Abstract
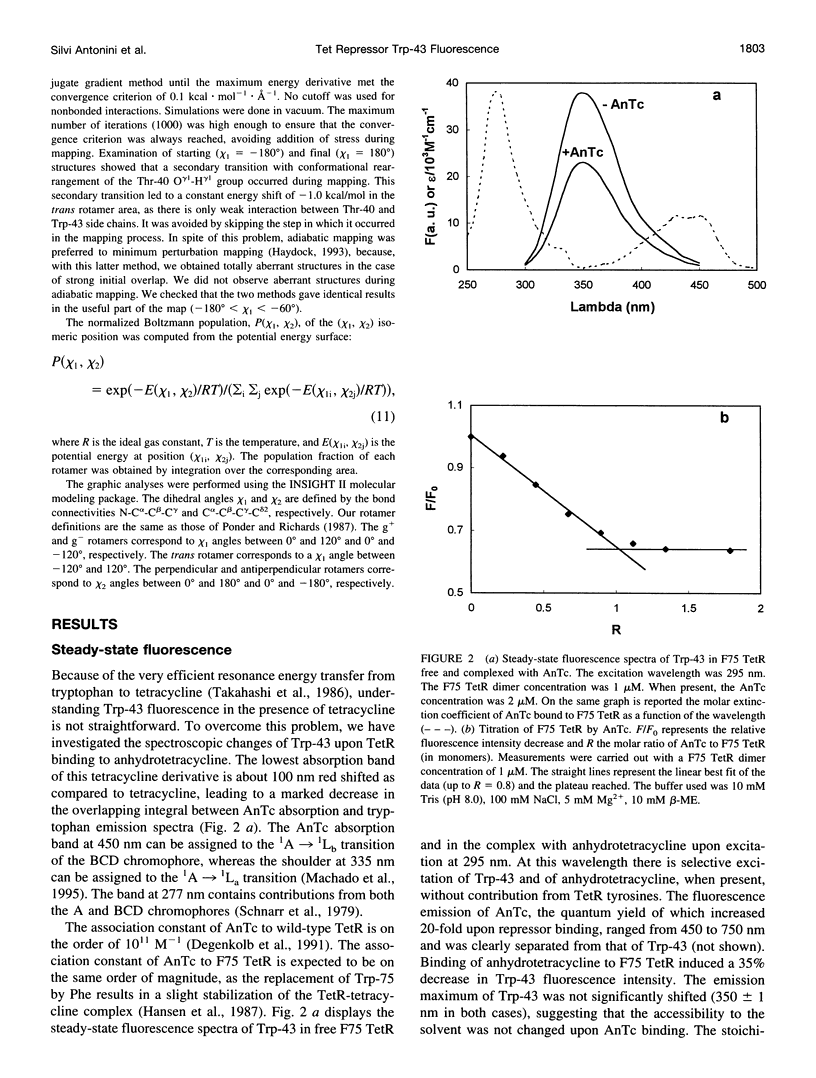
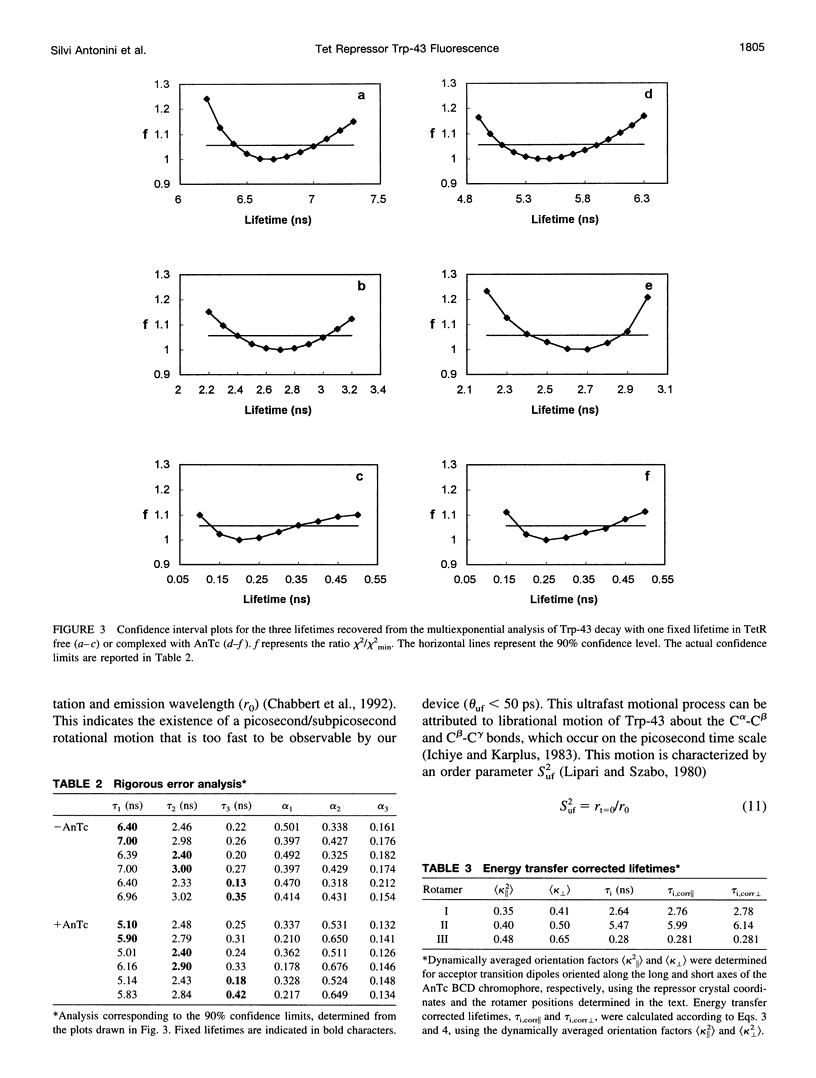
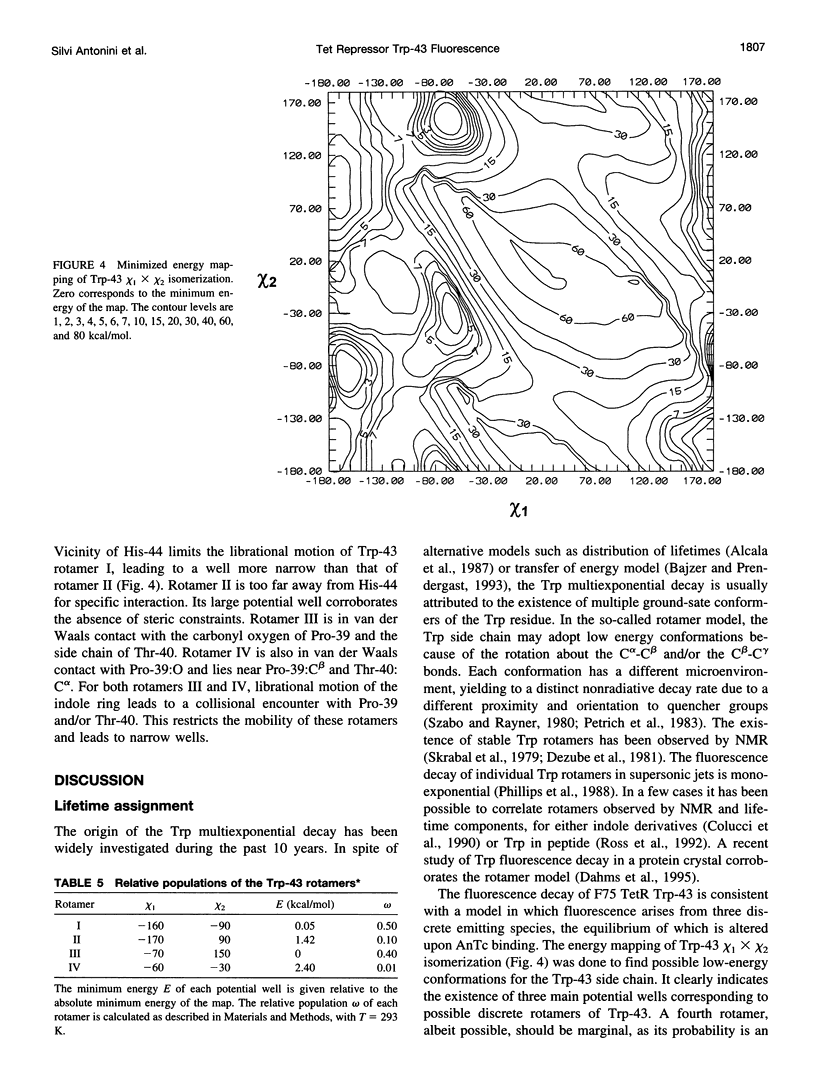
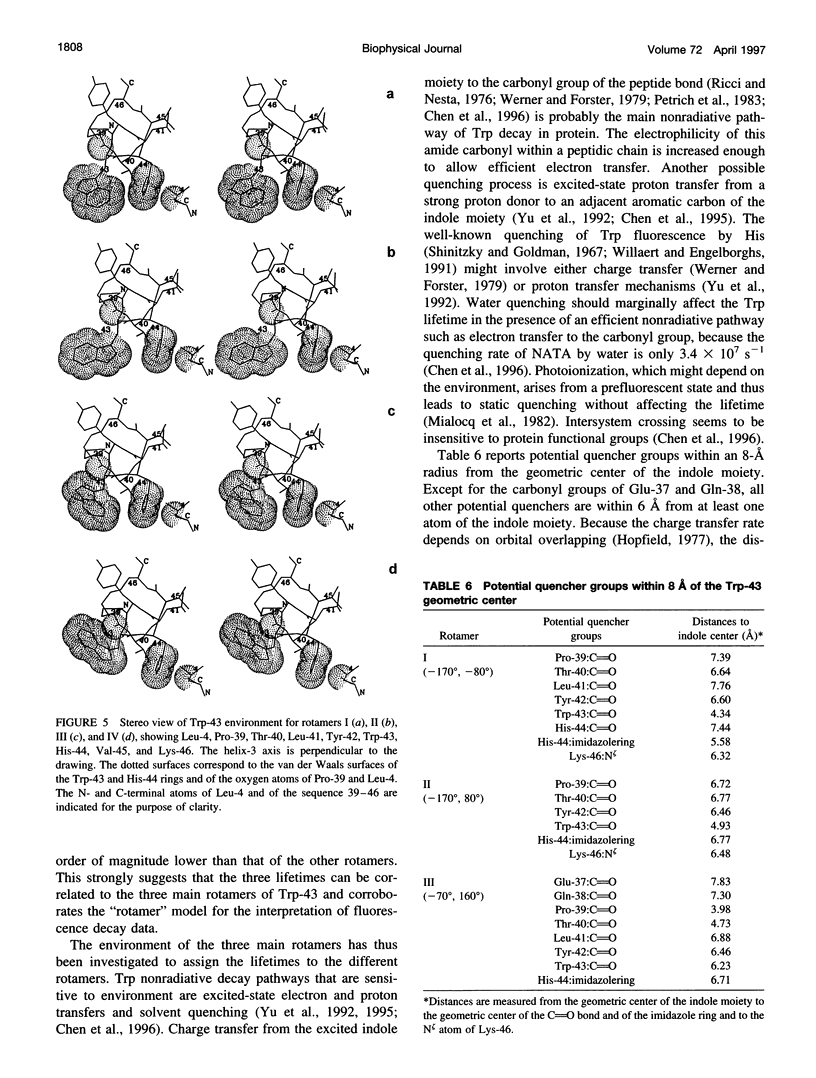
A 35% decrease in the fluorescence intensity of F75 TetR Trp-43 was observed upon binding of the tetracycline derivative 5a,6-anhydrotetracycline (AnTc) to the repressor. The fluorescence decay of Trp-43 in F75 TetR and in its complex with AnTc could be described by the sum of three exponential components, with lifetimes of about 6, 3, and 0.3 ns. The amplitudes, however, were markedly altered upon binding. The minimized energy mapping of Trp-43 chi 1 x chi 2 isomerization clearly indicated the existence of three main potential wells at positions (-160 degrees, -90 degrees) (rotamer I), (-170 degrees, 90 degrees) (rotamer II), and (-70, 150 degrees) (rotamer III). Our study of Trp-43 environment for each of the three rotamers suggests that the longest decay component may be assigned to rotamer II, the middle-lived component to rotamer I, and the subnanosecond component to rotamer III. The origin of the changes in the rotamer distribution upon AnTc binding is discussed. Anisotropy decays are also discussed within the framework of the rotamer model.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcala J. R., Gratton E., Prendergast F. G. Interpretation of fluorescence decays in proteins using continuous lifetime distributions. Biophys J. 1987 Jun;51(6):925–936. doi: 10.1016/S0006-3495(87)83420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrescu A. T., Shortle D. Backbone dynamics of a highly disordered 131 residue fragment of staphylococcal nuclease. J Mol Biol. 1994 Sep 30;242(4):527–546. doi: 10.1006/jmbi.1994.1598. [DOI] [PubMed] [Google Scholar]
