Abstract
TIA-1 related protein binds avidly to uridine-rich elements in mRNA and pre-mRNAs of a wide range of genes, including interleukin (IL)-8 and vascular endothelial growth factor (VEGF). The protein has diverse regulatory roles, which in part depend on the locus of binding within the transcript, including translational control, splicing and apoptosis. Here, we observed selective and potent inhibition of TIAR–RNP complex formation with IL-8 and VEGF 3′-untranslated regions (3′-UTRs) using thymidine-rich deoxyoligonucleotide (ODN) sequences derived from the VEFG 3′-UTR. We show by ultraviolet crosslinking and electrophoretic mobility shift assays that TIAR can bind directly to single-stranded, thymidine-rich ODNs but not to double-stranded ODNs containing the same sequence. TIAR had a nearly 6-fold greater affinity for DNA than RNA ( versus 9.4 × 10−9 M). Truncation of TIAR indicated that the high affinity DNA-binding site overlaps with the RNA-binding site involving RNA recognition motif 2 (RRM2). However, RRM1 alone could also bind to DNA. Finally, we show that TIAR can be displaced from single-stranded DNA by active transcription through the binding site. These results provide a potential mechanism by which TIAR can shuttle between RNA and DNA ligands.
INTRODUCTION
TIA-1 related protein (TIAR) is a high-affinity RNA-binding protein that promotes apoptosis (1–3). It possesses three RNA recognition motifs (RRMs) and shares 80% amino acid homology with TIA-1 (3). Both proteins have a predilection for binding uridylate (U)-rich sequences (4). Two relevant binding targets for TIAR and TIA-1 have been identified: the 3′-untranslated regions (3′-UTRs) of certain mRNAs including interleukin (IL)-8, human matrix metalloproteinase (MMP)-13, cyclooxygenase-2, and tumor necrosis factor (TNF)-α, and the U-rich sequence near 5′ splice sites of several pre-mRNAs, such as fibroblast growth receptor 2, Fas, human calcitonin/CGRP and TIAR itself (5–11). When binding to the 3′-UTR, such as with TNF-α or MMP-13, these proteins have been linked to translational silencing possibly by sequestering the mRNA away from the translational machinery to cytoplasmic stress granules (7,9,10,12–15). By binding to pre-mRNAs, on the other hand, TIAR and TIA-1 modulate splicing (5,8,12,16,17). In certain cell systems, TIA-1 may promote apoptosis by producing the alternatively spliced, membrane-bound isoform of Fas responsible for relaying the apoptotic signal (5). While RRM2 of TIAR and TIA-1 is essential for binding to U-rich sequences, little is known about the binding targets of RRM1 and RRM3. Here, we demonstrate that TIAR and TIA-1 can both bind thymidine-rich single-stranded DNA (ssDNA). A more in-depth analysis of TIAR indicates that it binds DNA targets with a higher affinity than RNA and, for the first time, that RRM1 can bind to these DNA targets. Using an in vitro model of transcription, we show that TIAR can be displaced from DNA by active transcription through the DNA-binding site. This finding provides a potential mechanism by which TIAR can shuttle between DNA and RNA ligands.
MATERIALS AND METHODS
Probes and constructs
All deoxyoligonucleotides (ODNs) were purchased from Sigma–Genosys (The Woodlands, TX) and the rVEGF-AS1 ribonucleotide from Integrated DNA Technologies (Coralville, IA). Sequences of ODNs and the RNA oligonucleotide are shown in the Table 1. Radiolabeled and biotinylated IL-8 and vascular endothelial growth factor (VEGF) 3′-UTR riboprobes were synthesized and quantified as described previously (11,18,19). All oligonucleotide probes were labeled with [γ-32P]ATP (Amersham, Piscataway, NJ) using T4 polynucleotide kinase according to the manufacturer's specifications (Promega, Madison, WI). Probes were purified using Quick Spin Oligo Columns (Roche, Indianapolis, IN). Plasmids containing TIAR-b (short) and TIA-1 cDNAs were generously provided by Dr Paul Anderson (4). For TIAR protein expression, the cDNA was subcloned into a pET21a vector containing a His tag at the C-terminus. Truncated TIAR constructs were generated by PCR and subcloned into the Nhe1 and Xho1 sites of pET21a using a common downstream primer (5′-GTTTACACTCGAGCTGTGTTTGGTAACTTGCCAT-3′) and the following upstream primers: RRM 2,3 (5′-CATAGCGGCTAGC TCCAATCACTTCCATGTGT-3′), and RRM 3 (5′-CATAGCGGCTAGC GATGTAGTAAACCAGTCAA-3′). The RRM1 construct was cloned into the BamH1 and Xho1 sites of pET21b using the following primers: upstream, 5′-AGATACAAGGATCCAATGGAAGACGACGGGCAG-3′ and downstream, 5′-GTTTACACTCGAGAGTATCTTTTTTCTGGCTA-3′. All constructs were verified by sequence and restriction analysis. Protein biosynthesis and purification were performed as described previously (20). All proteins were dialyzed and stored in 0.15 M KCl, 40 mM Tris, pH 7.5, 1 mM EDTA and 1 mM MgCl2. HuR and TIA-1 were expressed and purified as glutathione S-transferase (GST) fusion proteins as described previously (4,19). To ensure accurate quantification of His-tagged proteins, we utilized an enzyme-linked immunosorbent assay (ELISA) with two standard controls. The first standard was full-length TIAR and the second was an unrelated His-tagged protein, Eg5. Briefly, the standard and test proteins were diluted serially and incubated in an ELISA plate. The following day, the plate was washed, blocked and incubated with the Penta-His antibody (0.2 μg/ml) according to the manufacturer's specifications (Qiagen, Valencia, CA). On the next day, the plate was washed and incubated with an anti-mouse alkaline phosphatase-conjugated IgG (Pierce Endogen, Rockford, IL) and developed with p-nitrophenyl phosphate (Sigma–Aldrich, St Louis, MO). The OD (at 405 nm) was measured and plotted to determine the linear range of the standards. The mutant proteins were tested at three different dilutions and the concentration was extrapolated based on the dilution that fell within the linear range. The estimated concentration correlated with both standard proteins.
Table 1.
