-methyl cinnamic acid | C10H202 | 162.0681 | 59 |
| (71) 3-acetylaminoquinoline | C11H10N2O | 186.0793 | 34 |
| (72) 7-ethoxy-4-methylcoumarin | C12H12O3 | 204.0786 | 36 |
| (73) 4,6-dimethyl-8-tert-butylcoumarin | C15H18O2 | 230.1307 | 27 |
| (74) 1-O-hexyl-2,3,5-trimethylhydroquinone | C15H24O2 | 236.1776 | 179 |
| Phenolics | |||
| (75) Acetophenone | C8H8O | 120.0575 | 54 |
| (76) Alpha-hydroxyacetophenone | C8H8O2 | 136.0524 | 49 |
| (77) Nicotine | C10H14N2 | 162.1157 | 270 |
| (78) Swainsonine | C8H15N2 | 173.1052 | 500 |
| (79) (S)-6-hydroxynicotine | C10H14N2O | 178.1106 | 263 |
| Isoprenoid | |||
| (80) Nopinone | C9H14O | 138.1045 | 20 |
| (k) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (81) 2,3-cyclopentenopyridine | C8H9N | 119.0735 | 278 |
| (82) Dihydro-thymine | C6H5N2O2 | 128.0586 | 227 |
| (l) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (84) Dihydro-thymine | C6H5N2O2 | 128.0586 | 238 |
| (85) Uridine | C9H12N2O6 | 244.0695 | 400 |
| (m) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (86) Fumaric acid, monoethyl ester | C6H8O4 | 144.0423 | 56 |
| (n) | |||
|---|---|---|---|
| Formula | Massa | Changeb | |
| (87) Fumaric acid | C4H404 | 116.0110 | 270 |
| (88) DL-malic acid | C4H6O5 | 134.0215 | 270 |
| (89) Citric acid | C6H8O7 | 192.0270 | 385 |
| (90) Fumaric acid monoethyl ester | C6H8O4 | 144.0423 | 345 |
| (o) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (91) 3-hydroxy-1-pyrroline-delta-carboxylate | C5H7NO3 | 129.0426 | 133 |
| (p) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (92) Delta1-pyrroline 2-carboxylate | C5H7NO2 | 113.0477 | 217 |
| (93) 3-hydroxy-1-pyrroline-gamma-carboxylate | C5H7NO3 | 129.0426 | 244 |
| (q) | |||
|---|---|---|---|
| (94) N-nitrosopyrrolidine | C4H8N2O | 100.0637 | 152 |
| (95) 2-furylglyoxylonitrile | C6H3NO2 | 121.0164 | 182 |
| (96) L-threonate | C4H8O5 | 136.0372 | 370 |
| (97) 4-phenyl-2-thiazoleethanamide | C11H12N2S | 204.0721 | 47 |
| (98) Diethyl 1,4 piperazine dicarboxylate | C10H18N2O4 | 230.1267 | 54 |
| (99) Hopantenic acid | C10H18NO5 | 233.1263 | 34 |
| (100) Menthyl acetoacetate | C14H24O3 | 240.1725 | 23 |
| (101) N-methyl-5-allyl-cyclopentylbarbituric acid | C13H16N2O3 | 248.1161 | 208 |
| (102) 1-(3-benzoyloxyphenyl)-3-methyl-3-methoxyurea | C16H16N2O4 | 300.1110 | 192 |
| (103) 1-(3-benzyloxylphenyl)-3-methyl-3-methoxylurea | C16H20N2O4 | 304.1350 | 333 |
| (104) 1,4-Bis((2-((2-hydroxyethyl)amino)ethyl)amino)-9,10-anthracenedione diacetate | C26H32N4O6 | 496.2322 | 345 |
| (r) | |||
|---|---|---|---|
| Formula | Massa | Changeb | |
| (105) N-nitrosopyrrolidine | C4H8N2O | 100.0637 | 714 |
| (106) R-4-hydroxy-2-pyrrolidone | C4H7NO2 | 101.0477 | 435 |
| (107) 3-methoxy-1,2-propanediol | C4H10O3 | 106.0630 | 40 |
| (108) cis-2-hexenoic acid amide | C6H11NO | 113.0841 | 26 |