- Armstrong K. M., Fairman R., Baldwin R. L. The (i, i + 4) Phe-His interaction studied in an alanine-based alpha-helix. J Mol Biol. 1993 Mar 5;230(1):284–291. doi: 10.1006/jmbi.1993.1142. [DOI] [PubMed] [Google Scholar]
- Bajzer Z., Prendergast F. G. A model for multiexponential tryptophan fluorescence intensity decay in proteins. Biophys J. 1993 Dec;65(6):2313–2323. doi: 10.1016/S0006-3495(93)81325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Beechem J. M. Global analysis of biochemical and biophysical data. Methods Enzymol. 1992;210:37–54. doi: 10.1016/0076-6879(92)10004-w. [DOI] [PubMed] [Google Scholar]
- Berman H. A., Yguerabide J., Taylor P. Fluorescence energy transfer on acetylcholinesterase: spatial relationship between peripheral site and active center. Biochemistry. 1980 May 13;19(10):2226–2235. doi: 10.1021/bi00551a036. [DOI] [PubMed] [Google Scholar]
- Chabbert M., Hillen W., Hansen D., Takahashi M., Bousquet J. A. Structural analysis of the operator binding domain of Tn10-encoded Tet repressor: a time-resolved fluorescence and anisotropy study. Biochemistry. 1992 Feb 25;31(7):1951–1960. doi: 10.1021/bi00122a008. [DOI] [PubMed] [Google Scholar]
- Chen C., Feng Y., Short J. H., Wand A. J. The main chain dynamics of a peptide bound to calmodulin. Arch Biochem Biophys. 1993 Nov 1;306(2):510–514. doi: 10.1006/abbi.1993.1544. [DOI] [PubMed] [Google Scholar]
- Chen R. F., Knutson J. R., Ziffer H., Porter D. Fluorescence of tryptophan dipeptides: correlations with the rotamer model. Biochemistry. 1991 May 28;30(21):5184–5195. doi: 10.1021/bi00235a011. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Driscoll P. C., Wingfield P. T., Gronenborn A. M. Analysis of the backbone dynamics of interleukin-1 beta using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry. 1990 Aug 14;29(32):7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- Cross A. J., Fleming G. R. Analysis of time-resolved fluorescence anisotropy decays. Biophys J. 1984 Jul;46(1):45–56. doi: 10.1016/S0006-3495(84)83997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. E., Eisinger J., Blumberg W. E. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys J. 1979 May;26(2):161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P., Roberts V. A., Osguthorpe D. J., Wolff J., Genest M., Hagler A. T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1988;4(1):31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Dunbrack R. L., Jr, Karplus M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat Struct Biol. 1994 May;1(5):334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- Hansen D., Altschmied L., Hillen W. Engineered Tet repressor mutants with single tryptophan residues as fluorescent probes. Solvent accessibilities of DNA and inducer binding sites and interaction with tetracycline. J Biol Chem. 1987 Oct 15;262(29):14030–14035. [PubMed] [Google Scholar]
- Hillen W., Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- Hinrichs W., Kisker C., Düvel M., Müller A., Tovar K., Hillen W., Saenger W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994 Apr 15;264(5157):418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- Hopfield J. J. Photo-induced charge transfer. A critical test of the mechanism and range of biological electron transfer processes. Biophys J. 1977 Jun;18(3):311–321. doi: 10.1016/S0006-3495(77)85616-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiye T., Karplus M. Fluorescence depolarization of tryptophan residues in proteins: a molecular dynamics study. Biochemistry. 1983 Jun 7;22(12):2884–2893. doi: 10.1021/bi00281a017. [DOI] [PubMed] [Google Scholar]
- Kisker C., Hinrichs W., Tovar K., Hillen W., Saenger W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J Mol Biol. 1995 Mar 24;247(2):260–280. doi: 10.1006/jmbi.1994.0138. [DOI] [PubMed] [Google Scholar]
- Lipari G., Szabo A. Effect of librational motion on fluorescence depolarization and nuclear magnetic resonance relaxation in macromolecules and membranes. Biophys J. 1980 Jun;30(3):489–506. doi: 10.1016/S0006-3495(80)85109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenthal R., Sancho J., Fersht A. R. Histidine-aromatic interactions in barnase. Elevation of histidine pKa and contribution to protein stability. J Mol Biol. 1992 Apr 5;224(3):759–770. doi: 10.1016/0022-2836(92)90560-7. [DOI] [PubMed] [Google Scholar]
- Machado F. C., Demicheli C., Garnier-Suillerot A., Beraldo H. Metal complexes of anhydrotetracycline. 2. Absorption and circular dichroism study of Mg(II), Al(III), and Fe(III) complexes. Possible influence of the Mg(II) complex on the toxic side effects of tetracycline. J Inorg Biochem. 1995 Nov 15;60(3):163–173. doi: 10.1016/0162-0134(95)00017-i. [DOI] [PubMed] [Google Scholar]
- Mandel A. M., Akke M., Palmer A. G., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995 Feb 10;246(1):144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- Peviani C., Hillen W., Ettner N., Lami H., Doglia S. M., Piémont E., Ellouze C., Chabbert M. Spectroscopic investigation of Tet repressor tryptophan-43 upon specific and nonspecific DNA binding. Biochemistry. 1995 Oct 10;34(40):13007–13015. doi: 10.1021/bi00040a011. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Ricci R. W. Deuterium-isotope effect on the fluorescence yields and lifetimes of indole derivatives--including tryptophan and tryptamine. Photochem Photobiol. 1970 Jul;12(1):67–75. doi: 10.1111/j.1751-1097.1970.tb06039.x. [DOI] [PubMed] [Google Scholar]
