Abstract
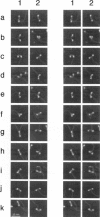
The motor and regulatory domains of the head and the 14-nm pitch of the alpha-helical coiled-coil of the tail of extended (6S) smooth-muscle myosin molecules were imaged with cryo atomic force microscopy at 80-85 K, and the effects of thiophosphorylation of the regulatory light chain were examined. The tail was 4 nm shorter in thiophosphorylated than in nonphosphorylated myosin. The first major bend was invariant, at approximately 51 nm from the head-tail junction (H-T), coincident with low probability in the paircoil score. The second major bend was 100 nm from the H-T junction in nonphosphorylated and closer to a skip residue than the bend (at 95 nm) in thiophosphorylated molecules. The shorter tail and distance between the two major bends induced by thiophosphorylation are interpreted to result from melting of the coiled-coil. An additional bend not previously reported occurred, with a lower frequency, approximately 24 nm from the H-T. The range of separation between the two heads was greater in thiophosphorylated molecules. Occasional high-resolution images showed slight unwinding of the coiled-coil of the base of the heads. We suggest that phosphorylation of MLC20 can affect the structure of extended, 6S myosin.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankrett R. J., Rowe A. J., Cross R. A., Kendrick-Jones J., Bagshaw C. R. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J Mol Biol. 1991 Jan 20;217(2):323–335. doi: 10.1016/0022-2836(91)90546-i. [DOI] [PubMed] [Google Scholar]
- Berger B., Wilson D. B., Wolf E., Tonchev T., Milla M., Kim P. S. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremo C. R., Sellers J. R., Facemyer K. C. Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J Biol Chem. 1995 Feb 3;270(5):2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- Elliott A., Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978 Aug 25;123(4):505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- Flicker P. F., Wallimann T., Vibert P. Electron microscopy of scallop myosin. Location of regulatory light chains. J Mol Biol. 1983 Sep 25;169(3):723–741. doi: 10.1016/s0022-2836(83)80167-3. [DOI] [PubMed] [Google Scholar]
- Hallett P., Offer G., Miles M. J. Atomic force microscopy of the myosin molecule. Biophys J. 1995 Apr;68(4):1604–1606. doi: 10.1016/S0006-3495(95)80333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Mou J., Sheng J., Yang J., Shao Z. Cryo atomic force microscopy: a new approach for biological imaging at high resolution. Biochemistry. 1995 Jul 4;34(26):8215–8220. doi: 10.1021/bi00026a001. [DOI] [PubMed] [Google Scholar]
- Harrington W. F. A mechanochemical mechanism for muscle contraction. Proc Natl Acad Sci U S A. 1971 Mar;68(3):685–689. doi: 10.1073/pnas.68.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y., Tanahashi K., Morita F. Regulatory mechanism by the phosphorylation of 20-kDa light chain of porcine aorta smooth muscle myosin. J Biochem. 1990 Dec;108(6):909–913. doi: 10.1093/oxfordjournals.jbchem.a123313. [DOI] [PubMed] [Google Scholar]
- Haystead C. M., Gailly P., Somlyo A. P., Somlyo A. V., Haystead T. A. Molecular cloning and functional expression of a recombinant 72.5 kDa fragment of the 110 kDa regulatory subunit of smooth muscle protein phosphatase 1M. FEBS Lett. 1995 Dec 18;377(2):123–127. doi: 10.1016/0014-5793(95)01318-0. [DOI] [PubMed] [Google Scholar]
- Houdusse A., Cohen C. Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation. Structure. 1996 Jan 15;4(1):21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- Huxley H. E. The mechanism of muscular contraction. Science. 1969 Jun 20;164(3886):1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- Ikebe M., Hartshorne D. J. Conformation-dependent proteolysis of smooth-muscle myosin. J Biol Chem. 1984 Oct 10;259(19):11639–11642. [PubMed] [Google Scholar]