Summary of the oligonucleotide sequences used in the analysis of TIAR binding binding properties
| Oligonucleotide | Sequence |
|---|---|
| C+ | ATTAGATTCCCCCCCCCCCCCCCC |
| GC+ | CACGCGGCCCCGCCTGGGCGC |
| Oligo(dT) | TTTTTTTTTTTTTTT |
| IL8-AS1 | TTAATATCTAAAATAAAATAAATT |
| VEGF-AS1 | AGAATTAAATTCTTTAATACAAAATGCTTTTTTTTTTTTA |
| VEGF-AS1a | AGAATTAAATTCTTTAATAC |
| VEGF-AS1b | AAAATGCTTTTTTTTTTTTA |
| VEGF-AS1b.1 | AAAATGCTTTTTTTTTTA |
| VEGF-AS1b.2 | AAAATGCTTTTTTT |
| VEGF-AS1b.3 | TTTTTTTTTA |
| rVEGF-AS1 | AAAAUGCUUUUUUUUUUUUA |
| VEGF-AS2 | AATAAATAAATGATAACAAATATTAAAATTAAAAAAG |
| VEGF-AS3 | AATATCTTTTCCCCACAATTATTACGGATAAAC |
| VEGF-AS4 | TAAAAATAAATATGTACTACGGAATATCTCGAAAAAC |
| VEGF-S2 | CTTTTTTAATTTTAATATTTGTTATCATTTATTTATT |
RNA-binding assays
All ultraviolet (UV) crosslinking reactions were carried out as previously described (21). For affinity binding assays, varying concentrations of recombinant protein (all in excess of probe) were mixed with labeled probe and UV crosslinked. The electrophoresed band was quantified on a phosphorimager. A non-linear regression curve and a were calculated using GraphPad Prism software v. 4.02 (GraphPad Software, San Diego, CA). For immunoprecipitation, 2 μl of an anti-TIAR antibody to the C-terminus (Santa Cruz Biotechnology, Inc. Santa Cruz, CA) were added after UV crosslinking in the presence of 1000 μl of immunoprecipitation (IP) buffer as described previously (21). The ELISA-based RNA-binding assay was carried out as described previously (22). The RNA-binding reactions for electrophoretic mobility shift assays (EMSAs) were carried out identically to the UV-crosslinking reaction. After the final 10 min incubation at room temperature, loading buffer was added (0.5× TBE, 10% glycerol, 0.25% bromophenol blue and 0.25% xylene cyanol) and the samples were electrophoresed in a native 6% polyacrylamide gel at 4°C based on a published protocol (23). For the supershift assay, the anti-TIAR antibody (or IgG control) was added after the binding reaction and allowed to incubate at room temperature for 30 min before electrophoresis.
Competition assays
With all competition assays, unlabeled competitor was added simultaneously with the probe prior to adding TIAR protein or cell extract. For double-stranded ODN competitions, the unlabeled complementary (or control) ODN was mixed with labeled probe in annealing buffer (10 mM Tris, pH 8.0, 50 mM NaCl and 1 mM EDTA), boiled and allowed to cool ∼1 h. The duplexed ODN was then used in the binding assay. For binding assays involving pre-UV crosslinked protein, TIAR was mixed with 5-fold excess unlabeled ODN or RNA oligonucleotide in binding buffer and then UV crosslinked. An aliquot of this mixture was then used in a subsequent binding reaction with labeled probe.
RNA transcription assay
The following ODNs were synthesized and polyacrylamide-gel purified (Sigma–Genosys, The Woodlands, TX): T7tiar, 5′-AAGATGAGAATAGTATTTTTTTTTTTTCGTATTGGAACAATATAAACAATACTATCCCTAT AGTGAGTCGTATTA-3′ and T7, 5′-TAATACGACTCACTATAGGG-3′. The ODNs were annealed as described above to form a duplex ODN in the T7 recognition sequence. The partially duplexed ODN was used as a template (1.3 pmol) in a T7 transcription reaction using a kit (Ambion, Austin, TX) and [32P]UTP with or without rCTP. Transcripts were electrophoresed on a 8 M urea/6% polyacrylamide gel and exposed to a phosphorimager. For TIAR binding and displacement experiments, the T7tiar ODN was labeled with [γ-32P]ATP as described above and then annealed to the T7 ODN. TIAR was incubated with the partially duplexed ODN in RNA-binding buffer for 20 min at room temperature and then added to the transcription mixture using unlabeled UTP with or without rCTP. T7 RNA polymerase was added and the reaction was carried out at room temperature for various time intervals, followed by UV crosslinking and immunoprecipitation with the anti-TIAR antibody as described above. All reactions were then electrophoresed on a SDS–acrylamide gel.
Cell culture and extract preparation
HS578t cells were purchased from the ATCC (Rockville, MD) and maintained in standard DMEM supplemented with 10% fetal bovine serum, 4 mM glutamine and 0.01 mg/ml bovine insulin in a humidified atmosphere with 10% CO2. Cells were grown to 75–90% confluency and then washed and scraped in phosphate-buffered saline (PBS) and pelleted. Nuclear extracts were prepared from cell pellets in the presence of protease inhibitors using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL) according to the manufacturer's protocol.
RESULTS
VEGF antisense ODNs block TIAR binding to IL-8 and VEGF 3′-UTR
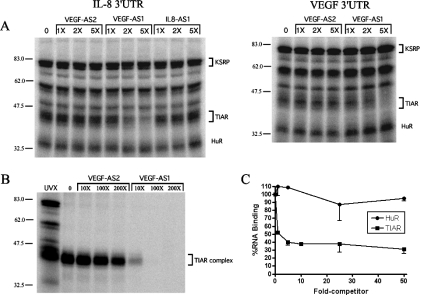
We initially constructed antisense (AS) ODNs complementary to the AU-rich elements (ARE) in the IL-8 and VEGF 3′-UTRs (Table 1) to disrupt RNP complexes by duplexing with the mRNA. We incubated the probe/ODN mixture with nuclear extract from HS578t breast cancer cells, followed by UV crosslinking and electrophoresis. We observed that one of the VEGF-specific ODNs (VEGF-AS1) effectively inhibited the formation of RNP complexes in the 42–45 kDa size range, with both the IL-8 and VEGF 3′-UTR RNA probes (Figure 1A). We had previously identified TIAR as the RNA-binding protein in these complexes (11). A near total loss of binding was observed at very low concentrations of ODN (5-fold excess), indicating a potent inhibition. Two control ODNs, VEGF-AS2 and IL-8-AS1, did not prevent the formation of these complexes, even up to 100-fold excess of probe (data not shown). The other RNP complexes, which include HuR and KSRP as identified, were not competed by VEGF-AS1. To confirm that this RNP complex included TIAR, we carried out the UV-crosslinking competition assay with the IL-8 3′-UTR probe and then immunoprecipitated the extract with a TIAR-specific antibody. As shown in Figure 1B, RNP complexes in the 42–45 kDa size range were immunoprecipitated with the anti-TIAR antibody (lanes 1 and 2), and the complex formation was blocked in the presence of VEGF AS1 but not AS2 ODNs. The loss of RNP complex, once again, occurred at low concentrations of ODN. We next confirmed this inhibitory effect using an ELISA-based RNA-binding assay previously described for ELAV proteins (22). This assay does not utilize UV crosslinking. Recombinant TIAR (b isoform) (3) and HuR proteins were adsorbed to the ELISA well and incubated with a biotinylated IL-8 3′-UTR probe. Potent inhibition of RNA binding was observed with TIAR but not HuR (Figure 1C), in a range of ODN concentrations similar to the UV-crosslinking competition (2- to 10-fold excess).
