Abstract
Based on comparative genomics, we created a bioinformatic package for computer prediction of small nucleolar RNA (snoRNA) genes in mammalian introns. The core of our approach was the use of the Mammalian Orthologous Intron Database (MOID), which contains all known introns within the human, mouse and rat genomes. Introns from orthologous genes from these three species, that have the same position relative to the reading frame, are grouped in a special orthologous intron table. Our program SNO.pl searches for conserved snoRNA motifs within MOID and reports all cases when characteristic snoRNA-like structures are present in all three orthologous introns of human, mouse and rat sequences. Here we report an example of the SNO.pl usage for searching a particular pattern of conserved C/D-box snoRNA motifs (canonical C- and D-boxes and the 6 nt long terminal stem). In this computer analysis, we detected 57 triplets of snoRNA-like structures in three mammals. Among them were 15 triplets that represented known C/D-box snoRNA genes. Six triplets represented snoRNA genes that had only been partially characterized in the mouse genome. One case represented a novel snoRNA gene, and another three cases, putative snoRNAs. Our programs are publicly available and can be easily adapted and/or modified for searching any conserved motifs within mammalian introns.
INTRODUCTION
Small nucleolar RNA (snoRNA) is a major component of small nucleolar ribonucleoprotein (snoRNP) particles that are located inside the nucleolus of eukaryotes and participate in post-transcriptional chemical modification or processing of different RNAs, including ribosomal RNA (rRNA) and small nuclear RNA (snRNA) [for reviews see (1–5)]. SnoRNAs are ancient genes, as they are widespread through the entire domain of Eukaryota, as well as Archaea. There are two types of these RNAs, called C/D-box and H/ACA-box snoRNAs, and they are characterized by distinct 3D structures and conserved sequence elements. The C/D-box RNA is associated with 2′-O-ribose methylation, and H/ACA-box RNA with pseudouridylation of substrate RNAs. The direct role of snoRNAs is determining the site for chemical modification, via complementary pairing of its specific sequence [known as the antisense element (ASE)] with the segment of substrate RNA undergoing modification. The major catalytic activity does not belong to snoRNA, but rather to a fibrillarin, a protein component of the snoRNP complex (1,6). Besides pseudouridylation and 2′-O-ribose methylation, some snoRNAs perform other documented and putative functions, such as (i) cleavage of the substrate precursor in rRNA (2), (ii) facilitation of rRNA folding (7), (iii) regulation of alternative splicing (8) and (iv) possible unknown functions carried out by so-called ‘orphan’ snoRNAs (2).
The first well-known computer program for genomic prediction of C/D-box snoRNA, named SNOSCAN, was created by Lowe and Eddy (9). SNOSCAN was used by several laboratories (10) to study compact genomes of eukaryotes such as yeast, Drosophila and Arabidopsis. SNOSCAN can predict classical C/D-box snoRNAs that guide modification of rRNA or snRNA molecules, and requires that these rRNA and snRNA sequences be available for the program before its invocation. In 2003, Vitali et al. (7) used this program on a restricted set of human and mouse ribosomal protein gene sequences, and found several novel C/D snoRNAs inside their introns. Recently, two computational approaches were developed to predict H/ACA snoRNAs from genomic sequences (11,12). Again, these programs can predict snoRNAs that are involved in chemical modifications of known rRNA and snRNA sequences, and they are suitable for searching compact eukaryotic genomes. However, it is problematic to apply the existing computational algorithms for studying vertebrate genomes in their entirety, because they are dozens of times larger than the genomes of yeast, insects or Arabidopsis. In the case of vertebrates, numerous false-positive signals may arise due to the computer processing of very large non-coding genomic sequences and thus computer prediction of novel genes may not be as efficient.
We have taken advantage of comparative genomics and present here a new algorithm for computational identification of mammalian C/D-box snoRNA genes with high efficiency. We exploit two well-known characteristics of mammalian genomic structure: (i) all known snoRNA genes of mammals are located inside introns (3) and (ii) the exon–intron gene structure of mammals is highly conserved. In fact, no well-documented case of intron gain exists between fully sequenced mammalian genomes, and there are only a few intron losses reported (13,14). Therefore, introns are ‘fossilized’ in mammalian genes, and the non-coding RNAs (ncRNAs, also known as small non-messenger RNAs, non-protein coding RNAs and untranslated RNAs) located within them could well be fixed inside the same intron since the origin of this taxon. For this reason, we created a database of orthologous introns comprising human, mouse and rat sequences. We define ‘orthologous introns’ as introns from orthologous genes that have the same position relative to the coding sequence. Hence, orthologous introns should have descended from the corresponding intronic sequence of the last common ancestor for the taxon.
