Abstract
Metazoan Rad51 plays a central role in homologous DNA recombination, and its activity is controlled by a number of Rad51 cofactors. These include five Rad51 paralogs, Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3. We previously hypothesized that all five paralogs participate collaboratively in repair. However, this idea was challenged by the biochemical identification of two independent complexes composed of either Rad51B/C/D/XRCC2 or Rad51C/XRCC3. To investigate if this biochemical finding is matched by genetic interactions, we made double mutants in either the same complex (rad51b/rad51d) or in both complexes (xrcc3/rad51d). In agreement with the biochemical findings the double deletion involving both complexes had an additive effect on the sensitivity to camptothecin and cisplatin. The double deletion of genes in the same complex, on the other hand, did not further increase the sensitivity to these agents. Conversely, all mutants tested displayed comparatively mild sensitivity to γ-irradiation and attenuated γ-irradiation-induced Rad51 foci formation. Thus, in accord with our previous conclusion, all paralogs appear to collaboratively facilitate Rad51 action. In conclusion, our detailed genetic study reveals a complex interplay between the five Rad51 paralogs and suggests that some of the Rad51 paralogs can separately operate in later step of homologous recombination.
INTRODUCTION
Rad51, a homolog of bacterial RecA, plays a central role in homologous recombination (HR) in metazoan cells (1,2). A defect in Rad51 causes the strongest phenotype among any known HR-deficient DT40 mutants and confers embryonic lethality to mice (1–3). Rad51 activity appears to be strictly regulated by a number of Rad51 cofactors including five Rad51 paralogs, namely Rad51B/Rad51L1/REC2, Rad51C/Rad51L2, Rad51D/Rad51L3, XRCC2 and XRCC3. These proteins share ∼20% amino acid identity with Rad51 and among each other [reviewed in (4)]. We have recently published a genetic study, analyzing mutants in each paralog gene in chicken DT40 cells (5–7). Each mutant exhibited a remarkably similar phenotype, including spontaneous chromosomal aberrations (CAs), similar levels of high sensitivity to a DNA cross-linking agent, mild sensitivity to γ-irradiation, reduced targeting efficiencies, significantly attenuated Rad51 focus formation after ionizing radiation (IR) (6,7) and a shift of immunoglobulin variable gene diversification from HR to non-templated single base substitutions (5). Similar to yeast rad51 paralog mutants (8,9), these defects were suppressed by overproduction of Rad51 (5–7). Thus, we previously hypothesized that in vivo all Rad51 paralogs work collaboratively in an early step of HR, facilitating the assembly of Rad51 protein at damaged DNA (7). Rodent xrcc2 and xrcc3 mutants display a similar hypersensitivity to cross-linking agents and mild sensitivity to IR (10–12), indicating that Rad51 paralogs play an important role for recombination reactions triggered by arrested DNA replication.
Biochemical analysis of paralog proteins challenged this simple interpretation of the phenotypic data. Interaction studies suggested that human Rad51 paralogs form several different complexes in the cells, i.e. Rad51B–Rad51C–Rad51D–XRCC2 (hereafter called BCDX2 complex), Rad51B–Rad51C (BC sub-complex), Rad51D–XRCC2 (DX2 sub-complex) and Rad51C–XRCC3 (CX3 complex) (13–21). The BC and DX2 sub-complexes bound to single-stranded DNA and double-stranded DNA, and hydrolyzed ATP (16,18,22), and BC sub-complex supports the strand-exchange reaction mediated by the Rad51 and RPA proteins (16), suggesting an early role for both complexes in recombination. In addition, recent biochemical studies have suggested that some of the Rad51 paralogs participate in branch migration and resolution of Holliday junction (HJ) recombination intermediates. For example, DX2 stimulates the disruption of HJ by the Blm RecQ DNA helicase (23), RAD51B binds to HJ in vitro (24,25), and RAD51C and XRCC3 are associated with HJ processing (26). These data raise the question, whether or not some of the Rad51 paralogs can contribute to HR independently of the other paralogs, acting at different points during the HR reaction.