- Ross J. B., Wyssbrod H. R., Porter R. A., Schwartz G. P., Michaels C. A., Laws W. R. Correlation of tryptophan fluorescence intensity decay parameters with 1H NMR-determined rotamer conformations: [tryptophan2]oxytocin. Biochemistry. 1992 Feb 18;31(6):1585–1594. doi: 10.1021/bi00121a002. [DOI] [PubMed] [Google Scholar]
- Schrauber H., Eisenhaber F., Argos P. Rotamers: to be or not to be? An analysis of amino acid side-chain conformations in globular proteins. J Mol Biol. 1993 Mar 20;230(2):592–612. doi: 10.1006/jmbi.1993.1172. [DOI] [PubMed] [Google Scholar]
- Shinitzky M., Goldman R. Fluorometric detection of histiine-tryptophan complexes in peptides and proteins. Eur J Biochem. 1967 Dec;3(2):139–144. doi: 10.1111/j.1432-1033.1967.tb19508.x. [DOI] [PubMed] [Google Scholar]
- Silva N. D., Jr, Prendergast F. G. Tryptophan dynamics of the FK506 binding protein: time-resolved fluorescence and simulations. Biophys J. 1996 Mar;70(3):1122–1137. doi: 10.1016/S0006-3495(96)79706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Altschmied L., Hillen W. Kinetic and equilibrium characterization of the Tet repressor-tetracycline complex by fluorescence measurements. Evidence for divalent metal ion requirement and energy transfer. J Mol Biol. 1986 Feb 5;187(3):341–348. doi: 10.1016/0022-2836(86)90437-7. [DOI] [PubMed] [Google Scholar]
- Vix A., Lami H. Protein fluorescence decay: discrete components or distribution of lifetimes? Really no way out of the dilemma? Biophys J. 1995 Mar;68(3):1145–1151. doi: 10.1016/S0006-3495(95)80290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A. J., Kemple M. D., Prendergast F. G. Fluorescence and 13C NMR determination of side-chain and backbone dynamics of synthetic melittin and melittin analogues in isotropic solvents. Biochemistry. 1989 Oct 17;28(21):8624–8639. doi: 10.1021/bi00447a053. [DOI] [PubMed] [Google Scholar]