| (109) 7-oxabicyclo[2.2.1]hept-5-ene-2,3-dione | C6H4O3 | 124.0160 | 41 |
| (110) 2-methoxy-3-methyl-pyrazine | C6H8N2O | 124.0637 | 51 |
| (111) Phthalic anhydride | C8H4O3 | 148.0160 | 24 |
| (112) Gamma-nonanolactone | C9H16O2 | 156.1150 | 43 |
| (113) 1,5-diazatricyclo [4.2.2.2(2,5)]dodecane | C10H18N2 | 166.0994 | 625 |
| (114) 2-decenoic acid | C10H18O2 | 170.1307 | 56 |
| (115) 2,2,6,6-tetramethyl-N-nitrosopiperidine | C9H18N2O | 170.1419 | 29 |
| (116) 1-acetyl-4-piperidinecarboxylic acid | C8H13NO3 | 171.0895 | 270 |
| (117) Decanamide | C10H21NO | 171.1623 | 435 |
| (118) Sulfuric acid dipropyl ester | C6H14N2O8 | 182.0613 | 56 |
| (119) o,o′-iminostilbene | C4H11N | 193.0892 | 13 |
| (120) Cyclohexanepropionic acid, 4-oxo-, ethyl ester | C11H18O3 | 198.1256 | 25 |
| (121) Cyclooctyl-1,1-dimethylurea | C11H22N2O | 198.1732 | 24 |
| (122) Sebacic acid | C10H18O4 | 202.1205 | 16 |
| (123) cis-2,6-di-tert-butylcyclohexanone | C14H26O | 210.1984 | 35 |
| (124) 6-[2-(5-nitrofuranyl)ethenyl]-2-pyridinemethanol | C12H10N2O4 | 224.0797 | 213 |
| (125) 5-allyl-5-butylbarbituric acid | C11H16N2O3 | 224.1161 | 22 |
| (s) | |||
|---|---|---|---|
| Empirical formula | Molecular massa | Percent changeb | |
| (126) Isothiocyanic acid 1,4-cyclohexylene-dimethylene ester | C15H24O2 | 226.0598 | 31 |
| (127) Tetradecanamide | C14H29NO | 227.2249 | 23 |
| (128) Cedrol methyl ether | C16H28O | 236.2140 | 21 |
| (129) Cyclohexadecanone | C16H30O | 238.2297 | 18 |
| (130) 1,3-di-o-tolylguanidine | C15H17N3 | 239.1422 | 400 |
| (131) Menthyl acetoacetate | C14H24O3 | 240.1725 | 13 |
| (132) Methocarbamol | C11H15NO3 | 241.0950 | 244 |
| (133) N-[2,6-bis(isopropyl)phenyl]-2-imidazolidineimine | C15H23N3 | 245.1892 | 345 |
| (134) (-)-ptilocaulin | C15H25N3 | 247.2048 | 294 |
| (135) 1-Lauryl-2-pyrrolidone | C16H31NO | 253.2406 | 29 |
| (136) Hexadecanamide | C16H33NO | 255.2562 | 12 |
| (137) Dodecylmalonic acid | C15H28O4 | 272.1988 | 46 |
| (138) 4-amino-N-(6-methoxy-4-pyrimidyl)-benzenesulfonamide | C11H12N4O3S | 280.0630 | 20 |
| (139) Rocastine | C13H19N3OS | 281.1198 | 276 |
| (140) Palmoxiric acid | C17H32O3 | 284.2351 | 35 |
| (141) Propionic acid, 3-dodecyloxy-2-ethoxy-, methyl ester | C18H36O4 | 316.2614 | 556 |
| (142) Benzenesulfonic acid dodecylester | C18H30O3S | 326.1916 | 63 |
| (143) Di(2-ethylhexyl) itaconate | C21H38O4 | 354.2770 | 40 |
| (144) 2,2′-ethyledene bis (4,6-di-t-butyl) | C30H45O2 | 438.3498 | 12 |
ACKNOWLEDGMENTS
We thank Dr. Rafiqa Ameziane for valuable discussions and help with experiments. Plant materials were developed with a grant from the Herman Frasch Foundation. Analyses were supported by grants from the Illinois-Missouri Biotechnology Alliance and the Council for Food and Agricultural Research. We thank all at Phenomenome for technical assistance with FT-ICR-MS.