In this study, we analyze xrcc3/rad51d DT40 cells to ask from a genetic perspective, whether the BCDX2 and CX3 complexes possess unique functions. Similarly, we generated rad51b/rad51d cells to study the complementary relationship between BC and DX2 sub-complexes in the BCDX2 complex. Using these double mutants as well as each single gene disrupted clones, we studied cellular response to IR-induced double-strand breaks (DSBs), as well as DSBs that arise as a consequence of replication fork damage by either cisplatin (CDDP), a cross-linking agent [reviewed in (27)], or camptothecin (CPT), a topoisomerase I inhibitor (28,29). By analyzing the CDDP, and CPT sensitivity of the various paralog single and double mutants, we found genetic evidence for the existence of the BCDX2 and CX3 complexes; rad51b/rad51d double mutants were epistatic, while xrcc3/rad51d double mutant cells showed an additive sensitivity when compared to the single mutant cells. On the other hand, we also precisely examined the kinetics of Rad51 and Rad54 foci formation following DNA damage in each genotype. In this assay, all mutant cells showed very comparable defects in foci formation. Thus, besides more complex functions associated with replication dependent damage, all Rad51 paralogs appear to operate collaboratively to facilitate the assembly of Rad51 at DSB site. Taken together, the data presented here shed light on a complex interplay among Rad51 paralogs, where each Rad51 paralog has distinct as well as overlapping roles, depending on the type of DNA damage and stage of HR.
MATERIALS AND METHODS
Generation of gene disrupted cells
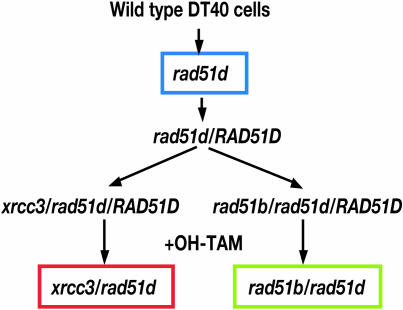
As a starting material for the creation of double mutant cell lines, we used rad51d cells expressing a mouse RAD51D cDNA together with the Cre-loxP system (30,31) (resulting cells are hereafter called rad51d/RAD51D cells) (Figure 1). This approach was necessary because of the great reduction in gene-targeting frequencies in Rad51 paralog mutants. The rad51d-deficient clones were transfected with pCR3-loxP-mRAD51D/IRES-EGFP-loxP expression vector together with pANMerCreMer-neo (30,31), followed by selection with G418 (2 mg/ml). Among stable transfected cells, clones that expressed enhanced green fluorescent protein (EGFP) were identified by FACScaliber (Becton Dickinson, Mountain View, CA) and isolated (rad51d/RAD51D clones). We exposed these clones to 4-hydroxy-tamoxifen (OH-TAM) to delete the mouse RAD51D transgene. The lack of functional Rad51D was confirmed by measuring CDDP sensitivity in OH-TAM-treated cell populations from rad51d/RAD51D clones. Two rad51d/RAD51D clones were transfected with gene disruption constructs to obtain rad51b/rad51d/RAD51D and xrcc3/rad51d/RAD51D clones. After 3 days treatment with OH-TAM, we isolated rad51b/rad51d and xrcc3/rad51d mutant clones. In this experiment, the deletion of the RAD51D transgene was identified by the absence of green fluorescent protein expression, and confirmed by genome PCR and RT–PCR (Supplementary Material). Both rad51b/rad51d and xrcc3/rad51d cells had growth properties similar to that of rad51d single mutants (Supplementary Figure S1D and Supplementary Table S1). Cell lines rad51b/rad51d/RAD51B and xrcc3/rad51d/XRCC3 were obtained by reconstituting rad51b/rad51d and xrcc3/rad51d cells with human RAD51B and XRCC3 expression vectors, respectively.
Figure 1.
Experimental strategy. For the creation of double mutant cell lines, we used rad51d cells expressing a mouse RAD51D cDNA together with the Cre-loxP system (30,31) (rad51d/RAD51D cells). Two rad51d/RAD51D clones were transfected with gene disruption constructs to obtain rad51b/rad51d/RAD51D and xrcc3/rad51d/RAD51D clones. After 3 days treatment of OH-TAM, we isolated rad51b/rad51d and xrcc3/rad51d mutant clones.
Genomic DNA was prepared from rad51b/rad51d/RAD51D and xrcc3/rad51d/RAD51D clones 3 days after the addition of OH-TAM. PCR was performed with the upstream CMV primer, 5′-CACTGCTTACTGGCTTATCG-3′ and the downstream 51D primer, 5′-TCTGCTGACCTCCCAGAAGT-3′ or the SP6 5′-TTTAGGTGACACTATAGAATAG-3′ primer, indicated in Supplementary Figure S1A. For the RT–PCR, total RNA was extracted using Sepazol (Nacalai Tesuque, Kyoto, Japan) following the manufacturer's instructions. A first strand cDNA was prepared using SuperScript First-Strand Synthesis System (Invitrogen). PCR was performed with the following primers, 5′-ATGGGCATGCTCAGGGCAGGGCTGTGCCCG-3′ and 5′-TCTGCTGACCTCC CAGAAGT-3′ for RAD51D transcripts; 5′-GATGATGATATTGCTGCGCTCGTTGTTGAC-3′ and 5′-GATTCATCGTACTCCTGCTTGCTGATCCAC-3′ for beta-actin transcripts (Supplementary Figure S1C).