Figure 1.
TIAR binding to VEGF or IL-8 3′-UTR RNA is competed by a T-rich antisense ODN (VEGF-AS1) derived from the VEGF 3′-UTR. (A) UV crosslinking of HS578t nuclear extract with IL-8 or VEGF 3′-UTR RNA probes in the presence of varying concentrations of competitor ODNs. (B) Immunoprecipitation of UV-crosslinked extracts with an anti-TIAR antibody after binding competition with ODNs and the IL-8 3′-UTR RNA probe. (C) ELISA RNA-binding assay with recombinant HuR or TIAR and a biotinylated IL-8 3′-UTR probe in the presence of VEGF-AS1 competitor ODN. Molecular markers (kDa) are shown to the left of the blots.
TIAR binds directly to thymidine-rich ssDNA
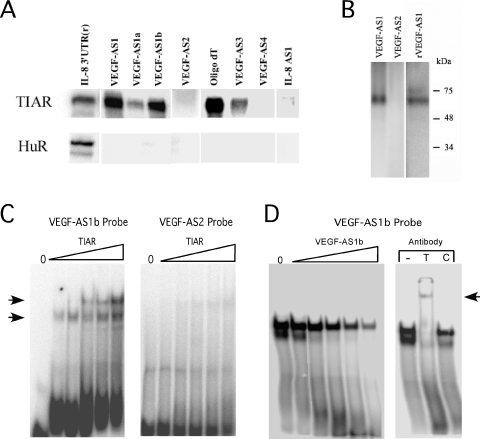
The lack of sequence homology between VEGF-AS1 and the IL-8 3′-UTR suggested that the binding inhibition was not due to duplexing between the DNA and RNA strands. We next tested the possibility that the inhibition was due to direct binding of the ODN to TIAR. We radiolabeled VEGF-AS1 and AS2 ODNs and performed an UV-crosslinking assay with recombinant TIAR (Figure 2). With TIAR, we observed a strong crosslinked band with VEGF-AS1 but not with VEGF-AS2 ODN (Figure 2A, upper panel). The IL-8 3′-UTR riboprobe was used as a control. We performed the same assay with recombinant HuR and observed binding only with the IL-8 3′-UTR riboprobe (lower panel). The concentration of HuR was ∼6-fold greater than that used for TIAR (127 nM versus 22 nM) and ∼40-fold greater than the Kd reported for RNA binding (24). To delineate sequences important for binding, we divided the VEGF-AS1 ODN into 5′ and 3′ fragments of equal length (VEGF-AS1a and VEGF-AS1b, see Table 1). We observed a strong band with VEGF-AS1b versus a weak one with VEGF-AS1a. The most striking sequence difference between the two ODNs was the presence of a 12 nt stretch of thymidines in VEGF-AS1b. We determined whether this sequence pattern alone was sufficient for binding, or whether binding was dependent on secondary structure (e.g. with the adenines at the 5′ end of the VEGF-AS1b). We tested oligo(dT), an iteration of 15 thymidines, and found strong binding, supporting the former possibility. The modest binding observed with VEGF-AS1a (containing three consecutive thymidines) suggested that binding efficiency depends on the length of the thymidine stretch. ODNs that had one or two consecutive thymidines showed no binding (VEGF-AS2, AS4 and IL-8 AS1), while those with stretches of three to four thymidines showed modest binding (VEGF-AS1a and VEGF-AS3). In no instance was there any binding observed with HuR other than the control RNA probe. Using the same crosslinking assay, we found that the closely related TIA-1 protein (fused to GST) could also bind VEGF-AS1 and the RNA homolog, rVEGF-AS1, but not VEGF-AS2 (Figure 2B). As an alternative analysis of DNA binding, we performed EMSA with the VEGF-AS1b ODN and found a prominent shifted complex (Figure 2C). At higher concentrations of TIAR, a second shifted complex was observed consistent with homodimerization of the protein. No complexes were observed with the negative control ODN (VEGF-AS2), and the shifted complex could readily be competed by unlabeled VEGF-AS1b (Figure 2D). Addition of the anti-TIAR antibody, but not control IgG, obliterated the shifted complex and also produced a supershifted complex.
Figure 2.
TIAR binds directly to ODNs. (A) UV-crosslinking results with recombinant TIAR (22 nM) or HuR (127 nM) and radiolabeled oligonucleotides as indicated. A radiolabeled IL-8 3′-UTR riboprobe was used as a control for both proteins. (B) UV crosslinking of recombinant GST-TIA-1 with oligonucleotide probes as indicated. (C) Results of EMSA with recombinant TIAR and oligonucleotide probes as indicated. The range of TIAR concentrations was from 35–210 nM. Shift complexes are indicated by arrows. (D) EMSA of recombinant TIAR and VEGF-AS1b probe with the addition of increasing amounts of unlabeled VEGF-AS1b (1×–100×). The addition of an anti-TIAR antibody (T), but not control IgG (C) obliterated the shifted complexes with VEGF-AS1b and produced a supershifted complex (indicated by an arrow in the right panel).
