Abstract
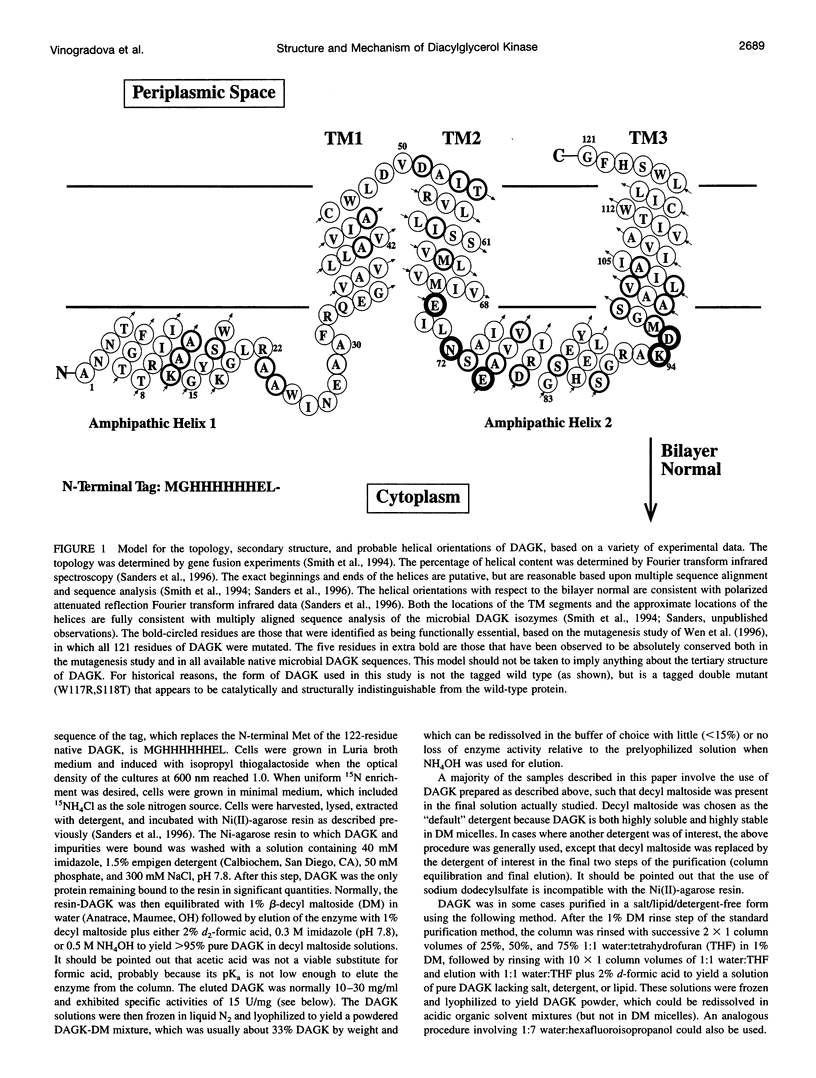
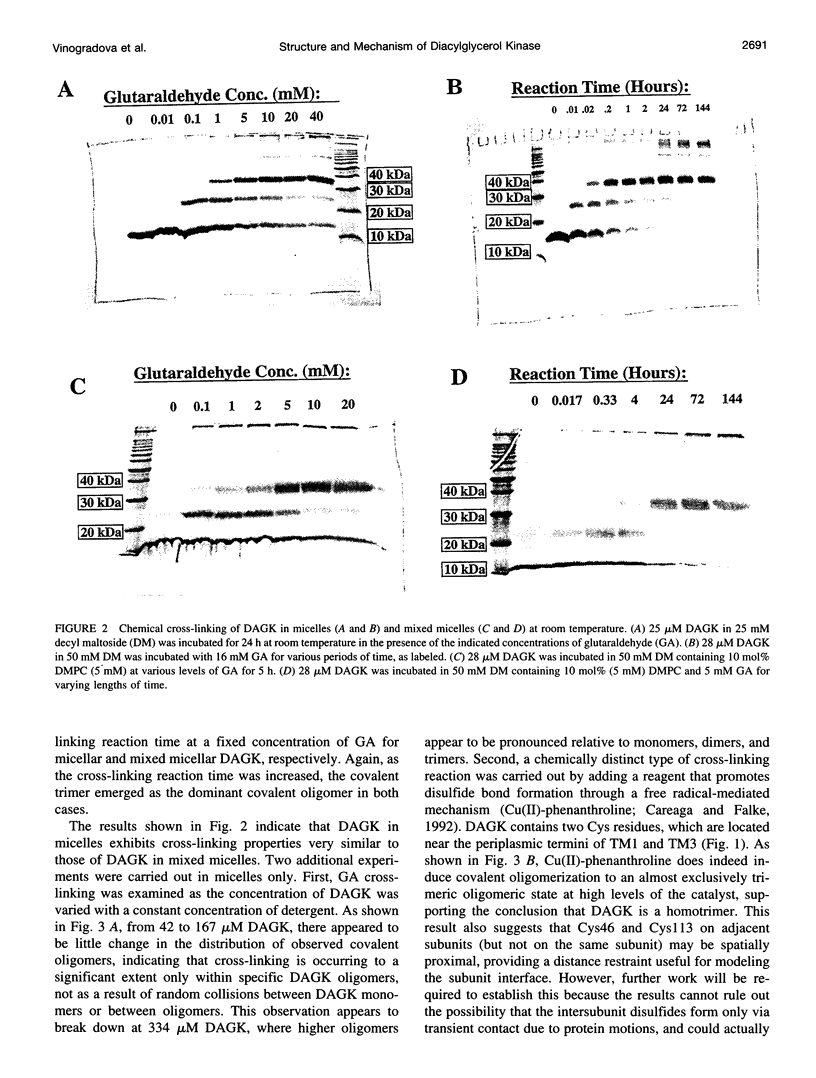
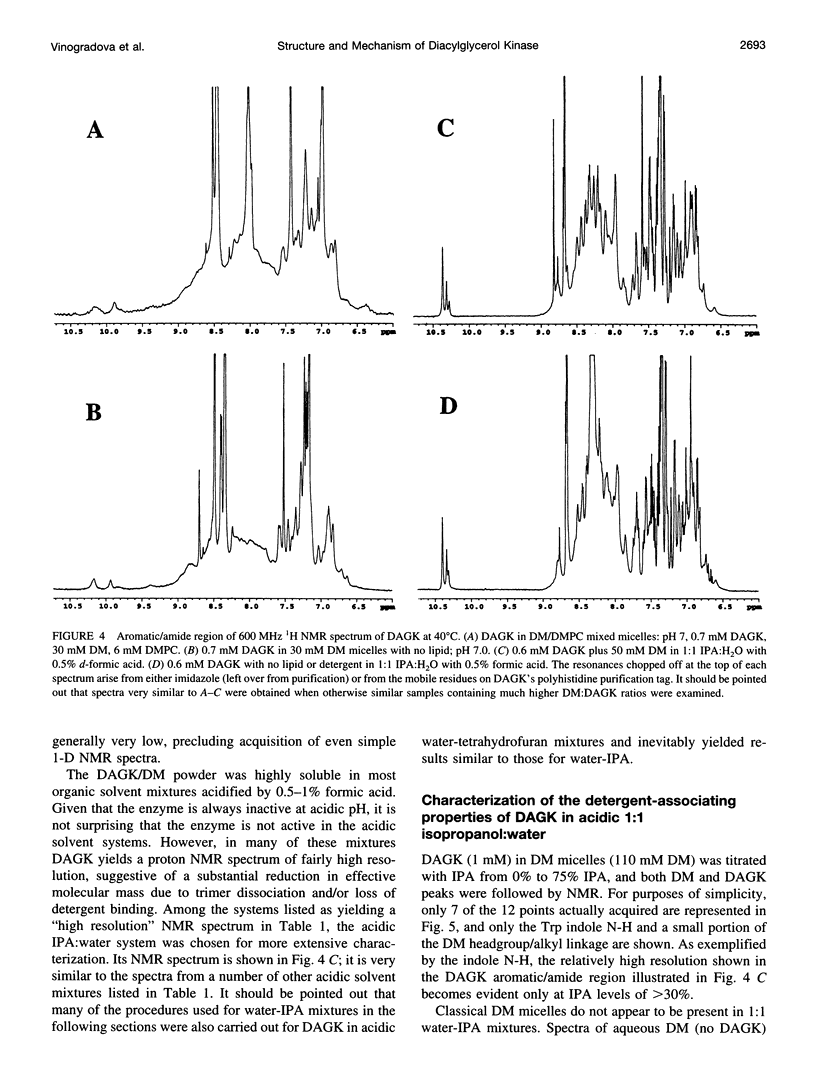
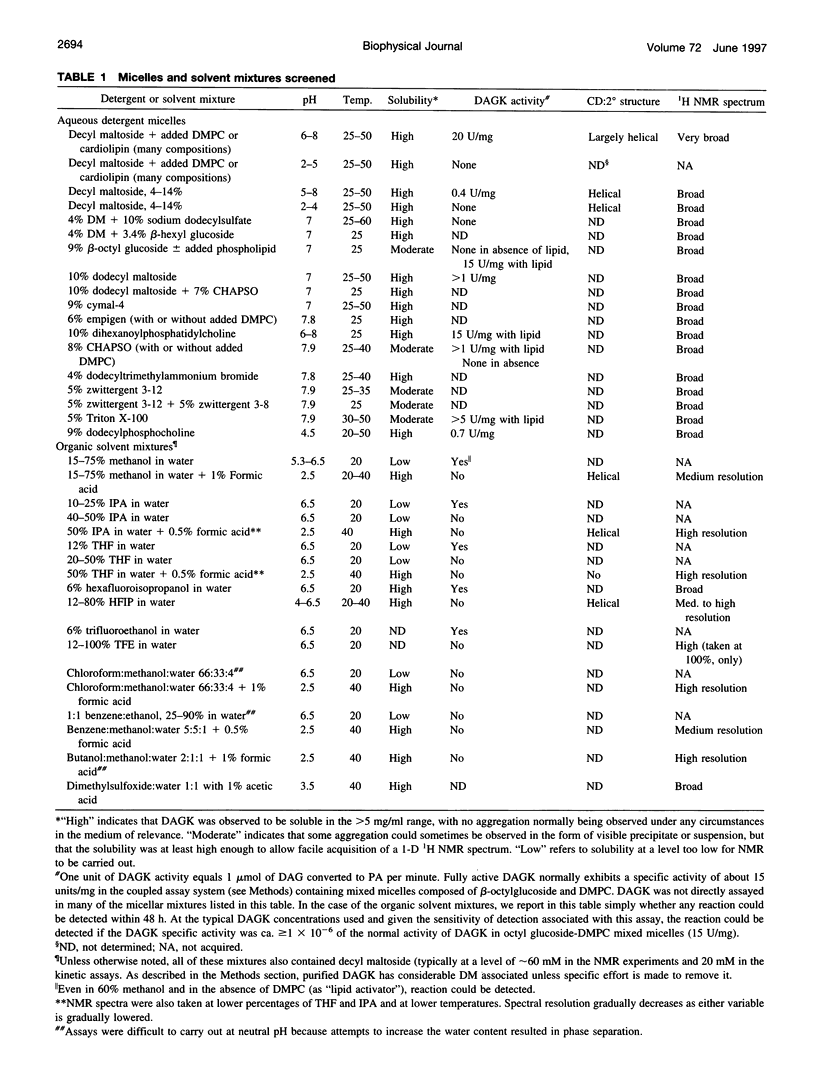
Diacylglycerol kinase (DAGK) is a 13-kDa integral membrane protein that spans the lipid bilayer three times and which is active in some micellar systems. In this work DAGK was purified using metal ion chelate chromatography, and its structural properties in micelles and organic solvent mixtures studies were examined, primarily to address the question of whether the structure of DAGK can be determined using solution NMR methods. Cross-linking studies established that DAGK is homotrimeric in decyl maltoside (DM) micelles and mixed micelles. The aggregate detergent-protein molecular mass of DAGK in both octyl glucoside and DM micelles was determined to be in the range of 100-110 kDa-much larger than the sum of the molecular weights of the DAGK trimers and the protein-free micelles. In acidic organic solvent mixtures, DAGK-DM complexes were highly soluble and yielded relatively well-resolved NMR spectra. NMR and circular dichroism studies indicated that in these mixtures the enzyme adopts a kinetically trapped monomeric structure in which it irreversibly binds several detergent molecules and is primarily