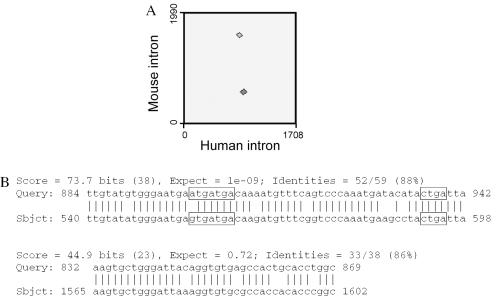
The evolutionary divergence of primate and rodent lineages occurred ∼70–90 million years ago, and during this period non-functional DNA segments representing a major portion of human and rodent orthologous introns lost almost all sequence similarity. For example, Figure 1 demonstrates an alignment of the third introns of the ribosomal protein S3a gene from human and mouse genomes that are orthologous to each other and are 1708 and 1990 nt long, respectively. It is known that these two introns contain U73b snoRNA sequences of human and mouse (NG_000961 and Z83331, respectively). A BLAST-2 alignment with default parameters reveals 88% sequence identity of a 58 nt long fragment of these introns containing snoRNA, and no sequence similarity outside this short functional intronic fragment except one 37 nt long segment (Figure 1). This second conserved segment could also represent a functional sequence not yet known. Therefore, when conserved snoRNA structures are identified within orthologous intron triplets from all three species (human, mouse and rat), it strongly suggests that the conserved sequence is functional. Here, we present our Mammalian Orthologous Intron Database (MOID) for public usage and two programs that search for conserved C/D-box snoRNA motifs inside the entire MOID and characterize putative ASEs of snoRNAs based on comparative genomics of mammalian species.
Figure 1.
Results of an online Blast2 alignment of the 1708 nt long third intron of the human ribosomal protein S3a gene, and its mouse ortholog (1990 nt long third intron of the mouse rps3a gene). (A) Dot plot figure of the intron comparison obtained by bl2seq online program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) with default parameters. (B) Alignment of human and mouse introns. The sequences of snoRNA C- and D-boxes inside these introns are boxed.
Mammalian introns bear all types of snoRNA genes (3), about three-fourth of microRNA genes (15), and also many conserved regulatory motifs (16). Our programs can be easily adapted to search for all kinds of functional sequences using MOID. We are further developing this approach by extending our Orthologous Intron Database with other mammalian and vertebrate species.
MATERIALS AND METHODS
Intron databases
Using the previously published program package for the generation of the Exon–Intron Database (EID) (17), we created three species-specific EIDs containing all known genes of mammals with entirely sequenced genomes. These EID for human, mouse and rat genomes and their documentation are freely available from our website (http://www.meduohio.edu/bioinfo/eid/index.html). We then applied the CIP.pl program (18) for the comparison of intron positions in all orthologous genes from human, mouse and rat genomes. Based on this comparison, we generated an MOID, which is also publicly available from the same web page (documentation provided). This first release of our MOID contains 100 000 orthologous introns from human, mouse and rat genomes. The next release of MOID, with a complete set of orthologous introns, is in preparation.
Programs for computations of snoRNA
We created a SNO.pl program for the identification of snoRNA-like sequences within MOID. This program was written in the PERL language and is publicly available from our web page http://www.meduohio.edu/bioinfo/eid/index.html. SNO.pl scans all human introns for the characteristic conserved C/D-box snoRNA elements defined by PERL regular expressions. Then, for those human introns with snoRNA-like structures, their orthologous mouse and rat intron sequences were extracted from MOID and were analyzed for the presence of the same snoRNA conserved elements. snoRNA-like sequences that were present in all three orthologous introns within human, mouse and rat genomes were output in a special file and are provided as a supplementary document. It takes only 4 min for a desktop computer (AMD Athlon 2200+ processor) to run this program. Further, simple modifications of the regular expressions inside this PERL script should allow the search for any type of conserved motifs within orthologous introns.
Another program written by using PERL, TARGET.pl, was designed to search for all putative targets for snoRNA ASE within databases of rRNA, snRNA and mRNA sequences. No mismatches were allowed in this ASE-target pairing, except non-Watson–Crick (non-WC) G–T pairs. The maximal number of G–T pairs were defined by the user. The execution time of this program was <2 min and it is also available from the same website.