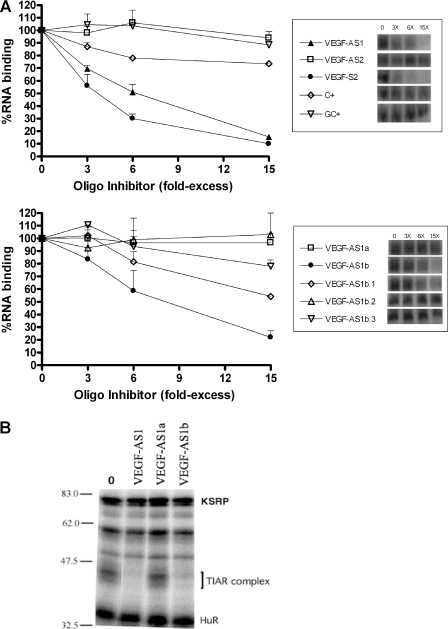
We next analyzed more precisely the sequence requirements for optimal inhibition of RNA binding to TIAR. In this assay, a competitor ODN was added to the reaction simultaneously with the IL-8 3′-UTR riboprobe prior to UV crosslinking. The resultant band was quantitated and compared with the control reaction with no competitor added. Consistent with the binding data shown in Figure 2, there was potent inhibition with VEGF-AS1 but not with VEGF-AS2 (Figure 3A, upper panel). Interestingly, VEGF-S2, the reverse complement of the negative control ODN VEGF-AS2, contains stretches of thymidines throughout and blocked binding to the RNA probe as effectively as VEGF-AS1. ODNs rich in cytidines or cytidines/guanidines (C+, GC+) produced little or no inhibition of binding. Next, we truncated VEGF-AS1 to better determine the components that were necessary for optimal competition of this ODN. As shown in the lower panel, the 3′ end possessed sequences necessary for complete inhibition of binding (VEGF-AS1b) versus no inhibition with the 5′ end (VEGF-AS1a). When the thymidine stretch was truncated to 10 residues, there was some decrease in binding inhibition (VEGF-AS1b.1). As the thymidines were further truncated, there was progressive reduction in inhibition (VEGF-AS1b.3), with a complete reversal at seven residues (VEGF-AS1b.2). The potency of inhibition cannot be totally explained by the number of consecutive thymidines since VEGF-S2 has thymidine stretches interspersed with adenines, with the longest stretch being 6 nt. To confirm that this binding inhibition was limited to the TIAR–RNP complex, we performed UV crosslinking of HS578t nuclear extracts in the presence of VEGF-AS1a and VEGF-AS1b at 10-fold excess of the IL-8 RNA probe. There was complete obliteration of the TIAR complex with VEGF-AS1b, similar to the full-length VEGF-AS1 ODN (Figure 3B). No significant changes were observed with HuR, KSRP or other unidentified RNP complexes, indicating the specificity of inhibition. As with the UV crosslinking of recombinant TIAR, VEGF-AS1a, which lacked any extended thymidine stretches, did not inhibit TIAR–RNP complex formation.
Figure 3.
TIAR binding to IL-8 3′-UTR is competed by thymidine-rich ODNs. (A) Competitor ODNs were added at the same time as the IL-8 3′-UTR probe prior to UV crosslinking. The resultant band was quantitated by phosphorimager and compared with the band intensity when no competitor was added. The results represent an average of three experiments. A representative blot is shown on the right. (B) UV crosslinking of HS578t breast cancer cell extract to IL-8 3′-UTR riboprobe in the presence of unlabeled ODNs (10-fold excess over probe) as indicated.
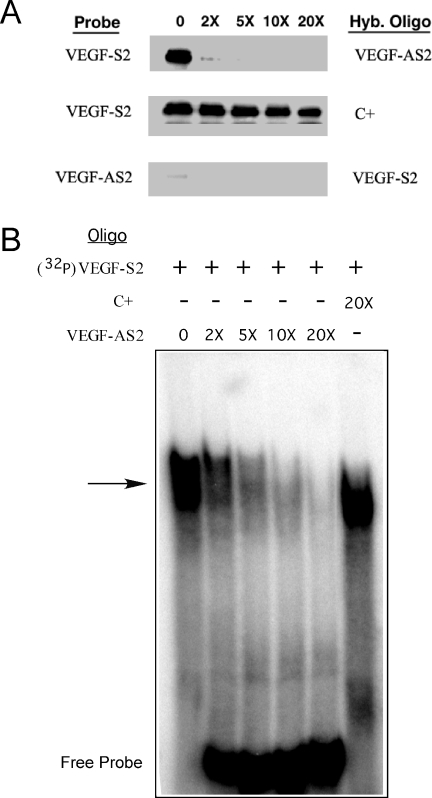
We evaluated whether the DNA-binding capacity of TIAR was limited to ssDNA. We utilized the negative control, VEGF-AS2, and its compliment, VEGF-S2, which potently inhibited IL-8 3′-UTR RNA binding to TIAR. We pre-hybridized the labeled VEGF-S2 ODN with varying amounts of unlabeled complementary ODN (VEGF-AS2), and then UV crosslinked the mixture to TIAR (Figure 4A). As expected, VEGF-S2 bound to TIAR (upper panel). When the probe was hybridized to VEGF-AS2, even at 2-fold excess, there was near complete obliteration of binding. However, when a non-complementary ODN was added (C+), there was continued binding even at 20-fold excess (middle panel). No appreciable binding was observed with radiolabeled VEGF-AS2, alone or when hybridized to VEGF-S2 (lower panel). We confirmed these observations using EMSA (Figure 4B). The VEGF-S2 ODN produced a strong shifted complex, which was gradually obliterated when the probe was pre-hybridized with increasing amounts of unlabeled VEGF-AS2. The C+ ODN at 20-fold excess did not affect the shifted band. Both assays indicated that the TIAR binds only to ssDNA.
Figure 4.
TIAR binds only to ssDNA. (A) Labeled VEGF-S2 ODN probe was pre-hybridized to its complementary ODN (VEGF-AS2) at varying concentrations and then incubated with recombinant TIAR and UV crosslinked (upper panel). A non-complementary ODN (C+) was used as a control hybridization ODN (middle panel). The same experiment was repeated with labeled VEGF-AS2 as a probe and VEGF-S2 as the hybridization ODN (lower panel). (B) EMSA of the VEGF-S2 ODN probe after duplexing with the complementary ODN (VEGF-AS2) or C+ at the concentrations (relative to probe) indicated.
