Abstract
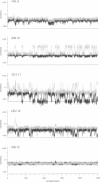
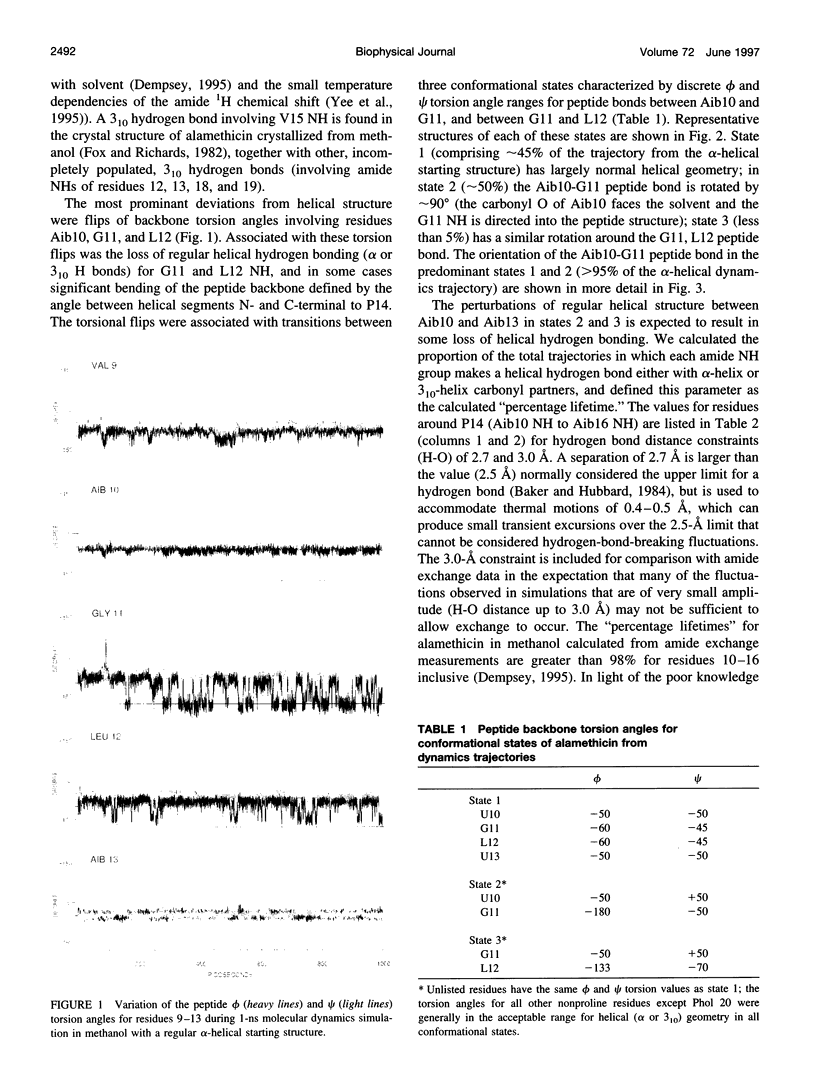
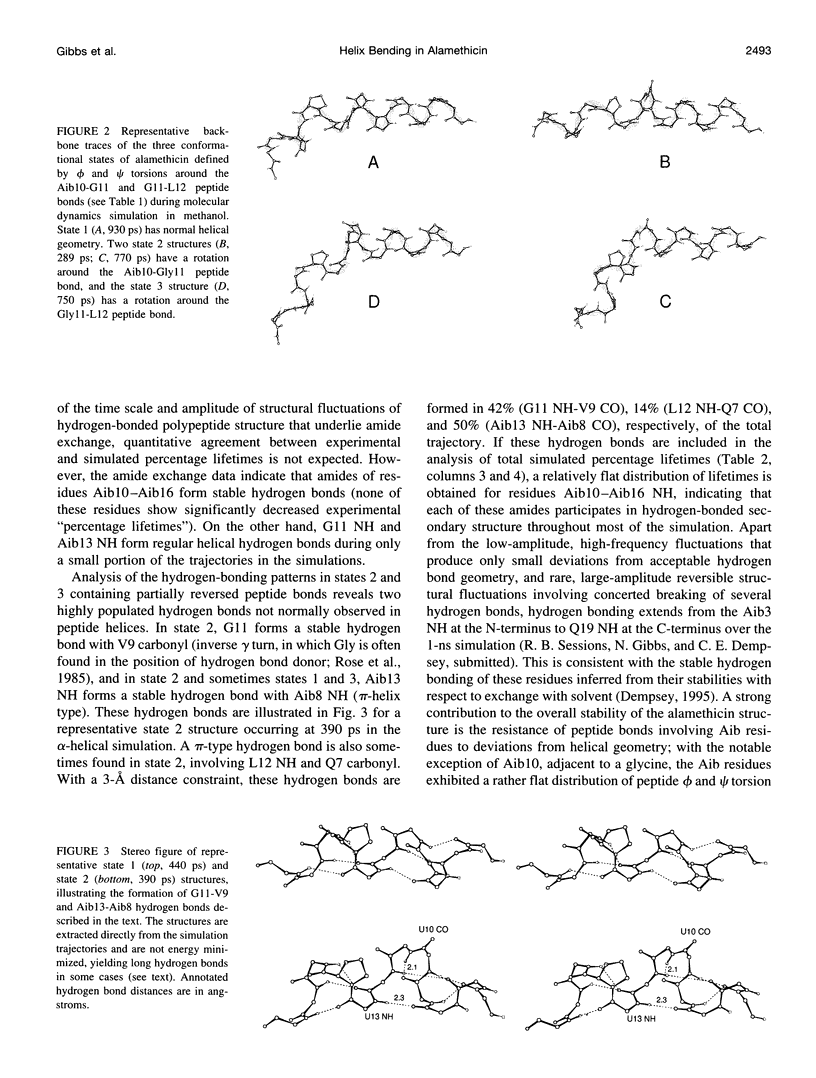
Molecular dynamics simulations of alamethicin in methanol were carried out with either a regular alpha-helical conformation or the x-ray crystal structure as starting structures. The structures rapidly converged to a well-defined hydrogen-bonding pattern with mixed alpha-helical and 3(10)-helical hydrogen bonds, consistent with NMR structural characterization, and did not unfold throughout the 1-ns simulation, despite some sizable backbone fluctuations involving reversible breaking of helical hydrogen bonds. Bending of the helical structure around residues Aib10-Aib13 was associated with reversible flips of the peptide bonds involving G11 (Aib10-G11 or G11-L12 peptide bonds), yielding discrete structural states in which the Aib10 carbonyl or (rarely) the G11 carbonyl was oriented away from the peptide helix. These peptide bond reversals could be accommodated without greatly perturbing the adjacent helical structure, and intramolecular hydrogen bonding was generally maintained in bent states through the formation of new (non-alpha or 3[10]) hydrogen bonds with good geometries: G11 NH-V9 CO (inverse gamma turn), Aib13 NH-Aib8 CO (pi-helix) and, rarely, L12 NH- Q7 NH (pi-helix). These observations may reconcile potentially conflicting NMR structural information for alamethicin in methanol, in which evidence for conformational flexibility in the peptide sequence before P14 (G11-Aib13) contrasts with the stability of backbone amide NH groups to exchange with solvent. Similar reversible reorientation of the Thr11-Gly12 peptide bond of melittin is also observed in dynamics simulations in methanol (R. B. Sessions, N. Gibbs, and C. E. Dempsey, submitted). This phenomenon may have some role in the orientation of the peptide carbonyl in solvating the channel lumen in membrane ion channel states of these peptides.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker E. N., Hubbard R. E. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. Helix geometry in proteins. J Mol Biol. 1988 Jun 5;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Brumfeld V., Miller I. R. Electric field dependence of alamethicin channels. Biochim Biophys Acta. 1990 May 9;1024(1):49–53. doi: 10.1016/0005-2736(90)90207-5. [DOI] [PubMed] [Google Scholar]