RESULTS AND DISCUSSION
Computation of C/D-box snoRNA-like structures in human and rodents
Using our program SNO.pl, we scanned the entire set of 238 014 human introns from the EID looking for the following conserved structures characteristic for C/D-box snoRNA: (6 nt 5′-stem)-(N)-(C-box)-(loop 40–100 nt)-(D-box)-(6 nt 3′-stem). The C-box is RTGATGA, N stands for any nucleotide, the D-box is CTGA, and the loop can include any nucleotide sequence. In addition, the 5′-stem must have complementarity with the 3′-stem and a score of at least 4 points (each Watson–Crick pair adds 1 point, non-WC G–T pair adds 0.5 points; mismatch: 0 points). In total, 3693 of such snoRNA-like structures were found and selected within 3382 human introns. Then, for each selected human intron, its orthologous counterpart in mouse and rat were searched in the MOID. As a result, 1441 mouse orthologous intron sequences and 1079 rat orthologous intron sequences were obtained and were used to search for the same conserved C/D-box snoRNA elements within these orthologous introns. Finally, 224 snoRNA-like sequences were found in 193 mouse orthologous introns and 124 were found in 108 rat orthologous introns. Intersection of these data represents 57 orthologous intron triplets from human, mouse and rat genomes. Every intron from these 57 triplets contains computed snoRNA-like structures that are described in Table 1. All of the sequences and the supporting materials described in this table are available in the Supplementary File ‘57snoRNA.doc’.
Table 1.
Description of computed snoRNA and snoRNA-like sequences detected inside each intron of 57 orthologous intron triplets from human, mouse and rat genomes
| Category | Number of orthologous intron triplets with snoRNA-like sequences | Description of computed snoRNA-like structures |
|---|---|---|
| Known snoRNA | 15 | U103 (AY349604); U103 (AY349604); U38a (NR_001456); U20 (Z34290); Z25 (HSAJ10666); U73b (NG_000961); U73 (Z83330); U14 (NR_001452); mgU2-19/30 (BK005567.1); Z32 (HSAJ9638); U59 (X96659); Z17a (HSA224024); U41 (X96640); U32 (NR_000021); U33 (NR_000020); U61 (X96661) |
| Partially characterized snoRNA in mouse | 6 | MBII-316 (AF357335); MBII-295 (AF357354); MBII-166 (AF357343); MBII-82 (AF357319); MBII-115 (AF357349); MBII-55(AF357318) |
| Novel snoRNA | 1 | (37..78) segment has 88% sequence identity to human U53 snoRNA (X96652) and (59..79) segment is identical to mouse MBII-35 (AF357377) |
| Putative snoRNA | 3 | Inside middle-size introns with conserved sequence motifs |
| Extra-long introns with false-positive snoRNA-like sequences | 32 | These snoRNA-like structures were found in introns longer than 100 000 nt in all three species |
As shown in Table 1, we found 16 known snoRNA sequences inside 15 orthologous intron triplets (the 19th intron of the human predicted gene KIAA1731 contains two snoRNA molecules, mgU2-19/30 and Z32). Another six snoRNA-like structures found in the set of 57 orthologous intron triplets represent real snoRNA molecules because all six computer-detected snoRNA-like sequences from mouse were identical to partially characterized murine snoRNA sequences that were detected in a large-scale experimental approach (19). A thorough description of these six cases is presented in Table 2, while their sequences are illustrated in Figure 2. One snoRNA-like structure presumably represents an unknown snoRNA gene. It has 88% sequence identity with the human U53 snoRNA and its 59–79 nt fragment is identical to mouse MBII-35 snoRNA. The predicted 12 nt long ASE-2 of this novel gene has a target on the 28S rRNA with one G–T non-WC base pair (Table 2 and Figure 2). Another three snoRNA-like sequences represent putative snoRNA genes with less certainty in respect to their functionality. With one exception, we did not find convincing targets for their ASEs among rRNA, ncRNA from RNAdb and mRNA sequences (Table 2 and Figure 2). Finally, the remaining 56% of introns with snoRNA-like structures (32 out of 57 orthologous triplets) most probably manifested false-positive results because they were found in extra-long introns and in most cases they did not share significant sequence similarity that we observed in real and putative snoRNAs. All of these cases were easy to detect and could be filtered out since our SNO.pl program output the length of each intron with the snoRNA-like structure. Yet, one should keep in mind that long introns could bear novel snoRNA molecules that do not have homologs in other species. The Supplementary File 57snoRNA.doc presents complete information on the 57 predicted sequences and shows that many extra-long introns contain several snoRNA-like sequences per intron.