A DNA-binding locus is present outside of the RNA-binding site
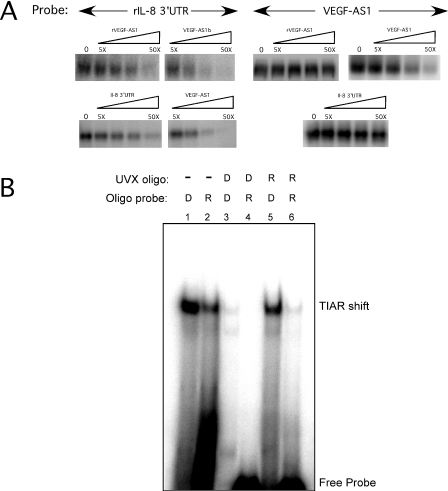
We next determined whether the DNA-binding capacity of TIAR was solely related to the U-rich RNA-binding site. To address this question, we performed a series of competition assays utilizing IL-8 3′-UTR and VEGF-AS1 probes (Figure 5A). We analyzed binding to TIAR in the presence of four different unlabeled competitors, VEGF-AS1, VEGF-AS1b, rVEGF-AS1 and an IL-8 3′-UTR in vitro transcript. TIAR bound to IL-8 3′-UTR, and the binding was competed by all four competitors. Interestingly, the DNA competitors blocked binding at lower concentrations than either the IL-8 3′-UTR transcript or rVEGF-AS1, suggesting that the DNA molecule had a higher affinity for TIAR. On the other hand, neither RNA competitor could compete with binding to VEGF-AS1 even at 50-fold excess. This finding suggested two possibilities. First, that there is an additional DNA-binding site(s) outside of the RNA-binding site. A previous report identified RRM2 as the locus for U-rich binding (4), but did not demonstrate any RNA target for RRM1. The second possibility is that the binding affinity to DNA is significantly greater than to RNA. The latter possibility was tested by increasing the amount of rVEGF-AS1 competitor to 200-fold. At this concentration, there was only a modest diminution of binding (data not shown), suggesting the presence of an additional DNA-binding site(s) within TIAR, distinct from the RNA-binding locus. As an alternative approach to test this possibility, we pre-crosslinked TIAR with unlabeled rVEGF-AS1 or VEGF-AS1b ODN, and then tested the protein for binding to the DNA or RNA probe by EMSA (Figure 5B). Lanes 1 and 2 show shifted complexes in the control reactions with labeled DNA and RNA oligonucleotides using uncrosslinked TIAR. When TIAR is pre-crosslinked with unlabeled VEGF-AS1, there is a loss of shifted complex with both the DNA probe (lane 3) and the RNA probe (lane 4). On the other hand, when the protein is pre-crosslinked with RNA, a shifted complex is still produced with the DNA probe similar to the control (compare lanes 1 and 5). The RNA probe, however, fails to produce any substantial shift (lane 6). Thus, both the UV crosslinking and EMSA data strongly suggest the presence of an additional DNA-binding site(s) outside of the RNA-binding locus.
Figure 5.
A DNA-binding locus can be separated from the RNA-binding site. (A) UV-crosslinking reactions with VEGF-AS1 ODN or rIL-8 3′-UTR (RNA) probes and recombinant TIAR were performed in the presence of competitors as indicated. (B) TIAR was pre-crosslinked with rVEGF-AS1 (R) or VEGF-AS1b (D) oligonucleotides (as indicated by UVX) and then assayed by EMSA using radiolabeled R or D probes.
TIAR possesses two distinct loci for DNA-binding activity
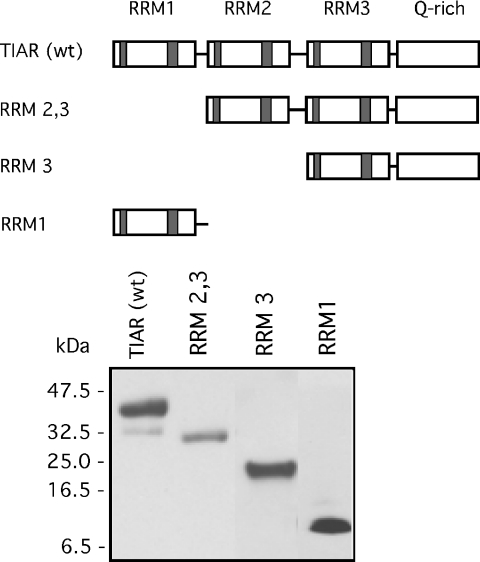
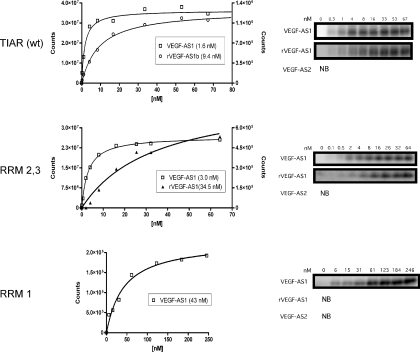
We next determined the binding loci within TIAR for DNA ligands using deletion mutagenesis. We created a series of truncated mutations spanning the TIAR protein (Figure 6). Each mutant was made with a C-terminal His tag similar to the wild-type protein. The protein product was verified by western blotting using an anti-His tag antibody. To determine binding affinity, we used the UV-crosslinking method with recombinant protein in excess of probe. As shown in Figure 7 (upper panel), the full-length TIAR bound to both DNA (VEGF-AS1) and RNA (rVEGF-AS1) oligonucleotides with high affinity, but not to a control ODN (VEGF-AS2). The affinity for DNA was ∼6-fold greater than RNA ( versus 9.4 × 10−9 M). The affinity for rVEGF-AS1 was similar to the reported elsewhere (8.0 × 10−9 M) for TIAR and U-rich sequences using a filter binding assay (4). To define further the domains involved with DNA binding, we evaluated a truncated protein lacking the first RRM (RRM2,3, middle panel). This mutant retained both DNA and RNA-binding capacities although at a several fold lower affinity than the full-length protein ( for the DNA probe and 3.5 × 10−8 M for the RNA probe). However, when both RRM1 and RRM2 were deleted, no RNA or DNA binding was observed (RRM 3, data not shown). These data are consistent with previous observations that RRM2 was essential for binding of U-rich RNA (4). We next tested a mutant expressing only RRM1 (lower panel). This mutant bound to VEGF-AS1 but not the RNA counterpart, rVEGF-AS1. The binding affinity () was lower than full-length protein or the RRM2,3 mutant, and no binding was observed with VEGF-AS2. Thus, a second T-rich DNA-binding domain resided in RRM1.
Figure 6.
Wild-type and truncated TIAR mutant constructs used in binding analysis to DNA and RNA ligands. (A) Schematic representation of the TIAR protein and the mutant constructs. (B) Western blot of the constructs using an anti-His tag antibody.
Figure 7.
Binding affinity analysis of TIAR and mutants. Proteins were in molar excess of probe (indicated in legend) and were UV crosslinked as described in Materials and Methods. Bound probe was quantitated densitometrically by phosphorimaging (shown in the right panels). Affinity curves are shown on the left and the shown in the legend was extrapolated from the curve. The y-axis for DNA probes is on the left and on the right for RNA probes. The curves shown are representative of a single experiment, but the analysis was repeated at least two times with similar results.