- Cafiso D. S. Alamethicin: a peptide model for voltage gating and protein-membrane interactions. Annu Rev Biophys Biomol Struct. 1994;23:141–165. doi: 10.1146/annurev.bb.23.060194.001041. [DOI] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P., Roberts V. A., Osguthorpe D. J., Wolff J., Genest M., Hagler A. T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1988;4(1):31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Dempsey C. E., Bazzo R., Harvey T. S., Syperek I., Boheim G., Campbell I. D. Contribution of proline-14 to the structure and actions of melittin. FEBS Lett. 1991 Apr 9;281(1-2):240–244. doi: 10.1016/0014-5793(91)80402-o. [DOI] [PubMed] [Google Scholar]
- Dempsey C. E., Handcock L. J. Hydrogen bond stabilities in membrane-reconstituted alamethicin from amide-resolved hydrogen-exchange measurements. Biophys J. 1996 Apr;70(4):1777–1788. doi: 10.1016/S0006-3495(96)79741-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclohier H., Molle G., Dugast J. Y., Spach G. Prolines are not essential residues in the "barrel-stave" model for ion channels induced by alamethicin analogues. Biophys J. 1992 Sep;63(3):868–873. doi: 10.1016/S0006-3495(92)81637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Carver J. A., Boyd J., Campbell I. D. High-resolution 1H NMR study of the solution structure of alamethicin. Biochemistry. 1987 Feb 24;26(4):1043–1050. doi: 10.1021/bi00378a010. [DOI] [PubMed] [Google Scholar]
- Fox R. O., Jr, Richards F. M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-A resolution. Nature. 1982 Nov 25;300(5890):325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Franklin J. C., Ellena J. F., Jayasinghe S., Kelsh L. P., Cafiso D. S. Structure of micelle-associated alamethicin from 1H NMR. Evidence for conformational heterogeneity in a voltage-gated peptide. Biochemistry. 1994 Apr 5;33(13):4036–4045. doi: 10.1021/bi00179a032. [DOI] [PubMed] [Google Scholar]
- Fraternali F. Restrained and unrestrained molecular dynamics simulations in the NVT ensemble of alamethicin. Biopolymers. 1990;30(11-12):1083–1099. doi: 10.1002/bip.360301109. [DOI] [PubMed] [Google Scholar]
- He K., Ludtke S. J., Worcester D. L., Huang H. W. Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys J. 1996 Jun;70(6):2659–2666. doi: 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh L. P., Ellena J. F., Cafiso D. S. Determination of the molecular dynamics of alamethicin using 13C NMR: implications for the mechanism of gating of a voltage-dependent channel. Biochemistry. 1992 Jun 9;31(22):5136–5144. doi: 10.1021/bi00137a007. [DOI] [PubMed] [Google Scholar]