Table 2.
Description of partially characterized, novel and putative snoRNAs, detected by the SNO.pl program
| snoRNA name (gene) | Species, chromosome | Intron identifier in MOID | Genomic location (within GenBank contig) | Putative targets for ASEs |
|---|---|---|---|---|
| MBII-316 (KIAA0007) | Hs, chr 2 | INTRON_5 2295_NT_022184 | NT_022184 (7952461..7952549) | AS1: gagtcgggg 28S rRNA (3 G–T) (1340–1348) |
| AS2: cacagccaaggga 28S rRNA (1 G–T) (3843–3855) | ||||
| Mm, chr 17 | INTRON_5 17444_NT_039658 | NT_039658 (5183792..5183880) | AS1? | |
| AS2: cacagccaaggga 28S rRNA (1 G–T) (3520–3532) | ||||
| Rn, chr 6 | INTRON_5 7091_NW_047756 | NW_047756 (comp6117735..6117827) | AS1: gagtcaggg 28S rRNA (2 G–T) (1252–1260) | |
| AS2: cacagccaaggga 28S rRNA (1 G–T) (3589–3601) | ||||
| MBII-295 (MNAB) | Hs, chr 9 | INTRON_4 9675_NT_008470 | NT_008470 (comp32963706..32963791) | AS1: gtctgccctat 18S rRNA (1 G–T) (351–361) |
| Mm, chr 2 | INTRON_7 1548_NT_039206 | NT_039206 (comp14828672..14828757) | AS1: gtctgccctat 18S rRNA (1 G–T) (352–362) | |
| Rn, chr 3 | INTRON_5 3929_NW_047653 | NW_047653 (comp3192607..3192692) | AS1: gtctgccctat 18S rRNA (1 G–T) (353–363) | |
| MBII-166 (ch-TOG) | Hs, chr 11 | INTRON_30 11040_NT_009237 | NT_009237 (comp45571179..45571289) | AS1: tggcccttg 28S rRNA (1 G–T) (2721–2729) |
| Mm, chr 2 | INTRON_31 1817_NT_039207 | NT_039207 (32430849..32430959) | AS1: tggcccttg 28S rRNA (1 G–T) (2487–2495) | |
| Rn, chr 3 | INTRON_27 4231_NW_047657 | NW_047657 (17383684..17383793) | AS1: tggcccttg 28S rRNA (1 G–T) (2575–2583) | |
| MBII-82 (SF3B3) | Hs, chr 16 | INTRON_6 15357_NT_010498 | NT_010498 (24186109..24186198) | AS1: gaagagacatgaga 28S rRNA (1 G–T) (3920–3933) |
| Mm, chr 8 | INTRON_6 9497_NT_078575 | NT_078575 (comp35865888..35865972) | AS1: gaagagacatgag 28S rRNA (1 G–T) (3597–3609) | |
| Rn, chr 19 | INTRON_2 16923_NW_047536 | NW_047536 (comp3195818..3195902) | AS1: gaagagacatgag 28S rRNA (2 G–T) (3666–3678) | |
| MBII-115 (GLTSCR2) | Hs, chr 19 | INTRON_10 17981_NT_011109 | NT_011109 (20527299..20527410) | AS2? |
| Mm, chr 7 | INTRON_10 7232_NT_039395 | NT_039395 (comp177705..177815) | AS2: ccccgggcg 28S rRNA (2 G–T) (1022–1030) | |
| Rn, chr 1 | INTRON_10 500_NW_047555 | NW_047555 (comp21675179..21675289) | AS2: ccccgggcg 28S rRNA (2 G–T) (1086–1084) | |
| MBII-55 (NOL5A) | Hs, chr 20 | INTRON_3 18370_NT_011387 | NT_011387 (2574856..2574934) | AS1: ggattgacagatt 18S rRNA (0 G–T) (1285–1297) |
| Mm, chr 2 | INTRON_2 2112_NT_039207 | NT_039207 (70965250..70965321) | AS1: ggattgacagat 18S rRNA (0 G–T) (1285–1296) | |
| Rn, chr 3 | INTRON_3 4524_NW_047658 | NW_047658 (8066063..8066135) | AS1: ggattgacagatt 18S rRNA (0 G–T) (1289–1301) | |
| Novel 1 (KIAA0007) | Hs, chr 2 | INTRON_11 2295_NT_022184 | NT_022184 (7966780..7966861) | AS2: cagccaagggaa 28S rRNA (1G–T) (3845–3856) |
| Mm, chr 17 | INTRON_11 17444_NT_039658 | NT_039658 (5194095..5194174) | AS2: cagccaagggaa 28S rRNA (1G–T) (3522–3533) | |