Recombinant plasmid construction
Two XRCC3 disruption constructs, XRCC3-hygro and XRCC3-puro/loxP, were generated from genomic PCR products (7). To improve the targeting efficiency of previous RAD51B disruption construct (6), we constructed new RAD51B disruption constructs, RAD51B-hygro and RAD51B-puro/loxP, in which both the left (upstream) and right (downstream) arms were replaced. A new left arm is a 7 kb fragment generated by PCR amplification with the following primers, 5′-CGGGGTACCATTCCAGGGAATCTCTGCTAC-3′ (KpnI site denoted by underline) and 5′-CGGGGTACCCTAGTCCTCCCCATGCTTACGG-3′, while a new right arm (5 kb) is PCR amplified with the 5′-GACCCGTAGCATTCATCTG TATCGAG-3′ and 5′-CTGCGTACAGTGTTGTTACATACAGCAACG-3′ primers. The right arm was cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsband, CA), and the resulting recombinant plasmid was inserted with selection marker cassettes flanked by the BamHI sites, followed by insertion with the KpnI fragment containing the left arm.
We constructed a mouse RAD51D expression vector, pCR3-loxP-mRAD51D/IRES/EGFP-loxP, in which both a mouse RAD51D cDNA and the EGFP genes are flanked by the loxP sequences, by inserting the cDNA into the EcoRI–BamHI sites of pCR3-loxP-MCS/IRES-EGFP-loxP (Supplementary Figure S1A) (32). The human XRCC3 and human RAD51B cDNAs were separately cloned into pCR3-loxP-MCS/IRES-EGFP-loxP as well. These expression plasmids were transfected into rad51b/rad51d and xrcc3/rad51d cells together with a marker plasmid, pBSIIKS(+)-puro.
Cell culture and DNA transfection
Wild-type and mutant DT40 cells were maintained in RPMI-1640 medium (Sigma, St Louis, MO) supplemented with 100 μM beta-mercaptoethanol, 10% fetal calf serum (FCS) and 1% chicken serum at 39.5°C. DNA transfection and selection were performed as described previously (33).
Flow cytometric analysis
To measure the growth kinetics, cells were counted daily using flow cytometric analysis in comparison with a fixed number of 25 μm microspheres (Polyscience Inc., Warrington, PA). Cells were split each day to keep them ∼105/ml. To determine the proportion of dead cells, we added 5 μg/ml of propidium iodide to cultured cells, and immediately analyzed by FACScalibur as described previously (34).
Measurement of sensitivity of cells to killing by gamma-rays, cisplatin and camptothecin
Colonogenic survival was monitored by colony formation assay, as described previously (34). To measure sensitivities to cisplatin (Nihon-Kayaku, Tokyo, Japan) and camptothecin (Topogene, Columbus, OH), cells were incubated at 39.5°C in the complete medium containing the compound for 1 h. Following the genotoxic treatments, appropriate numbers of cells were plated into six-well cluster plates containing the complete medium supplemented with 1.5% methylcellulose (Aldrich, Milwaukee, WI). Colony numbers were counted at 7–14 days, and the percent survival was determined, as those relative to the number of colonies of untreated cells. To measure IR sensitivity, serially diluted cells were plated in the medium containing methylcellulose, irradiated with a 137Cs γ-ray source, and then incubated.
Chromosome analysis
To see the response to CDDP and CPT, the cells were treated with CDDP for 1 h at a concentration of 5 μM and washed by phosphate-buffered saline twice. The cells treated with CDDP were then harvested 9 h after treatment for making air-drying chromosome preparations after treatment with 0.1 μg/ml colcemid for the final 2 h. The cells treated with CPT for 9 h at a concentration of 10 nM were harvested in the same manner. The samples were prepared and then stained with conventional Giemsa staining method for CA analysis. A minimum of 100 cells were examined for CAs. The frequencies were analyzed only in the 12 macrochromosomes including one Z chromosome (3).
Immunofluorescent visualization of nuclear foci
Cells were harvested at various time points after 137Cs irradiation (4Gy) or exposure to CDDP (15 μM) for 1 h. Cytospin slides were prepared using Cytospin3 (Shandon, Pittsburgh, PA). Staining and visualization of Rad51 or Rad54 foci were performed as described previously using the anti-Rad51 rabbit polyclonal antibody (EMD Biosciences, Darmstadt, Germany) (35) or Rad54 anti-sera (a gift from Dr C. Morrison, Galway, Ireland) (36). Only brightly fluorescent foci were counted as positive focus formation. At least 50 morphologically intact cells were examined at each time point.
Statistical analysis
The statistical analyses were carried out by using StatView software (SAS Institute, Inc., Cary, NC). A Student's t-test was used to analyze differences between clones. The differences were considered statistically significant at 95% confidence limit (P < 0.05).