TIAR binding to DNA is displaced by active transcription
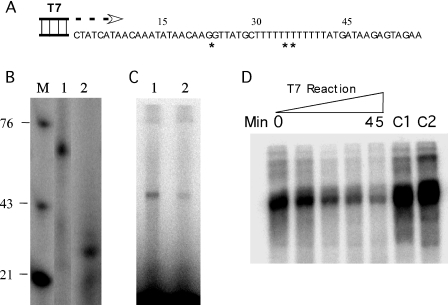
One potential role for the dual binding capacity of TIAR would be to shuttle between DNA and RNA molecules, potentially even bridging them. One of the main loci where ssDNA and RNA interface is that of active transcription. Splicing, for example, which is one of the defined roles of TIAR and TIA-1 with respect to RNA binding, is known to occur concurrently with transcription at the same locus (25,26). Effective shuttling to RNA would require overcoming the nearly 6-fold greater affinity of TIAR for DNA ligands. Since our competition data indicated that TIAR binding to DNA could not be displaced by excessive RNA concentrations, one possible scenario would be displacement of DNA binding by active transcription. To test this possibility in vitro, we synthesized an ODN (Figure 8A) that contained a single-stranded TIAR binding site (double asterisk) and a T7 promoter site. We chose most of the VEGF-AS1b sequence for the TIAR binding locus. The ODN was also designed such that when rCTP was not added to the reaction, the transcript would stall prior to the TIAR binding site (denoted by the single asterisk). We first tested whether this single-stranded template (duplexed at the T7 promoter site) could direct transcription. A full-length transcript was formed (Figure 8B, lane 1) with rCTP in the transcription reaction. When rCTP was not added, a truncated transcript was formed (lane 2). Thus, the partially duplexed ODN was an adequate template for RNA transcription. With the full-length transcript, we were able to demonstrate binding to TIAR by UV crosslinking (Figure 8C, lane 1). The shortened transcript, which is relatively U-rich, produced a faint band suggestive of binding as well (lane 2). We next radiolabeled the template strand of this partially duplexed ODN and pre-incubated it with TIAR prior to transcription. T7 RNA polymerase was then added to the reaction for various time points up to 45 min. The reaction was subjected to UV crosslinking and then immunoprecipitated with the anti-TIAR antibody (Figure 8D). We observed a progressive loss of TIAR binding to the ODN over the 45 min incubation period. When no polymerase was added, TIAR binding remained strong at 45 min (C1). When polymerase was added in the absence of rCTP, TIAR also remained bound to the ODN similar to the control (C2). The intensification of crosslinked TIAR in the controls compared with the time just prior to adding polymerase (0 min) is likely due to the increased time for labeled ODN to bind TIAR. These results indicate that active transcription through the TIAR binding site was necessary for displacement of the protein from its DNA-binding site.
Figure 8.
TIAR binding to DNA is displaced by transcription. (A) Partially duplexed ODN used as a transcription template for T7 RNA polymerase. The T7 promoter site is duplexed as shown. Single asterisk indicates the transcript synthesized if no rCTP is added, and double asterisk indicates the TIAR DNA-binding site. (B) Transcription reaction using the partially duplexed ODN in the presence (lane 1) or in the absence (lane 2) of rCTP. Markers (labeled ODNs) are shown on the left. (C) Results of UV crosslinking of TIAR with the labeled transcripts shown in (B). (D) TIAR was prebound to the partially duplexed, radiolabeled (template strand) ODN, and the complex was incubated with T7 RNA polymerase in the presence of rNTPs for various time intervals (0–45 min). The complex was then subjected to UV crosslinking and immunoprecipitated with an anti-TIAR antibody (shown in the blot). C1 refers to a control reaction in which no T7 polymerase is added, and C2 is a control reaction in which no rCTP is added.
DISCUSSION
TIAR was first identified as an RNA-binding protein with three homologous RRMs in tandem and a glutamine-rich C-terminal domain (3). The protein, which is closely related to TIA-1, has a predilection for binding U-rich sequence, and its role in alternative splicing and translational silencing has been linked to binding U-rich regions near 5′ splice sites or in 3′-UTRs (4,5). The potent and selective disruption of TIAR binding to IL-8 or VEGF-3′-UTR riboprobes by VEGF-AS1 ODN suggested that the DNA and RNA ligands were competing for the same binding locus (Figure 1). Previous analysis of TIAR and TIA-1 revealed that RRM2 alone conferred specificity of binding to U-rich sequence (4). The overlap with DNA binding was confirmed by mutation analysis whereby RRM2,3, but not RRM3 alone, bound to both RNA and DNA ligands at affinities approaching that of full-length protein (Figure 7). Several observations from the competition studies, however, supported the possibility that TIAR has an additional DNA-binding locus. First, VEGF-AS1 ODN effectively competed with U-rich RNA probes for TIAR binding, but neither rVEGF-AS1 nor IL-8 3′-UTR transcript could abrogate DNA binding even at high concentrations (Figure 5). Second, when TIAR was pre-crosslinked to rVEGF-AS1, it could still readily produce a shifted complex with VEGF-AS1 probe. Finally, with mutational studies, it became clear that RRM1 could independently bind T-rich DNA ligands (Figure 7). In an earlier study of TIAR and TIA-1, no RNA target was identified for RRM1 even using labeled total cellular RNA as a probe (4). Our construct, which used a different fusion tag, also did not bind to U-rich RNA ligands. These combined findings suggest that RRM1 only recognizes DNA ligands; however, it is possible that the RNA-binding capacity is preserved only in the context of full-length protein. Binding properties (affinity and specificity) of RRMs can change substantially when analyzed in isolation or in different combinations. For example, the four RRMs of poly(A) binding protein, when expressed individually, do not bind poly(A) RNA efficiently or specifically (27). Similarly, with hnRNPA1, the specificity in target RNA ligands differed for the individual two RRMs of this protein compared with full-length protein. Synergy that occurs between RRMs to promote RNA binding may also explain the lower affinity of the RRM1 construct for T-rich ligands (28–30). In the original binding studies of TIAR, the lack of RNA binding by RRM1 was postulated to be related to an atypical RNP1 motif in RRM1 (4). Usually the first position of this motif is occupied by a basic amino acid residue, whereas in RRM1 of TIAR there is a negatively charged amino acid (aspartic acid). RNP motifs are the most highly conserved elements in the RRM and, based on structural analysis, are present in two adjacent β-sheets that provide a surface for RNA binding through ring stacking (31–34). Positively charged amino acid residues in position 1 may serve to form salt bridges with the negatively charged phosphate backbone of the RNA and stabilize binding. This residue was essential for U1A binding, for example, as substitution of arginine with uncharged glutamine totally disrupted RNA binding (35). However, the amino acid requirements are not absolute, as substitutions of aspartic acid for hydrophobic residues in RNP1 did not disrupt binding of HSH49, a yeast splicing factor, to its RNA target (36). Thus, it is not apparent from primary sequence or known structure as to why RRM1 of TIAR would bind DNA rather than RNA. RRM1 is also the locus of alternative splicing for TIAR, whereby a ‘long’ form is created with a 17 amino acid insertion between RNP1 and 2 (3). In this study, we analyzed the ‘short’ form in recombinant protein experiments and cannot comment on what additional effects on specificity or affinity of binding this insertion would confer.