| Rn, chr 6 | INTRON_12 7091_NW_047756 | NW_047756 (comp6107760..6107839) | AS2: cagccaagggaa 28S rRNA (1G–T) (3591–3602) | |
| Putative 1 (LOC131368) | Hs, chr 3 | INTRON_5 4087_NT_005612 | NT_005612 (8672246..8672320) | ? |
| Mm, chr 16 | INTRON_6 16457_NT_096987 | NT_096987 (comp20526051..20526125) | ? | |
| Rn, chr 11 | INTRON_7 12205_NW_047355 | NW_047355 (9207232..9207306) | ? | |
| Putative 2 (APTX) | Hs, chr 9 | INTRON_5 9220A_NT_008413 | NT_008413 (comp32977364..32977439) | AS1: ccatgaacgag 18S rRNA (3 G–T) (1627–1637) |
| Mm, chr 4 | INTRON_4 3776_NT_039260 | NT_039260 (comp15064752..15064844) | AS1: ccatgaacgag 18S rRNA (3 G–T) (1627–1637) | |
| Rn, chr 5 | INTRON_4 6027_NW_047711 | NW_047711 (comp32384161..32384253) | AS1: ccatgaacgag 18S rRNA (3 G–T) (1627–1637) | |
| Putative 3 (AP4E1) | Hs, chr 15 | INTRON_15 14222_NT_010194 | NT_010194 (22053882..22053953) | ? |
| Mm, chr 2 | INTRON_15 2063_NT_039207 | NT_039207 (67744852..67744930) | ? | |
| Rn, chr 3 | INTRON_4 4479_NW_047658 | NW_047658 (4776101..4776177) | ? |
The first column represents names of partially characterized snoRNAs followed by the GenBank identifier of the human gene inside which snoRNAs were found. The last column represents putative targets for snoRNA ASEs that were detected by the program TARGET.pl. The number of non-WC G–T pairs is shown in parentheses followed by the position of the target within the rRNA sequence. Cases in which no ASE targets were detected are denoted by ‘?’.
Figure 2.
Sequences and conserved motifs of partially characterized, novel and putative snoRNAs that were detected by the SNO.pl program. C-, C′-, D- and D′-boxes are boxed. ASE-1s are underlined by a single line, ASE-2s are underlined by a double line. Hypothetical ASEs, which do not have strong targets, are underlined by a dotted line. All ASE targets are listed in Table 2.
Computation of snoRNA targets
As described by Huttenhofer et al. (20), the C/D-box snoRNA ASEs are 9–20 nt long sequences with 1 nt upstream of the D-boxes and are complementary to the ncRNA molecules that they are guiding for modification. Up to three G–T non-WC pairs are allowed within this ASE-target pairing and the rest of the bases should have perfect complementarity. We have generated and applied the TARGET.pl program to the search for putative targets for our predicted snoRNA-like structures in the databases of rRNA, ncRNA and mRNA sequences. For searching the putative targets within rRNA, we required that the length of ASE be at least 9 nt (L ≥ 9) and that the maximal number of non-WC G–T pairs should not exceed three (GT ≤ 3). In the search for targets among the ncRNA database, we required (L ≥ 12, GT ≤ 3), and among the mRNA database (L ≥ 16, GT ≤ 3). All calculated putative targets are described in Table 2 and their corresponding ASE sequences are underlined in Figure 2.