alpha-helical, but in which its tertiary structure is largely disordered. Although these results provide new information regarding the native oligomeric state of DAGK and the structural properties of complex membrane proteins in micelles and organic solvent mixtures, the results discourage the notion that the structure of DAGK can be readily determined at high resolution with solution NMR methods.
Full text
PDF


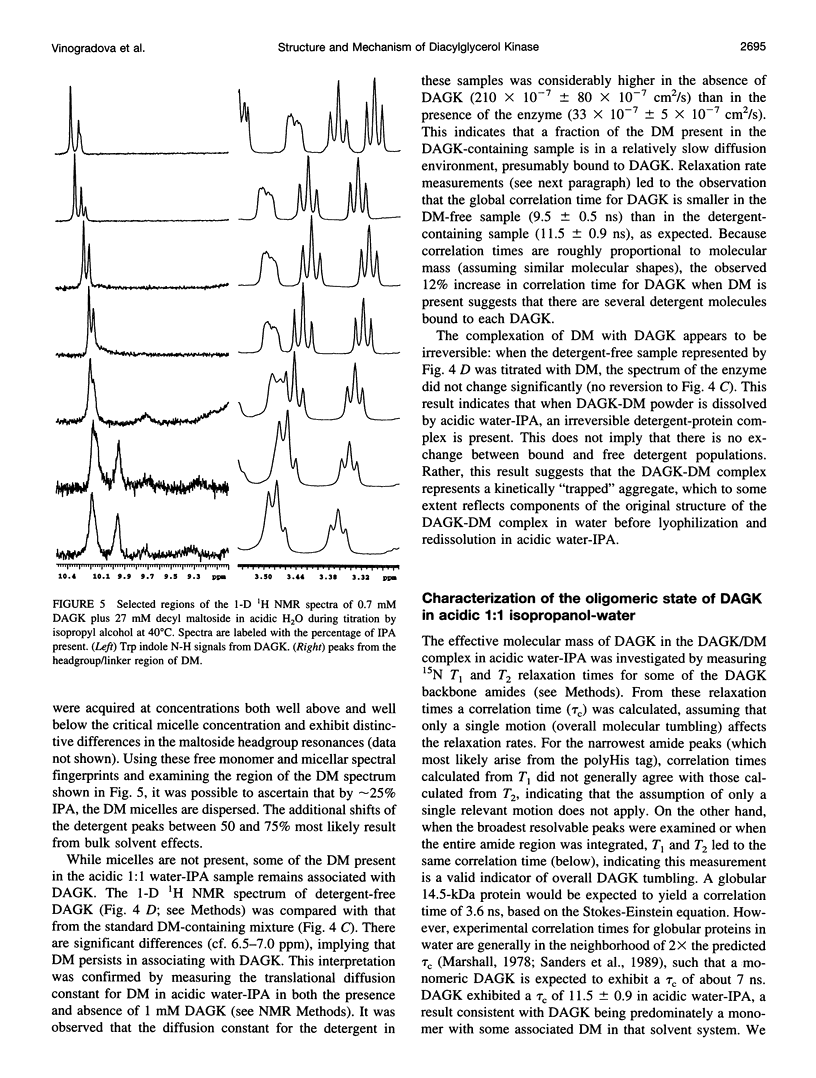

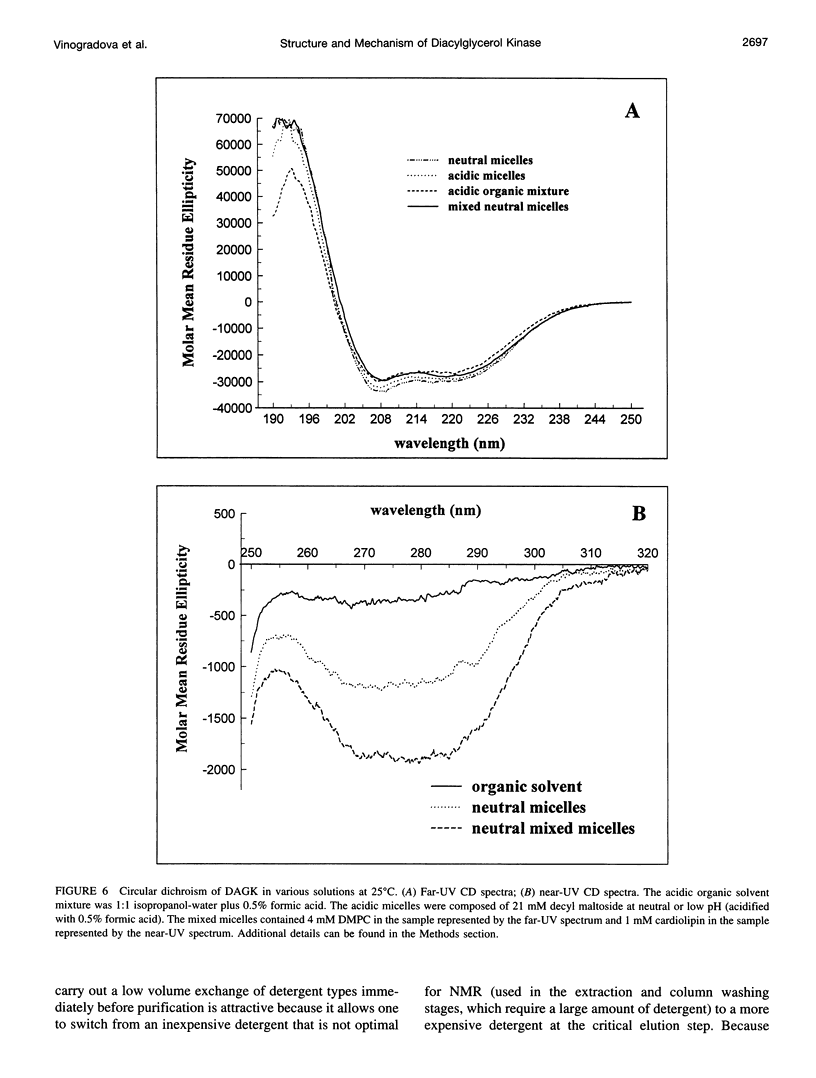
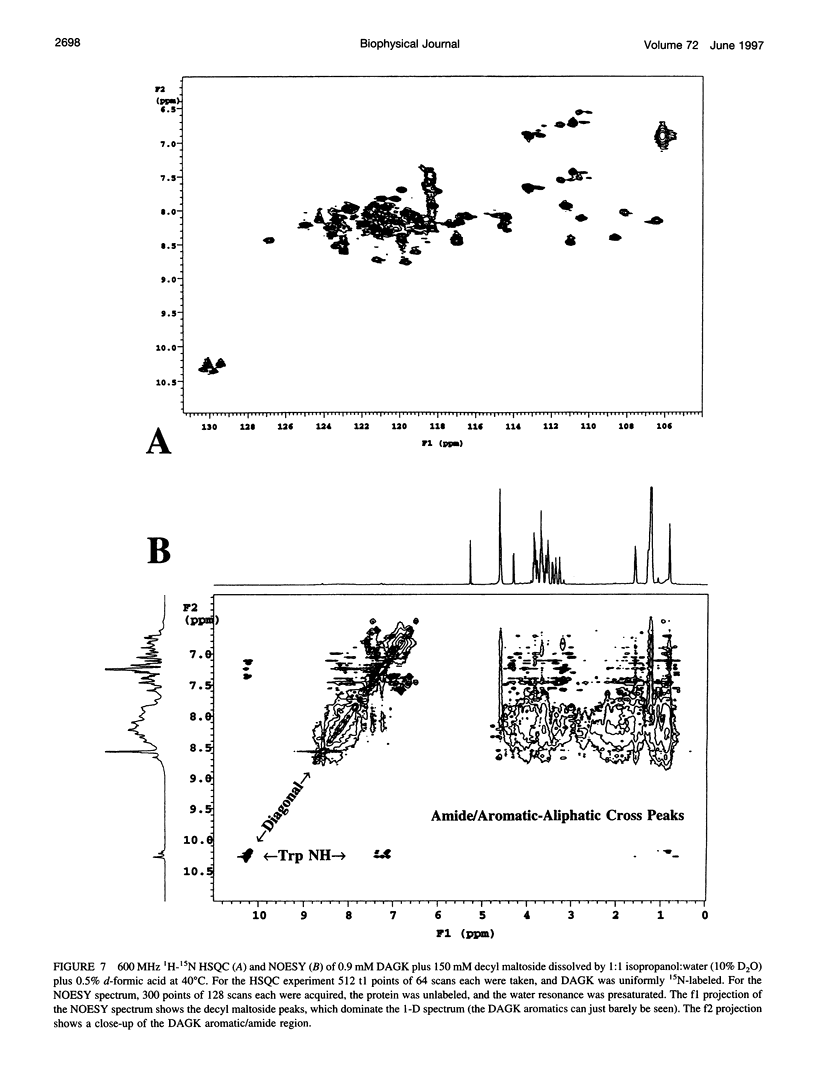



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexandrescu A. T., Evans P. A., Pitkeathly M., Baum J., Dobson C. M. Structure and dynamics of the acid-denatured molten globule state of alpha-lactalbumin: a two-dimensional NMR study. Biochemistry. 1993 Feb 23;32(7):1707–1718. doi: 10.1021/bi00058a003. [DOI] [PubMed] [Google Scholar]
- Bohnenberger E., Sandermann H., Jr Lipid dependence of diacylglycerol kinase from Escherichia coli. Eur J Biochem. 1983 May 16;132(3):645–650. doi: 10.1111/j.1432-1033.1983.tb07412.x. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga C. L., Falke J. J. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J Mol Biol. 1992 Aug 20;226(4):1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994 May 17;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Gaffney B. J. Chemical and biochemical crosslinking of membrane components. Biochim Biophys Acta. 1985 Dec 9;822(3-4):289–317. doi: 10.1016/0304-4157(85)90012-7. [DOI] [PubMed] [Google Scholar]
- Garavito R. M., Picot D., Loll P. J. Strategies for crystallizing membrane proteins. J Bioenerg Biomembr. 1996 Feb;28(1):13–27. [PubMed] [Google Scholar]
- Girvin M. E., Fillingame R. H. Determination of local protein structure by spin label difference 2D NMR: the region neighboring Asp61 of subunit c of the F1F0 ATP synthase. Biochemistry. 1995 Feb 7;34(5):1635–1645. doi: 10.1021/bi00005a020. [DOI] [PubMed] [Google Scholar]
- Girvin M. E., Fillingame R. H. Helical structure and folding of subunit c of F1F0 ATP synthase: 1H NMR resonance assignments and NOE analysis. Biochemistry. 1993 Nov 16;32(45):12167–12177. doi: 10.1021/bi00096a029. [DOI] [PubMed] [Google Scholar]
- Goffeau A. Life with 482 genes. Science. 1995 Oct 20;270(5235):445–446. doi: 10.1126/science.270.5235.445. [DOI] [PubMed] [Google Scholar]
- Haltia T., Freire E. Forces and factors that contribute to the structural stability of membrane proteins. Biochim Biophys Acta. 1995 Jul 17;1241(2):295–322. doi: 10.1016/0304-4157(94)00161-6. [DOI] [PubMed] [Google Scholar]