- Mak D. O., Webb W. W. Molecular dynamics of alamethicin transmembrane channels from open-channel current noise analysis. Biophys J. 1995 Dec;69(6):2337–2349. doi: 10.1016/S0006-3495(95)80103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall G. R., Hodgkin E. E., Langs D. A., Smith G. D., Zabrocki J., Leplawy M. T. Factors governing helical preference of peptides containing multiple alpha,alpha-dialkyl amino acids. Proc Natl Acad Sci U S A. 1990 Jan;87(1):487–491. doi: 10.1073/pnas.87.1.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North C. L., Barranger-Mathys M., Cafiso D. S. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys J. 1995 Dec;69(6):2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North C. L., Franklin J. C., Bryant R. G., Cafiso D. S. Molecular flexibility demonstrated by paramagnetic enhancements of nuclear relaxation. Application to alamethicin: a voltage-gated peptide channel. Biophys J. 1994 Nov;67(5):1861–1866. doi: 10.1016/S0006-3495(94)80667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osguthorpe D. J., Dauber-Osguthorpe P. FOCUS: a program for analyzing molecular dynamics simulations, featuring digital signal-processing techniques. J Mol Graph. 1992 Sep;10(3):178-84, 164. doi: 10.1016/0263-7855(92)80053-g. [DOI] [PubMed] [Google Scholar]
- Piela L., Némethy G., Scheraga H. A. Proline-induced constraints in alpha-helices. Biopolymers. 1987 Sep;26(9):1587–1600. doi: 10.1002/bip.360260910. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Gierasch L. M., Smith J. A. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- Sansom M. S. Structure and function of channel-forming peptaibols. Q Rev Biophys. 1993 Nov;26(4):365–421. doi: 10.1017/s0033583500002833. [DOI] [PubMed] [Google Scholar]
- Sessions R. B., Dauber-Osguthorpe P., Osguthorpe D. J. Filtering molecular dynamics trajectories to reveal low-frequency collective motions: phospholipase A2. J Mol Biol. 1989 Dec 5;210(3):617–633. doi: 10.1016/0022-2836(89)90136-8. [DOI] [PubMed] [Google Scholar]
- Woolley G. A., Wallace B. A. Model ion channels: gramicidin and alamethicin. J Membr Biol. 1992 Aug;129(2):109–136. doi: 10.1007/BF00219508. [DOI] [PubMed] [Google Scholar]
- Woolley G. A., Wallace B. A. Temperature dependence of the interaction of alamethicin helices in membranes. Biochemistry. 1993 Sep 21;32(37):9819–9825. doi: 10.1021/bi00088a037. [DOI] [PubMed] [Google Scholar]
- Yee A. A., O'Neil J. D. Uniform 15N labeling of a fungal peptide: the structure and dynamics of an alamethicin by 15N and 1H NMR spectroscopy. Biochemistry. 1992 Mar 31;31(12):3135–3143. doi: 10.1021/bi00127a014. [DOI] [PubMed] [Google Scholar]
- Yun R. H., Anderson A., Hermans J. Proline in alpha-helix: stability and conformation studied by dynamics simulation. Proteins. 1991;10(3):219–228. doi: 10.1002/prot.340100306. [DOI] [PubMed] [Google Scholar]