RESULTS
Generation of rad51b/rad51d and xrcc3/rad51d cells
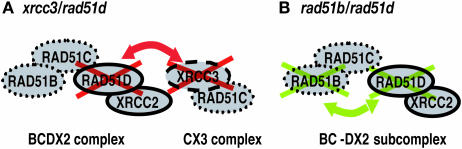
In this study, we undertook a genetic approach to clarify the previously described interaction between different Rad51 paralogs. Figure 2 presents the rationale for our genetic experiments. We aimed to target either both predicted complexes BCDX2 and CX3 (Figure 2A), or target proteins within the same complex, albeit belonging to different sub-complexes BC and DX2 (Figure 2B) (33,34). We disrupted the RAD51B or XRCC3 gene in rad51d/RAD51D cells, and later deleted the RAD51D transgene by adding OH-TAM to obtain rad51b/rad51d and xrcc3/rad51d cells (Figure 1). The lack of Rad51D expression was confirmed by RT–PCR and genome PCR as described previously (Supplementary Figure S1) (32).
Figure 2.
The reasoning of generating xrcc3/rad51d and rad51b/rad51d cells. (A) Deletion of Rad51D and XRCC3 may disrupt both BCDX2 and CX3 complexes. On the other hand, (B) deletion of RAD51B and RAD51D may disrupt both BC and DX2 sub-complexes as well as their physical interactions.
Rad51 paralogs contribute equally to the repair of IR-induced DSBs
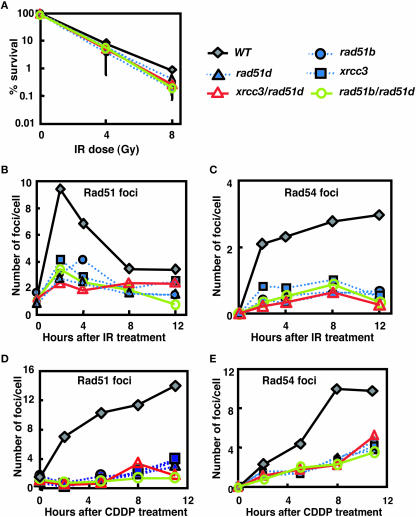
We investigated DNA repair capacity in the various single and double mutants by colony-survival assays. First, we re-evaluated the IR sensitivity of rad51 paralog single mutants, and also rad51b/rad51d and xrcc3/rad51d cells. The single and double mutants showed a very similar sensitivity to IR as judged by colony survival (Figure 3A and Supplementary Table S2). To gain insight into the molecular basis for the role of individual Rad51 paralogs in HR, we measured the kinetics of Rad51 and Rad54 foci formation following IR and CDDP treatments (36–42). Foci formation was severely compromised in all mutants tested, and a quantitative analysis over time showed no differences among the single and double knockout cells (Figure 3B–E and Supplementary Tables S3–S6). This finding supports our previous conclusion that all five Rad51 paralogs act collaboratively, each playing an indispensable role in the promotion of Rad51 assembly at DNA damage (6,7).
Figure 3.
(A) Percent colony survival assay after treatment with γ-irradiation. Comparison of wild-type cells with rad51b, rad51d, xrcc3, rad51b/rad51d and xrcc3/rad51d cells. The data shown are mean ± SD of at least three separate experiments. Two independently targeted clones of each genotype consistently showed the same sensitivity to genotoxic agents (data not shown). The kinetics of Rad51 and Rad54 foci formation after genotoxic treatments (B–E). The number of Rad51 foci per cell is displayed at the time point of 0, 2, 4, 8 and 12 h after (B) IR (4Gy) and 0, 2, 5, 8 and 11 h after (D) CDDP (15 μM, 1 h). Similarly, the number of Rad54 foci per cell is shown after exposure to (C) IR and (E) CDDP. Each result represents data of scoring at least 50 cells. Symbol of each mutant: WT (gray filled diamond, solid line), rad51b (blue filled circle, dotted line), rad51d (blue filled triangle, dotted line), xrcc3 (blue filled square, dotted line), rad51b/rad51d (green open circle, solid line) and xrcc3/rad51d (red open triangle, solid line).