- Henry G. D., Sykes B. D. Methods to study membrane protein structure in solution. Methods Enzymol. 1994;239:515–535. doi: 10.1016/s0076-6879(94)39020-7. [DOI] [PubMed] [Google Scholar]
- Hufnagel P., Schweiger U., Eckerskorn C., Oesterhelt D. Electrospray ionization mass spectrometry of genetically and chemically modified bacteriorhodopsins. Anal Biochem. 1996 Dec 1;243(1):46–54. doi: 10.1006/abio.1996.0480. [DOI] [PubMed] [Google Scholar]
- Jones S., Thornton J. M. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis C. R., Walsh J. P., Bell R. M. sn-1,2-Diacylglycerol kinase of Escherichia coli. Purification, reconstitution, and partial amino- and carboxyl-terminal analysis. J Biol Chem. 1985 Apr 10;260(7):4091–4097. [PubMed] [Google Scholar]
- Miller K. J., McKinstry M. W., Hunt W. P., Nixon B. T. Identification of the diacylglycerol kinase structural gene of Rhizobium meliloti 1021. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):363–371. doi: 10.1094/mpmi-5-363. [DOI] [PubMed] [Google Scholar]
- Moréra S., Chiadmi M., LeBras G., Lascu I., Janin J. Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium. Biochemistry. 1995 Sep 5;34(35):11062–11070. [PubMed] [Google Scholar]
- Møller J. V., le Maire M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J Biol Chem. 1993 Sep 5;268(25):18659–18672. [PubMed] [Google Scholar]