The dual capacity to bind RNA and DNA ligands is not unique to TIAR, and occurs in a number of proteins with RRMs and other types of RNA-binding domains (e.g. KH and RGG) (37–39). However, the varying affinity for RNA versus DNA and double-stranded DNA (dsDNA) versus ssDNA suggests that binding properties of a RRM are specific rather than ‘generic’ for DNA. Even closely related hnRNPs, such as AUF1 and hnRNP DOB, both of which have homologous RRMs that bind AU-rich RNAs, have markedly different affinities for DNA (40). With TIAR, the nearly 6-fold higher affinity for DNA over RNA, however, was surprising since RRMs in general bind preferably to RNA (39). Several notable exceptions include Stage-specific activator protein (SSAP) from sea urchin and Gbp1p, a telomere-binding protein from Chlamydomonas reinhardtii (41–43). The former binds with high affinity to ssDNA derived from sequences in the enhancer element of late histone H1 gene, but very weakly to RNA counterparts. Gbp1, on the other hand, possesses two RRMs and binds strongly to G-strand telomeric ssDNA when dimerized, but only weakly to the cognate RNA sequence. Interestingly, while both proteins bind more avidly to DNA, SSAP binds both dsDNA and ssDNA, whereas Gbp1 binds only to ssDNA. Here, we found that TIAR binds only to ssDNA and that duplexing the ligand inhibits binding. This finding is in contrast to the double-stranded binding properties of the closely related T-cluster binding protein (TCBP) (44). This protein was identified through screening of a phage library with a concatenated double-stranded oligomer consisting of 26 thymidines. It is 100% homologous with TIAR in RRMs 2 and 3, but lacks a glutamine-rich C-terminus and a complete RRM1. Our mutant RRM2,3 most closely resembles this protein yet clearly binds ssDNA (Figure 7). While a glutamine-rich domain can affect binding activities of attached RRMs (28), it unlikely plays a role here in determining double-stranded versus single-stranded affinity since the RRM1 mutant of TIAR bound to ssDNA.
The capacity of TIAR (and TIA-1) to bind ssDNA potentially expands its functional role in nucleic acid metabolism. The predominantly nuclear localization of TIAR (under normal conditions) places it in proximity to nuclear DNA (2,15). Single-stranded conformations occur in chromatin undergoing active transcription, possibly related to negative torsional forces that create non-B-form DNA structure and local DNA melting (45–47). Regions with high AT content, which TIAR targets, are subject to greater bending and torsion, and thus may be more apt to have local DNA melting (48,49). These openings provide access for ssDNA-binding proteins as shown with two KH-type RNA-binding proteins, HnRNP K and FBP. These proteins bind to ssDNA conformations in far upstream elements from the c-myc promoter and enhance transcription (47,50–53). Reporter studies have also suggested that TCBP, the dsDNA-binding homolog of TIAR, may function as a transcriptional activator (44). Thus, TIAR may serve as a modulator of transcription.
In light of TIAR's role in alternative splicing, the dual binding capacity of TIAR could allow shuttling between RNA and DNA ligands. In our study, we found that TIAR could bind simultaneously to RNA and DNA targets (Figure 5), indicating that it may possibly bridge the two molecules. There is compelling evidence that pre-mRNA splicing (a function of TIAR) is linked spatially and temporally to transcription, whereby RNP particle formation and intron removal have been seen to occur on nascent transcripts (25,26,54,55). Thus, TIAR may bind to T-rich ssDNA forms that occur within the transcription template, either on the coding (e.g. VEGF-S2) or on the non-coding (e.g. VEGF-AS1) strand in preparation for shuttling to the pre-mRNA. To effectively shuttle, TIAR would have to be displaced from its DNA-binding site to permit binding to the pre-mRNA and participate in subsequent RNA processing events. The substantially greater affinity that we observed with DNA binding could be offset by two possibilities: an increase in local concentration of U-rich transcripts or the displacement of TIAR by active transcription. For the former possibility, local competition between RNA and ssDNA has been shown to occur with hnRNPA1/2 proteins that are involved in maintenance of telomeres (56–58). These hnRNPs possess two RRMs that can bind both the single-stranded RNA subunit of telomerase and single-stranded telomeric repeat DNA. These proteins normally have a higher affinity for telomeric RNA, but some investigators have proposed that this binding may be displaced in favor of telomeric DNA in the 3′ overhang as it is lengthened by telomerase (59). In this study, DNA binding, however, could not be displaced in vitro even with very high amounts of RNA, most likely due to the second DNA-binding locus in RRM1 outside of the RNA-binding locus. The second possibility, supported by our in vitro transcription assay (Figure 8), is that TIAR is displaced from its DNA-binding locus by active transcription, thus freeing it to bind the nascent RNA transcript. The displacement may result from RNA–DNA duplex formation during transcription or direct displacement by the polymerase. In our in vitro model, TIAR was also able to bind to the transcript that encompassed the DNA-binding site. Thus, the nascent RNA may help sequester TIAR once it is displaced from the DNA-binding site. In that scenario, the DNA-binding capacity may serve the purpose of correctly positioning TIAR for shuttling to the appropriate locus within the transcript (e.g. to U-rich sequences in the pre-mRNA or the 3′-UTR) for subsequent RNA processing, transport or translational regulation. For example, the recruitment of splicing factors to the nascent RNA has typically been linked to protein–protein interactions with RNA polymerase II [reviewed in (25,26,60)]. The sequence-directed targeting may confer a more specific and efficient way of recruiting TIAR or other splicing factors to the nascent RNA for processing. Clearly, additional studies will be necessary to confirm or exclude whether this shuttling occurs in vivo.