References
- 1.Glassbrook N, Ryals J. A systematic approach to biochemical profiling. Curr Opin Plant Biol. 2001;4(3):186–190. doi: 10.1016/s1369-5266(00)00159-x. [DOI] [PubMed] [Google Scholar]
- 2.Katona Z.F, Sass P, Molnár-Perl I. Simultaneous determination of sugars, sugar alcohols, acids and amino acids in apricots by gas chromatography mass spectrometry. J Chromatogr A. 1999;266(1):77–84. [Google Scholar]
- 3.Adams M.A, Chen Z.L, Landman P, Colmer T.D. Simultaneous determination by capillary gas chromatography of organic acids, sugars, and sugar alcohols in plant tissue extracts as their trimethylsilyl derivatives. Anal Biochem. 1999;266(1):77–84. doi: 10.1006/abio.1998.2906. [DOI] [PubMed] [Google Scholar]
- 4.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey R.N, Willmitzer L. Metabolite profiling for plant functional genomics [published correction appears in Nat Biotechnol. 2000;19(2):173] Nat Biotechnol. 2000;18(11):1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 5.Angotti M, Maunit B, Muller J.F, Bezdetnaya L, Guillemin F. Characterization by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry of the major photoproducts of temoporfin (m-THPC) and bacteriochlorin (m-THPBC) J Mass Spectrom. 2001;36(7):825–831. doi: 10.1002/jms.186. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Tolic N, Zhao R, et al. High-throughput proteomics using high-efficiency multiple-capillary liquid chromatography with on-line high-performance ESI FTICR mass spectrometry. Anal Chem. 2001;73(13):3011–3021. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- 7.Marshall A.G, Hendrickson C.L, Shi S.D. Scaling MS plateaus with high-resolution FT-ICRMS. Anal Chem. 2002;74(9):252A–259A. doi: 10.1021/ac022010j. [DOI] [PubMed] [Google Scholar]
- 8.Aharoni A, Ric de Vos C.H, Verhoeven H.A, et al. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS. 2002;6(3):217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A, Karas M, Dulcks T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI? J Am Soc Mass Spectrom. 2003;14(5):492–500. doi: 10.1016/S1044-0305(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 10.Allen J, Davey H.M, Broadhurst D, et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21(6):692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 11.Roessner U, Willmitzer L, Fernie A.R. High-resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems. Identification of phenocopies. Plant Physiol. 2001;127(3):749–764. [PMC free article] [PubMed] [Google Scholar]
- 12.Ameziane R, Bernhard K, Lightfoot D.A. Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant and Soil. 2000;221(1):47–57. [Google Scholar]
- 13.Ameziane R, Bernhardt K, Lightfoot D.A. Expression of the bacterial gdhA gene encoding a glutamate dehydrogenase in tobacco and corn increased tolerance to the phosphinothricin herbicide. In: Martins-Loucao M.A, Lips S.H, editors. Nitrogen in a Sustainable Ecosystem: From the Cell to the Plant. Leiden, The Netherlands: Backhuys Publishers; 2000. pp. 339–343. [Google Scholar]
- 14.Mungur R. Metabolic Profiles of GDH transgenic crops [MS thesis] Carbondale, Ill: SIUC; 2002. 189 pp. [Google Scholar]
- 15.Mungur R, Glass A.D, Wood A.J, Lightfoot D.A. Increased water deficit tolerance in Nicotiana tabacum expressing the Escherichia coli glutamate dehydrogenase gene. Plant Cell Physiology. In press. [Google Scholar]
- 16.Raamsdonk L.M, Teusink B, Broadhurst D, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19(1):45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 17.Lightfoot D.A, Long L.M, Vidal M.E. Plants containing the gdhA gene and methods of use thereof. 5 998 700. US patent. 1999
- 18.Lightfoot D.A, Long L.M, Vidal M.E. Plants containing the gdhA gene and methods of use thereof. 6 329 573. US patent. 2001
- 19.Schmidt R.R, Miller P, inventors. Polypeptides and polynucleotides relating to alpha and beta subunits of lutamate dehydrogenase and methods of use. 5 879 941. US patent. 1999
- 20.Melo-Oliveira R, Oliveira I.C, Coruzzi G.M. Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci U S A. 1996;93(10):4718–4723. doi: 10.1073/pnas.93.10.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker T.W, Carrayol E, Hirel B. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta. 2000;211(6):800–806. doi: 10.1007/s004250000355. [DOI] [PubMed] [Google Scholar]