- Opella S. J., Kim Y., McDonnell P. Experimental nuclear magnetic resonance studies of membrane proteins. Methods Enzymol. 1994;239:536–560. doi: 10.1016/s0076-6879(94)39021-5. [DOI] [PubMed] [Google Scholar]
- Orekhov VYu, Abdulaeva G. V., Musina LYu, Arseniev A. S. 1H-15N-NMR studies of bacteriorhodopsin Halobacterium halobium. Conformational dynamics of the four-helical bundle. Eur J Biochem. 1992 Nov 15;210(1):223–229. doi: 10.1111/j.1432-1033.1992.tb17412.x. [DOI] [PubMed] [Google Scholar]
- Park K., Perczel A., Fasman G. D. Differentiation between transmembrane helices and peripheral helices by the deconvolution of circular dichroism spectra of membrane proteins. Protein Sci. 1992 Aug;1(8):1032–1049. doi: 10.1002/pro.5560010809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K. V., Orekhov VYu, Popov A. I., Musina LYu, Arseniev A. S. Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy. Eur J Biochem. 1994 Jan 15;219(1-2):571–583. doi: 10.1111/j.1432-1033.1994.tb19973.x. [DOI] [PubMed] [Google Scholar]
- Popot J. L., Engelman D. M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990 May 1;29(17):4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- Raetz C. R., Newman K. F. Diglyceride kinase mutants of Escherichia coli: inner membrane association of 1,2-diglyceride and its relation to synthesis of membrane-derived oligosaccharides. J Bacteriol. 