Differential contribution of Rad51 paralogs to the repair of DNA damage associated with replication
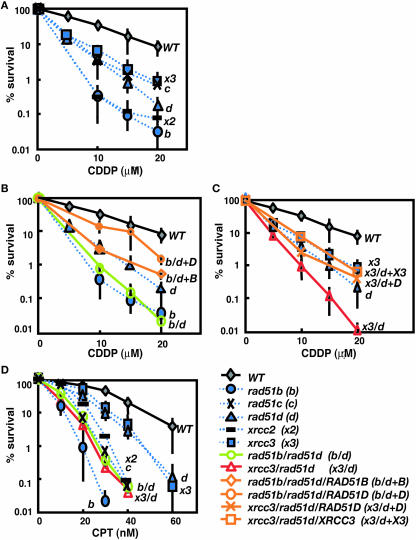
In contrast with the cellular response to IR, rad51b and xrcc2 mutants displayed higher sensitivity to CDDP than rad51c, rad51d and xrcc3 cells (Figure 4A and Supplementary Table S7). The more prominent phenotype of rad51b, rad51c and xrcc2 cells was also found in cellular tolerance to CPT (Figure 4D and Supplementary Table S8). These data suggest that Rad51B and Xrcc2 may play a more important role than the other Rad51 paralogs in processing DNA damage associated with replication fork block. Moreover, the relative sensitivity of rad51c mutant was slightly different depending on the type of DNA damaging agents. rad51c cells showed only moderate sensitivity to CDDP comparable with rad51d and xrcc3 cells (Figure 4A and Supplementary Table S7), while they were highly sensitive to CPT, comparable with rad51b and xrcc2 cells (Figure 4D and Supplementary Table S8). These observations imply complex functional interactions between different Rad51 paralogs.
Figure 4.
(A–C) Colony-survival assay after treatment with cisplatin (CDDP) and (D) camptothecin (CPT). Single gene disrupted clones are compared with the wild-type cells in the (A) CDDP sensitivity and (D) sensitivity to CPT. The CDDP sensitivity of (B) rad51b/rad51d and (C) xrcc3/rad51d cells is compared with the relevant single mutants. (D) The CPT sensitivity of the double mutants is compared with single mutants and wild-type cells. The data shown are mean ± SD of at least three separate experiments. Symbols are same as shown in Figure 3. Six symbols are added shown as rad51c (black cross, dotted line), xrcc2 (black bar, dotted line), rad51b/rad51d/RAD51B (orange open diamond, solid line), rad51b/rad51d/RAD51D (orange open circle, solid line), xrcc3/rad51d/RAD51D (orange cross, solid line) and xrcc3/rad51d/XRCC3 (orange open square, solid line).
We next addressed whether or not the BC and/or DX2 sub-complexes have distinct functions, which might have been masked in our previous study due to the functional redundancy between the two sub-complexes. To investigate such functional overlap, we compared the rad51b/rad51d phenotype with that of rad51b and rad51d single mutants. The double mutants displayed the same level of CDDP sensitivity as rad51b cells (Figure 4B and Supplementary Table S7). Furthermore, reconstitution of the rad51b/rad51d cells with a RAD51B transgene (rad51b/rad51d/RAD51B in Figure 4B and Supplementary Table S7) reversed their phenotype to the level of rad51d cells. Similar pattern of sensitivity was observed following treatment with CPT (Figure 4D and Supplementary Table S8). These findings do not support a specific function associated with BC or DX2 sub-complexes.
Additive increase in cisplatin- and camptothecin-sensitivity in rad51d and xrcc3 double mutants
To evaluate possible functional overlap between the BCDX2 and CX3 complexes, we measured sensitivity to killing by CDDP and CPT in xrcc3/rad51d cells. Interestingly, the double mutants showed an additive sensitivity to both agents (Figure 4C and D, Supplementary Tables S7 and 8). Expression of XRCC3 cDNA in xrcc3/rad51d cells (xrcc3/rad51d/XRCC3) reversed this phenotype to the level of rad51d cells. Similarly, xrcc3/rad51d/RAD51D cells showed the same sensitivity to CDDP as xrcc3 cells (Figure 4C and Supplementary Table S3). These experiments confirm the additive effect of rad51d and xrcc3 defects. A simple interpretation of this result is that Rad51D and XRCC3 dependent pathways may each act independently of each other and differentially enhance cellular survival following CDDP or CPT treatment. The additive effect of rad51d and xrcc3 is in agreement with the notion that BCDX2 and CX3 complexes are involved in different steps of HR. However, the relatively mild sensitivity of rad51c cells and the highly reduced survival rates of rad51b cells point to a more complex picture (Figure 4A and Supplementary Table S7), in which each individual Rad51 paralog might contribute differently to replication stress and DNA cross-links.