- Ikebe M., Hartshorne D. J. Proteolysis of smooth muscle myosin by Staphylococcus aureus protease: preparation of heavy meromyosin and subfragment 1 with intact 20 000-dalton light chains. Biochemistry. 1985 Apr 23;24(9):2380–2387. doi: 10.1021/bi00330a038. [DOI] [PubMed] [Google Scholar]
- Ikebe M., Hartshorne D. J. Reverse reaction of smooth muscle myosin light chain kinase. Formation of ATP from phosphorylated light chain plus ADP. J Biol Chem. 1986 Jun 25;261(18):8249–8253. [PubMed] [Google Scholar]
- Kitazawa T., Gaylinn B. D., Denney G. H., Somlyo A. P. G-protein-mediated Ca2+ sensitization of smooth muscle contraction through myosin light chain phosphorylation. J Biol Chem. 1991 Jan 25;266(3):1708–1715. [PubMed] [Google Scholar]
- Knight P. J. Dynamic behaviour of the head-tail junction of myosin. J Mol Biol. 1996 Jan 19;255(2):269–274. doi: 10.1006/jmbi.1996.0022. [DOI] [PubMed] [Google Scholar]
- Levine R. J., Kensler R. W., Yang Z., Sweeney H. L. Myosin regulatory light chain phosphorylation and the production of functionally significant changes in myosin head arrangement on striated muscle thick filaments. Biophys J. 1995 Apr;68(4 Suppl):224S–224S. [PMC free article] [PubMed] [Google Scholar]
- Matsu-ura M., Ikebe M. Requirement of the two-headed structure for the phosphorylation dependent regulation of smooth muscle myosin. FEBS Lett. 1995 Apr 24;363(3):246–250. doi: 10.1016/0014-5793(95)00326-5. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982 Sep 16;299(5880):226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J Mol Biol. 1983 Mar 15;164(4):605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- Offer G., Knight P. The structure of the head-tail junction of the myosin molecule. J Mol Biol. 1996 Mar 1;256(3):407–416. doi: 10.1006/jmbi.1996.0096. [DOI] [PubMed] [Google Scholar]
- Offer G. Skip residues correlate with bends in the myosin tail. J Mol Biol. 1990 Nov 20;216(2):213–218. doi: 10.1016/S0022-2836(05)80309-2. [DOI] [PubMed] [Google Scholar]
- Olney J. J., Sellers J. R., Cremo C. R. Structure and function of the 10 S conformation of smooth muscle myosin. J Biol Chem. 1996 Aug 23;271(34):20375–20384. doi: 10.1074/jbc.271.34.20375. [DOI] [PubMed] [Google Scholar]
- Onishi H., Wakabayashi T. Electron microscopic studies of myosin molecules from chicken gizzard muscle I: the formation of the intramolecular loop in the myosin tail. J Biochem. 1982 Sep;92(3):871–879. doi: 10.1093/oxfordjournals.jbchem.a134001. [DOI] [PubMed] [Google Scholar]
- Onishi H., Wakabayashi T., Kamata T., Watanabe S. Electron microscopic studies of myosin molecules from chicken gizzard muscle II: The effect of thiophosphorylation of the 20K-dalton light chain on the ATP-induced change in the conformation of myosin monomers. J Biochem. 1983 Oct;94(4):1147–1154. doi: 10.1093/oxfordjournals.jbchem.a134459. [DOI] [PubMed] [Google Scholar]
- Quinlan R. A., Stewart M. Crystalline tubes of myosin subfragment-2 showing the coiled-coil and molecular interaction geometry. J Cell Biol. 1987 Jul;105(1):403–415. doi: 10.1083/jcb.105.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993 Jul 2;261(5117):58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rayment I., Smith C., Yount R. G. The active site of myosin. Annu Rev Physiol. 1996;58:671–702. doi: 10.1146/annurev.ph.58.030196.003323. [DOI] [PubMed] [Google Scholar]
- Rimm D. L., Sinard J. H., Pollard T. D. Location of the head-tail junction of myosin. J Cell Biol. 1989 May;108(5):1783–1789. doi: 10.1083/jcb.108.5.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Yang J., Somlyo A. P. Biological atomic force microscopy: from microns to nanometers and beyond. Annu Rev Cell Dev Biol. 1995;11:241–265. doi: 10.1146/annurev.cb.11.110195.001325. [DOI] [PubMed] [Google Scholar]