1979 Feb;137(2):860–868. doi: 10.1128/jb.137.2.860-868.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R., Newman K. F. Neutral lipid accumulation in the membranes of Escherichia coli mutants lacking diglyceride kinase. J Biol Chem. 1978 Jun 10;253(11):3882–3887. [PubMed] [Google Scholar]
- Russ E., Kaiser U., Sandermann H., Jr Lipid-dependent membrane enzymes. Purification to homogeneity and further characterization of diacylglycerol kinase from Escherichia coli. Eur J Biochem. 1988 Jan 15;171(1-2):335–342. doi: 10.1111/j.1432-1033.1988.tb13795.x. [DOI] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Czerski L., Vinogradova O., Badola P., Song D., Smith S. O. Escherichia coli diacylglycerol kinase is an alpha-helical polytopic membrane protein and can spontaneously insert into preformed lipid vesicles. Biochemistry. 1996 Jul 2;35(26):8610–8618. doi: 10.1021/bi9604892. [DOI] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Tian G. C., Tsai M. D. Mechanism of adenylate kinase. Is there a relationship between local substrate dynamics, local binding energy, and the catalytic mechanism? Biochemistry. 1989 Nov 14;28(23):9028–9043. doi: 10.1021/bi00449a011. [DOI] [PubMed] [Google Scholar]