Acknowledgments
The authors thank Ms. Yuanyuan Huang for technical assistance and Dr Ching-yi Chen for critical review of this manuscript. This work was supported by a Merit Review Grant from the Department of Veterans Affairs (P.H.K.). Funding to pay the Open Access publication charges for this article was provided by a Merit Review grant from the Department of Veterans Affairs.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tian Q., Streuli M., Saito H., Schlossman S.F., Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 2.Taupin J., Tian Q., Kedersha N., Robertson M., Anderson P. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl Acad. Sci. USA. 1995;92:1629–1633. doi: 10.1073/pnas.92.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawakami A., Tian Q., Duan X., Streuli M., Schlossman S.F., Anderson P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl Acad. Sci. USA. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dember L.M., Kim N.D., Liu K.Q., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 5.Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 6.Cok S.J., Acton S.J., Morrison A.R. The proximal region of the 3′-untranslated region of Cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 2003;278:36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 7.Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 8.Le Guiner C., Lejeune F., Galiana D., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 9.Piecyk M., Wax S., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-a. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Q., Cok S.J., Zeng C., Morrison A.R. Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR) J. Biol. Chem. 2003;278:1579–1584. doi: 10.1074/jbc.M203526200. [DOI] [PubMed] [Google Scholar]
- 11.Suswam E.A., Nabors L.B., Huang Y., Yang X., King P.H. IL-1β induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′ untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int. J. Cancer. 2005;113:911–919. doi: 10.1002/ijc.20675. [DOI] [PubMed] [Google Scholar]
- 12.Forch P., Valcarcel J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis. 2001;6:463–468. doi: 10.1023/a:1012441824719. [DOI] [PubMed] [Google Scholar]
- 13.Anderson P., Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperons. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2a to the assembly of mammalian stress granules. J. Cell. Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.-C., Stevenin J., Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla S., Dirksen W.P., Joyce K.M., Le Guiner-Blanvillain C., Breathnach R., Fisher S.A. TIA proteins are necessary but not sufficient for the tissue-specific Splicing of the myosin phosphatase targeting subunit 1. J. Biol. Chem. 2004;279:13668–13676. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- 18.Nabors L.B., Suswam E., Huang Y., Yang X., Johnson M.J., King P.H. Tumor necrosis factor alpha induces angiogenic factor up-regulation in malignant glioma cells: a role for RNA stabilization and HuR. Cancer Res. 2003;63:4181–4187. [PubMed] [Google Scholar]
- 19.Nabors L.B., Gillespie G.Y., Harkins L., King P.H. HuR, an RNA stability factor, is expressed in malignant brain tumors and binds to adenine and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 20.King P.H., Redden D., Palmgren J.S., Nabors L.B., Lennon V.A. Hu antigen specificities of ANNA-I autoantibodies in paraneoplastic neurological syndromes. J. Autoimmun. 1999;13:435–443. doi: 10.1006/jaut.1999.0337. [DOI] [PubMed] [Google Scholar]
- 21.Dixon D.A., Tolley N.D., King P.H., Nabors L.B., McIntyre T.M., Zimmerman G.A., Prescott S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King P.H. RNA-binding analyses of HuC and HuD with the VEGF and c-myc 3′-untranslated regions using a novel ELISA-based assay. Nucleic Acids Res. 2000;28:e20. doi: 10.1093/nar/28.7.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garabedian M.J., LaBaer J., Liu W., Thomas J.R. Analysis of protein–DNA interactions. In: Hames B.D., Higgins S.J., editors. Gene Transcription: A Practical Approach. Oxford: Oxford University Press; 1993. pp. 243–293. [Google Scholar]
- 24.Ma W.J., Cheng S., Campbell C., Wright A., Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 25.Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 26.Szentirmay M.N., Sawadogo M. Spatial organization of RNA polymerase II transcription in the nucleus. Nucleic Acids Res. 2000;28:2019–2025. doi: 10.1093/nar/28.10.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burd C.G., Matunis E.L., Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda M., Arai K., Masai H. CTBP1/RBP1, a Saccharomyces cerevisiae protein which binds to T-rich single-stranded DNA containing the 11-bp core sequence of autonomously replicating sequence, is a poly(deoxypyrimidine)-binding protein. Eur. J. Biochem. 1996;238:38–47. doi: 10.1111/j.1432-1033.1996.0038q.x. [DOI] [PubMed] [Google Scholar]
- 29.Caceres J.F., Krainer A.R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J., Hayashi M.K., Zhang Y., Manche L., Krainer A.R., Xu R.-M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenan D.J., Query C.C., Keene J.D. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman D.W., Query C.C., Golden B.L., White S.W., Keene J.D. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc. Natl Acad. Sci. USA. 1991;88:2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 34.Nagai K., Oubridge C., Jessen T.H., Li J., Evans P.R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 35.Jessen T.H., Oubridge C., Teo C.H., Pritchard C., Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igel H., Wells S., Perriman R., Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 37.Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP Proteins and the Biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 38.Krecic A.M., Swanson M.S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell. Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 39.Weighardt F., Biamonti G., Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 40.Tolnay M., Vereshchagina L.A., Tsokos G.C. Heterogeneous nuclear ribonucleoprotein D0B is a sequence-specific DNA-binding protein. Biochem. J. 1999;338:417–425. [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston S.D., Lew J.E., Berman J. Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol. Cell. Biol. 1999;19:923–933. doi: 10.1128/mcb.19.1.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petracek M.E., Konkel L.M., Kable M.L., Berman J. A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J. 1994;13:3648–3658. doi: 10.1002/j.1460-2075.1994.tb06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeAngelo D., DeFalco J., Rybacki L., Childs G. The embryonic enhancer-binding protein SSAP contains a novel DNA- binding domain which has homology to several RNA-binding proteins. Mol. Cell. Biol. 1995;15:1254–1264. doi: 10.1128/mcb.15.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doi T., Minami T., Itoh M., Aburatani H., Kawabe Y., Kodama T., Kondo N., Satoh Y., Asayama T., Imanishi T. An alternative form of nucleolysin binds to a T-cluster DNA in the silencer element of platelet factor 4 gene. Biochem. Biophys Res. Commun. 1997;235:625–630. doi: 10.1006/bbrc.1997.6817. [DOI] [PubMed] [Google Scholar]
- 45.Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;29:609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 46.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncan R., Bazar L., Michelotti G., Tomonaga T., Krutzsch H., Avigan M., Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 48.Kahn J.D., Yun E., Crothers D.M. Detection of localized DNA flexibility. Nature. 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 49.Vlahovicek K., Munteanu M.G., Pongor S. Sequence-dependent modelling of local DNA bending phenomena: curvature prediction and vibrational analysis. Genetica. 1999;106:63–73. doi: 10.1023/a:1003724710997. [DOI] [PubMed] [Google Scholar]
- 50.Tomonaga T., Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 51.Davis-Smyth T., Duncan R.C., Zheng T., Michelotti G., Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 52.Duncan R., Collins I., Tomonaga T., Zhang T., Levens D. A unique transactivation sequence motif is found in the carboxyl- terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol. 1996;16:2274–2282. doi: 10.1128/mcb.16.5.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michelotti E.F., Michelotti G.A., Aronsohn A.L., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beyer A.L., Osheim Y.N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 55.Zhang G., Taneja K.L., Singer R.H., Green M.R. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 56.Ishikawa F., Matunis M., Dreyfuss G., Cech T. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiset S., Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29:2268–2275. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ford L.P., Wright W.E., Shay J.W. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- 59.Moran-Jones K., Wayman L., Kennedy D.D., Reddel R.R., Sara S., Snee M.J., Smith R. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33:486–496. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neugebauer K.M., Roth M.B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]