Simultaneously computing the snoRNA structures for three different species assists in validating the ASE targets and also sheds light on the coevolution of the ASEs and their target sequences. For instance, the tttcgactc ASE1 for the human counterpart of the MBII-316 snoRNA has a computed gagtcGggg target on the 28S rRNA (positions 1340–1348) with three non-WC G–T pairs (as shown in the first row of Table 2 and in Figure 2). In mouse and rat, this target on the 28S rRNA has a single nucleotide G→A change in the middle of the corresponding segment gagtcAggg on the 28S rRNA when compared with human (this difference between human and rodents is shown in uppercase). The corresponding mouse putative ASE1 tttcgactc is identical to human and, therefore, owing to this mutation in the rRNA sequence, it most probably cannot guide modification of this site on the 28S rRNA. At the same time, there is a compensatory mutation of 2 nt for the rat ASE1 ttctgactc that restores the complementarity of the rat ASE1 with the corresponding site gagtcaggg on the 28S rRNA (positions 1252–1260). Such coevolution of the rat ASE1 and its computed target on the 28S rRNA supports the assumption that this putative ASE1 is a functional guiding sequence for chemical modification of the target on the human and rat 28S rRNA.
In contrast to the results with rRNAs, our results for possible ASE targets in mRNA, obtained from the mRNA database using our TARGET.pl program, were not as strong as the one described by Cavaille et al. (8) for ASE1 of MBII-52. All of our computed targets within mRNA corresponded to different genes in human, mouse and rat; thus, we were not inclined to presume that these targets were functional.
In conclusion, many snoRNA molecules have variations in the conserved C- and D-boxes and also in the terminal stem structures; thus, only a small fraction of snoRNA-like sequences from human and rodent genomes have been detected in this search. Modifications of the search pattern in the SNO.pl program should reveal many more putative snoRNAs. Different types of ncRNAs (C/D-box snoRNA, ACA/H-box snoRNA, microRNA and probably others) are also located inside introns. Hence, our approach can be easily adapted for searching all kinds of ncRNAs and functional motifs inside introns.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Dr Robert Blumenthal, Medical University of Ohio, for discussion and valuable suggestions on our manuscript. Support for this work was provided by the Medical University of Ohio Foundation and the Stranahan Foundation, through the Program in Bioinformatics and Proteomics/Genomics. Funding to pay the Open Access publication charges for this article was provided by start-up fund of A.F.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fatica A., Tollervey D. Insights into the structure and function of a guide RNP. Nature Struct. Biol. 2003;10:237–239. doi: 10.1038/nsb0403-237. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 3.Huttenhofer A., Brosius J., Bachellerie J.P. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell E.S., Fournier M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein L.B., Steitz J.A. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell. Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 6.Aittaleb M., Rashid R., Chen Q., Palmer J.R., Daniels C.J., Li H. Structure and function of archaeal box C/D sRNP core proteins. Nature Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 7.Vitali P., Royo H., Seitz H., Bachellerie J.P., Huttenhofer A., Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe T.M., Eddy S.R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 10.Accardo M.C., Giordano E., Riccardo S., Digilio F.A., Iazzetti G., Calogero R.A., Furia M. A computational search for box C/D snoRNA genes in the Drosophila melanogaster genome. Bioinformatics. 2004;20:3293–3301. doi: 10.1093/bioinformatics/bth394. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z.P., Zhou H., Liang D., Qu L.H. Different expression strategy: multiple intronic gene clusters of box H/ACA snoRNA in Drosophila melanogaster. J. Mol. Biol. 2004;341:669–683. doi: 10.1016/j.jmb.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 12.Schattner P., Decatur W.A., Davis C.A., Ares M., Jr, Fournier M.J., Lowe T.M. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorov A., Roy S., Fedorova L., Gilbert W. Mystery of intron gain. Genome Res. 2003;13:2236–2241. doi: 10.1101/gr.1029803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S.W., Fedorov A., Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl Acad. Sci. USA. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Fedorova L., Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. [PubMed] [Google Scholar]
- 17.Saxonov S., Daizadeh I., Fedorov A., Gilbert W. EID: the Exon–Intron Database—an exhaustive database of protein-coding intron-containing genes. Nucleic Acids Res. 2000;28:185–190. doi: 10.1093/nar/28.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorov A., Merican A.F., Gilbert W. Large-scale comparison of intron positions between plant, animal and fungal genes. Proc. Natl Acad. Sci. USA. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huttenhofer A., Kiefmann M., Meier-Ewert S., O'Brien J., Lehrach H., Bachellerie J.P., Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttenhofer A., Cavaille J., Bachellerie J.P. Experimental RNomics: a global approach to identifying small nuclear RNAs and their targets in different model organisms. Methods Mol. Biol. 2004;265:409–428. doi: 10.1385/1-59259-775-0:409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.