- Sattler M., Fesik S. W. Use of deuterium labeling in NMR: overcoming a sizeable problem. Structure. 1996 Nov 15;4(11):1245–1249. doi: 10.1016/s0969-2126(96)00133-5. [DOI] [PubMed] [Google Scholar]
- Schneider E. G., Kennedy E. P. Partial purification and properties of diglyceride kinase from Escherichia coli. Biochim Biophys Acta. 1976 Aug 23;441(2):201–212. doi: 10.1016/0005-2760(76)90163-6. [DOI] [PubMed] [Google Scholar]
- Seigneuret M., Kainosho M. Localisation of methionine residues in bacteriorhodopsin by carbonyl 13C-NMR with sequence-specific assignments. FEBS Lett. 1993 Jul 19;327(1):7–12. doi: 10.1016/0014-5793(93)81027-w. [DOI] [PubMed] [Google Scholar]
- Smith R. L., O'Toole J. F., Maguire M. E., Sanders C. R., 2nd Membrane topology of Escherichia coli diacylglycerol kinase. J Bacteriol. 1994 Sep;176(17):5459–5465. doi: 10.1128/jb.176.17.5459-5465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E. H. Aromatic contributions to circular dichroism spectra of proteins. CRC Crit Rev Biochem. 1974 Jan;2(1):113–175. doi: 10.3109/10409237409105445. [DOI] [PubMed] [Google Scholar]
- Walsh J. P., Bell R. M. Diacylglycerol kinase from Escherichia coli. Methods Enzymol. 1992;209:153–162. doi: 10.1016/0076-6879(92)09019-y. [DOI] [PubMed] [Google Scholar]