- Somlyo A. P. Myosin isoforms in smooth muscle: how may they affect function and structure? J Muscle Res Cell Motil. 1993 Dec;14(6):557–563. doi: 10.1007/BF00141552. [DOI] [PubMed] [Google Scholar]
- Stafford W. F., 3rd, Szentkiralyi E. M., Szent-Györgyi A. G. Regulatory properties of single-headed fragments of scallop myosin. Biochemistry. 1979 Nov 27;18(24):5273–5280. doi: 10.1021/bi00591a002. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Stafford W. F., 3rd, Slayter H. S., Seidel J. C. A conformational transition in gizzard heavy meromyosin involving the head-tail junction, resulting in changes in sedimentation coefficient, ATPase activity, and orientation of heads. J Biol Chem. 1985 Nov 25;260(27):14810–14817. [PubMed] [Google Scholar]
- Thomson N. H., Fritz M., Radmacher M., Cleveland J. P., Schmidt C. F., Hansma P. K. Protein tracking and detection of protein motion using atomic force microscopy. Biophys J. 1996 May;70(5):2421–2431. doi: 10.1016/S0006-3495(96)79812-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K. M., Huiatt T. W., Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K. M., Lowey S. Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984 Jul 10;259(13):8564–8571. [PubMed] [Google Scholar]
- Trybus K. M. Regulation of expressed truncated smooth muscle myosins. Role of the essential light chain and tail length. J Biol Chem. 1994 Aug 19;269(33):20819–20822. [PubMed] [Google Scholar]
- Tyler J. M., Branton D. Rotary shadowing of extended molecules dried from glycerol. J Ultrastruct Res. 1980 May;71(2):95–102. doi: 10.1016/s0022-5320(80)90098-2. [DOI] [PubMed] [Google Scholar]
- Uyeda T. Q., Abramson P. D., Spudich J. A. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesenka J., Manne S., Giberson R., Marsh T., Henderson E. Colloidal gold particles as an incompressible atomic force microscope imaging standard for assessing the compressibility of biomolecules. Biophys J. 1993 Sep;65(3):992–997. doi: 10.1016/S0006-3495(93)81171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibert P., Cohen C. Domains, motions and regulation in the myosin head. J Muscle Res Cell Motil. 1988 Aug;9(4):296–305. doi: 10.1007/BF01773873. [DOI] [PubMed] [Google Scholar]
- Walker M., Knight P., Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985 Aug 5;184(3):535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- Warshaw D. M., Fay F. S. Cross-bridge elasticity in single smooth muscle cells. J Gen Physiol. 1983 Aug;82(2):157–199. doi: 10.1085/jgp.82.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M., Wilson-Kubalek E. M., Smith J. E., Faust L., Milligan R. A., Sweeney H. L. A 35-A movement of smooth muscle myosin on ADP release. Nature. 1995 Dec 14;378(6558):748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- Winkelmann D. A., Lowey S. Probing myosin head structure with monoclonal antibodies. J Mol Biol. 1986 Apr 20;188(4):595–612. doi: 10.1016/s0022-2836(86)80009-2. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M., Hamada Y., Katsuragawa Y., Imamura M., Mikawa T., Masaki T. Complete primary structure of vertebrate smooth muscle myosin heavy chain deduced from its complementary DNA sequence. Implications on topography and function of myosin. J Mol Biol. 1987 Nov 20;198(2):143–157. doi: 10.1016/0022-2836(87)90302-0. [DOI] [PubMed] [Google Scholar]
- Yang J., Mou J., Yuan J. Y., Shao Z. The effect of deformation on the lateral resolution of atomic force microscopy. J Microsc. 1996 May;182(Pt 2):106–113. doi: 10.1046/j.1365-2818.1996.140422.x. [DOI] [PubMed] [Google Scholar]
- Yang J., Shao Z. Effect of probe force on the resolution of atomic force microscopy of DNA. Ultramicroscopy. 1993 Jul;50(2):157–170. doi: 10.1016/0304-3991(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sheng S., Shao Z. Imaging biological structures with the cryo atomic force microscope. Biophys J. 1996 Oct;71(4):2168–2176. doi: 10.1016/S0006-3495(96)79418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]