- 22.Wootton J.C. Re-assessment of ammonium-ion affinities of NADP-specific glutamate dehydrogenases. Activation of the Neurospora crassa enzyme by ammonium and rubidium ions. Biochem J. 1983;209(2):527–531. doi: 10.1042/bj2090527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keys A.J, Bird I.F, Cornelius M.J, Lea P.J, Wallsgrove R.M, Miflin B.J. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- 24.Aubert S, Bligny R, Douce R, Gout E, Ratcliffe R.G, Roberts J.K. Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by (13)C and (31)P nuclear magnetic resonance. J Exp Bot. 2001;52(354):37–45. [PubMed] [Google Scholar]
- 25.Coruzzi G.M, Zhou L. Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects.”. Curr Opin Plant Biol. 2001;4(3):247–253. doi: 10.1016/s1369-5266(00)00168-0. [DOI] [PubMed] [Google Scholar]
- 26.Lea P.J, Robinson S.A, Stewart G.R. The enzymology and metabolism of glutamine, glutamate and asparagine. In: Miflin B.J, Lea P.J, editors. The Biochemistry of Plants. New York, NY: Academic Press; 1990. pp. 121–159. [Google Scholar]
- 27.Stitt M, Muller C, Matt P, et al. Steps towards an integrated view of nitrogen metabolism. J Exp Bot. 2002;53(370):959–970. doi: 10.1093/jexbot/53.370.959. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Guegler K, LaBrie S.T, Crawford N.M. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12(8):1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Larkins B.A. Genetic analysis of amino acid accumulation in opaque-2 maize endosperm. Plant Physiol. 2001;125(4):1766–1777. doi: 10.1104/pp.125.4.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass A.D.M, Britto D.T, Kaiser B.N, et al. The regulation of nitrate and ammonium transport systems in plants. J Exp Bot. 2002;53(370):855–864. doi: 10.1093/jexbot/53.370.855. [DOI] [PubMed] [Google Scholar]
- 31.Geiger M, Walch-Liu P, Engels C, et al. Enhanced carbon dioxide leads to a modified diurnal rhythm of nitrate reductase activity in older plants, and a large stimulation of nitrate reductase activity and higher levels of amino acids in young tobacco plants. Plant Cell Environ. 1998;21(3):253–268. [Google Scholar]
- 32.Noctor G, Novitskaya L, Lea P.J, Foyer C.H. Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J Exp Bot. 2002;53(370):939–945. doi: 10.1093/jexbot/53.370.939. [DOI] [PubMed] [Google Scholar]
- 33.Chen T.H, Murata N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol. 2002;5(3):250–257. doi: 10.1016/s1369-5266(02)00255-8. [DOI] [PubMed] [Google Scholar]
- 34.Hoshida H, Tanaka Y, Hibino T, et al. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol. 2000;43(1):103–111. doi: 10.1023/a:1006408712416. [DOI] [PubMed] [Google Scholar]
- 35.Fell D.A, Wagner A. The small world of metabolism. Nat Biotechnol. 2000;18(11):1121–1122. doi: 10.1038/81025. [DOI] [PubMed] [Google Scholar]
- 36.Wink M. Special nitrogen metabolism. In: Dey P.M, Harborne J.B, editors. Plant Biochemistry. London, UK: Academic Press; 1997. pp. 439–486. [Google Scholar]
- 37.Balint R, Cooper G, Staebell M, Filner P. N-caffeoyl-4-amino-n-butyric acid, a new flower-specific metabolite in cultured tobacco cells and tobacco plants. J Biol Chem. 1987;262(23):11026–11031. [PubMed] [Google Scholar]
- 38.Baumert A, Mock H.P, Schmidt J, Herbers K, Sonnewald U, Strack D. Patterns of phenylpropanoids in non-inoculated and potato virus Y-inoculated leaves of transgenic tobacco plants expressing yeast-derived invertase. Phytochemistry. 2001;56(6):535–541. doi: 10.1016/s0031-9422(00)00422-2. [DOI] [PubMed] [Google Scholar]
- 39.Duncan S.H, Flint H.J, Stewart C.S. Inhibitory activity of gut bacteria against Escherichia coli O157 mediated by dietary plant metabolites. FEMS Microbiol Lett. 1998;164(2):283–288. doi: 10.1111/j.1574-6968.1998.tb13099.x. [DOI] [PubMed] [Google Scholar]
- 40.Born S.L, Caudill D, Fliter K.L, Purdon M.P. Identification of the cytochromes P450 that catalyze coumarin 3,4-epoxidation and 3-hydroxylation. Drug Metab Dispos. 2002;30(5):483–487. doi: 10.1124/dmd.30.5.483. [DOI] [PubMed] [Google Scholar]
- 41.Strack D. Phenolic metabolism. In: Dey P.M, Harborne J.B, editors. Plant Biochemistry. London, UK: Academic Press; 1997. pp. 387–416. [Google Scholar]
- 42.Rooprai H.K, Kandanearatchi A, Maidment S.L, et al. Evaluation of the effects of swainsonine, captopril, tangeretin and nobiletin on the biological behaviour of brain tumour cells in vitro. Neuropathol Appl Neurobiol. 2001;27(1):29–39. doi: 10.1046/j.0305-1846.2000.00298.x. [DOI] [PubMed] [Google Scholar]