- Walsh J. P., Bell R. M. sn-1,2-Diacylglycerol kinase of Escherichia coli. Mixed micellar analysis of the phospholipid cofactor requirement and divalent cation dependence. J Biol Chem. 1986 May 15;261(14):6239–6247. [PubMed] [Google Scholar]
- Walsh J. P., Bell R. M. sn-1,2-Diacylglycerol kinase of Escherichia coli. Structural and kinetic analysis of the lipid cofactor dependence. J Biol Chem. 1986 Nov 15;261(32):15062–15069. [PubMed] [Google Scholar]
- Wen J., Chen X., Bowie J. U. Exploring the allowed sequence space of a membrane protein. Nat Struct Biol. 1996 Feb;3(2):141–148. doi: 10.1038/nsb0296-141. [DOI] [PubMed] [Google Scholar]
- Yamashita Y., Takehara T., Kuramitsu H. K. Molecular characterization of a STreptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 1993 Oct;175(19):6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaluzec E. J., Gage D. A., Watson J. T. Matrix-assisted laser desorption ionization mass spectrometry: applications in peptide and protein characterization. Protein Expr Purif. 1995 Apr;6(2):109–123. doi: 10.1006/prep.1995.1014. [DOI] [PubMed] [Google Scholar]
- Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994 Nov;4(6):845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]