Increased breaks in both sister chromatids point to a role of Rad51 paralogs at later stages of HR
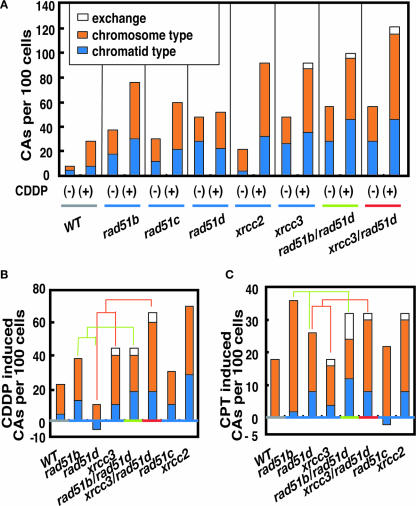
As shown before in Figure 3D and E, all different paralog mutants showed comparable defects in Rad51/54 foci formation induced by CDDP. Thus, the differential responses of different mutants may be caused by a defect at later stages of HR after the assembly of Rad51. To assess such late steps of HR, we measured CA induced by CDDP and CPT in the mutant cells (Figure 5 and Supplementary Tables S9 and 10). CDDP and CPT block the replication fork leading to DSBs (27,29). Since HR-mediated repair of the DSB is carried out by interactions between damaged and intact sister chromatids, failure at later stages of HR after strand invasion may result in breakage of the entire recombination structure involving both sister chromatids during separation of two sisters associated with chromosome condensation. This could ultimately lead to the appearance of chromosome type breaks, where two sister chromatids are broken at the same site (43). Remarkably, CDDP and CPT indeed induced substantial fractions of chromosome type breaks (Figure 5). Thus, Rad51 paralogs may also have a role in a late step of HR.
Figure 5.
(A) Total number of CAs per 100 cells before (−) and after (+) CDDP treatment (5 μM, 1 h). (B) CDDP-induced and (C) CPT-induced (10 nM, 9 h) CAs are determined by subtracting spontaneously occurring CAs from the number of induced ones.
Figure 5A shows that rad51 paralog mutants exhibit marked variations in the level of CAs induced by CDDP. Reproductive cell death is thought to ensue when DSBs remain unrepaired, or when they are misrepaired. Indeed, there is evidence in yeast that one unrepaired DNA DSB constitutes a lethal event (44,45). However, it is paradoxical that the level of unrepaired chromosomal breaks following CDDP treatment did not correlate with colony survival in the different mutants. For example, rad51d cells were almost as sensitive to CDDP as xrcc3 cells (Figure 4A), but had significantly lower levels of induced CAs (Figure 5B). Conceivably, the type of unrepaired damage in rad51d cells might differ from that of the other rad51 paralog mutants, and a majority of rad51d cells might die prior to the entry to the M phase. rad51b/rad51d double and rad51b single mutants, on the other hand, had higher levels of induced CAs than the rad51d single mutant in accordance with their increased sensitivity (Figure 5B). Similarly, both CAs and sensitivity were increased in xrcc3/rad51d cells when compared to each single mutant (Figure 5B). Moreover, CPT-induced CA followed a similar pattern as the sensitivity to CPT in each tested mutant (compare Figure 4D with Figure 5C). The chromosome breaks as well as variations in the level of induced CAs reveal a complex interplay of Rad51 paralog protein function in replication-linked DNA repair, where each Rad51 paralog may have individual as well as collaborative roles at early and late stages of HR.
DISCUSSION
All Rad51 paralogs act in concert during Rad51 accumulation at sites of DNA damage, but have individual roles in later steps of HR
In this study, we have set out to find genetic evidence for the biochemical model of Rad51 paralog interactions. Our genetic analysis, which directly addresses the function of each paralog in vivo, provides evidence for two different scenarios concerning Rad51 paralog function. In the case of DSBs induced by IR, all Rad51 paralogs seem to act in concert to allow the accumulation of Rad51 and Rad54 at the site of damage. Interestingly, paralog mutants are not very sensitive to IR, although Rad51 foci formation was substantially impaired after IR-induced DNA damage. There are two possible explanations. First, in the absence of Rad51 paralogs, Rad51 accumulation may occur at a reduced rate helped by other factors such as BRCA2, resulting in suboptimal HR reactions. However, this Rad51 accumulation may be too low to be detected by immunofluorescence microscopy. Second, another pathway, such as NHEJ, may play a major role in the repair of IR-induced DSBs (46), but not in the CPT (47) and CDDP (48) repair pathway. Similarly, all paralog mutants equally suffer from a reduction in Rad51/54 foci induced by CDDP. Thus, accumulation of Rad51 at DSBs appears to reflect paralog function that requires each individual paralog to a similar extent and does not necessarily involve different complexes of these proteins. In contrast with the concerted action of all Rad51 paralogs in an early step of HR, a comparison of CDDP and CPT sensitivities with IR sensitivity of different mutants reveals a more prominent and complex role of the paralogs in HR. These agents caused significantly different responses in individual paralog mutants as judged by the sensitivity assays, while displaying a very similar defect as judged by the foci formation assay. Taken together, these results point to a dual function of the paralogs during HR in connection with replication stress. Clearly, Rad51 paralogs play the same role in the early step of HR, as evidenced by the foci formation assay following exposure to IR and CDDP (Figure 3B–E). However, the sensitivity among these mutants to CDDP and CPT varies by an order of magnitude with rad51b and xrcc2 cells displaying the lowest levels of survival. Interestingly, there are also differences between rad51 paralog mutants in the responses to either of these agents. For example, Rad51C seems to play a more prominent role in the repair of CPT induced lesions than Rad51D and XRCC3 while rad51c, rad51d and xrcc3 cells seem to have similar sensitivity to CDDP. These data point to other more complex paralog functions in DNA repair that do not involve Rad51 accumulation, but other, possibly later steps of DNA repair. Arguably, the different phenotypes of the paralogs could derive from subtle differences in the ability to promote Rad51 accumulation that cannot be detected by the foci formation assay. However, our finding of increased levels of chromosome type breaks following CDDP and CPT treatment is a more direct evidence that Rad51 paralogs act at a stage when the sister chromatids are already entangled during a later stage of HR. These findings are in accord with other studies that suggest HJ binding and resolvase activity for Rad51 paralog proteins (23–26). We conclude that besides their common early role the paralogs have acquired novel functions at later stages of replication dependent repair induced by CDDP and CPT. They are likely to involve the HJ processing activity associated with RAD51C/XRCC3 (26). The comparatively high sensitivity of rad51b mutant, which does not appear to possess an in vitro HJ processing activity (24,25), also points to the existence of novel yet unknown activities for individual paralogs. Understanding the precise mechanism of Rad51 paralog function could thus shed light on special aspects of HR that is associated with DNA replication; a branch of DNA repair that we are only beginning to understand.
Genetic versus biochemical evidence for Rad51 paralog complexes
One of the aims of this study was to reinvestigate by genetic means the interactions between Rad51 paralogs that were proposed by biochemical methods (14–21,23). Our starting hypothesis was that partners in the same complex should show an epistatic interaction, while double mutants spanning different complexes should interact either additively or synergistically depending on the functional interaction of the two complexes. Our analysis of rad51b/rad51d and xrcc3/rad51d cells clearly supports the findings that the paralogs fall in the two biochemically defined interaction groups BCDX2 and CX3 (14–21,23). Thus, the rad51b/rad51d mutation was no more CDDP and CPT sensitive than rad51b single mutants, while the xrcc3/rad51d mutation led to an additive increase in the sensitivity to the same reagents. However, given the complex differences that our analysis of the single and double mutants reveals, this finding needs to be interpreted very carefully. First, if these complexes do indeed operate collaboratively in the same pathway, their specific activity does not involve the accumulation of Rad51 early in the HR reaction, but is restricted to the other functions probably at later steps. Second, the differences between each individual paralog in their CDDP and CPT sensitivities suggest that different paralogs contribute differentially within these complexes and are dependent on the type of damage. For example, Rad51C is present in both complexes but its deletion confers only a relatively weak sensitivity to CDDP. Hence, it might play only a minor role in the repair of damage induced by CDDP. Third, our data indicate that some of the paralogs might exert individual functions independently of the complexes. Thus, even a deletion of both RAD51D and XRCC3 does not result in a higher sensitivity to CPT and CDDP, when compared with rad51b and xrcc2 mutant cells. To elucidate the precise contribution of each Rad51 paralog to HR, more detailed phenotypic assays to characterize each step of the whole HR-mediated repair are necessary.
In summary, the present study reveals that Rad51 paralogs concerted and individual function differ depending on the type of DNA damage and on the stage of HR. The five molecules appear to operate together in promoting the assembly of Rad51 at DNA damage, while some of the paralog molecules have distinct roles perhaps in a later stage of HR dealing with lesions arising at stalled replication forks.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank M. Nakaoka, Y. Sato and T. Ogawa for technical assistance. Financial support was provided in part by CREST, Japan Science and Technology (Saitama, Japan); and the Center of Excellence (COE) grant for Scientific Research from the Ministry of Education, Culture, Sports and Technology; and by grants from The Uehara Memorial Foundation and The Naito Foundation. This work was also funded in part by grants from the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim. Funding to pay the Open Access publication charges for this article was provided by grants from the Ministry of Education, Culture, Sports and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lim D.S., Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonoda E., Sasaki M.S., Buerstedde J.-M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S. Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 5.Hatanaka A., Yamazoe M., Sale J.E., Takata M., Yamamoto K., Kitao H., Sonoda E., Kikuchi K., Yonetani Y., Takeda S. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol. Cell. Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takata M., Sasaki M.S., Sonoda E., Fukushima T., Morrison C., Albala J.S., Swagemakers S.M., Kanaar R., Thompson L.H., Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hays S.L., Firmenich A.A., Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson R.D., Symington L.S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N., Lamerdin J.E., Tebbs R.S., Schild D., Tucker J.D., Shen M.R., Brookman K.W., Siciliano M.J., Walter C.A., Fan W., et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 11.Caldecott K., Jeggo P. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
- 12.Jones N.J., Cox R., Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 13.Braybrooke J.P., Spink K.G., Thacker J., Hickson I.D. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- 14.Masson J.Y., Stasiak A.Z., Stasiak A., Benson F.E., West S.C. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA. 2001;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masson J.Y., Tarsounas M.C., Stasiak A.Z., Stasiak A., Shah R., McIlwraith M.J., Benson F.E., West S.C. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J.S., Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurumizaka H., Ikawa S., Nakada M., Eda K., Kagawa W., Takata M., Takeda S., Yokoyama S., Shibata T. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl Acad. Sci. USA. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurumizaka H., Ikawa S., Nakada M., Enomoto R., Kagawa W., Kinebuchi T., Yamazoe M., Yokoyama S., Shibata T. Homologous pairing and ring and filament structure formation activities of the human Xrcc2/Rad51D complex. J. Biol. Chem. 2002;277:14315–14320. doi: 10.1074/jbc.M105719200. [DOI] [PubMed] [Google Scholar]
- 19.Liu N., Schild D., Thelen M.P., Thompson L.H. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller K.A., Yoshikawa D.M., McConnell I.R., Clark R., Schild D., Albala J.S. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem. 2002;277:8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- 21.Wiese C., Collins D.W., Albala J.S., Thompson L.H., Kronenberg A., Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lio Y.C., Mazin A.V., Kowalczykowski S.C., Chen D.J. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 23.Braybrooke J.P., Li J.L., Wu L., Caple F., Benson F.E., Hickson I.D. Functional interaction between the Bloom's syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D) J. Biol. Chem. 2003;278:48357–48366. doi: 10.1074/jbc.M308838200. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama H., Kurumizaka H., Ikawa S., Yokoyama S., Shibata T. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama H., Sarai N., Kagawa W., Enomoto R., Shibata T., Kurumizaka H., Yokoyama S. Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 2004;32:2556–2565. doi: 10.1093/nar/gkh578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Masson J.Y., Shah R., O'Regan P., West S.C. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 27.Dronkert M.L., Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 28.Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 29.Li T.K., Liu L.F. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Riesterer C., Ayrall A.M., Sablitzky F., Littlewood T.D., Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Wienands J., Zurn C., Reth M. Induction of the antigen receptor expression on B lymphocytes results in rapid competence for signaling of SLP-65 and Syk. EMBO J. 1998;17:7304–7310. doi: 10.1093/emboj/17.24.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimori A., Tachiiri S., Sonoda E., Thompson L.H., Dhar P.K., Hiraoka M., Takeda S., Zhang Y., Reth M., Takata M. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buerstedde J.M., Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 34.Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi-Iwai Y., Sonoda E., Buerstedde J.M., Bezzubova O., Morrison C., Takata M., Shinohara A., Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison C., Sonoda E., Takao N., Shinohara A., Yamamoto K., Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage [Erratum (2000) EMBO J., 19, 786] EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ristic D., Wyman C., Paulusma C., Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petukhova G., Van Komen S., Vergano S., Klein H., Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 39.Sigurdsson S., Trujillo K., Song B., Stratton S., Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2000;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 40.Solinger J.A., Lutz G., Sugiyama T., Kowalczykowski S.C., Heyer W.D. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- 41.Solinger J.A., Heyer W.D. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl Acad. Sci. USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexiadis V., Kadonaga J.T. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.French C.A., Masson J.Y., Griffin C.S., O'Regan P., West S.C., Thacker J. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 2002;277:19322–19330. doi: 10.1074/jbc.M201402200. [DOI] [PubMed] [Google Scholar]
- 44.Game J.C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin. Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 45.Resnick M.A. The repair of double-strand breaks in DNA: a model involving recombination. J. Theor. Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Zeng Z.C., Bui T.A., Sonoda E., Takata M., Takeda S., Iliakis G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene. 2001;20:2212–2224. doi: 10.1038/sj.onc.1204350. [DOI] [PubMed] [Google Scholar]
- 47.Adachi N., So S., Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004;279:37343–37348. doi: 10.1074/jbc.M313910200. [DOI] [PubMed] [Google Scholar]
- 48.Diggle C.P., Bentley J., Knowles M.A., Kiltie A.E. Inhibition of double-strand break non-homologous end-joining by cisplatin adducts in human cell extracts. Nucleic Acids Res. 2005;33:2531–2539. doi: 10.1093/nar/gki528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.