Abstract
Fushi tarazu factor 1 (Ftz-F1, NR5A) is a zinc-finger transcription factor that belongs to the nuclear receptor superfamily and regulates genes that are involved in sterol and steroid metabolism in gonads, adrenals, liver and other tissues. To understand the evolutionary origins and developmental genetic relationships of the Ftz-F1 genes, we have cloned four homologous Ftz-f1 genes in zebrafish, called ff1a, ff1b, ff1c and ff1d. These four genes have different temporal and spatial expression patterns during development, indicating that they have distinct mechanisms of genetic regulation. Among them, the ff1a expression pattern is similar to mammalian Nr5a2, while the ff1b pattern is similar to that of mammalian Nr5a1. Genetic mapping experiments show that these four ff1 genes are located on chromosome segments conserved between the zebrafish and human genomes, indicating a common ancestral origin. Phylogenetic and conserved synteny analysis show that ff1a is the orthologue of NR5A2, and that ff1b and ff1d genes are co-orthologues of NR5A1 that arose by a gene-duplication event, probably a whole-genome duplication, in the ray-fin lineage, and each gene is located next to an NR6A1 co-orthologue as in humans, showing that the tandem duplication occurred before the divergence of human and zebrafish lineages. ff1c does not have a mammalian counterpart. Thus we have characterized the phylogenetic relationships, expression patterns and chromosomal locations of these Ftz-F1 genes, and have demonstrated their identities as NR5A genes in relation to the orthologous genes in other species.
Keywords: ftz-f1, LRH-1, NR5A, nuclear receptor, phylogeny, SF-1
Abbreviations: Ftz-F1, fushi tarazu factor 1; HS, Heat-Shock; MOP, Mother of Pearl; 3′/5′-RACE, rapid amplification of 3′/5′-cDNA ends; RT, reverse transcription
INTRODUCTION
Nuclear receptors form one of the largest families of transcription factors in the human genome, with 48 members [1]. In contrast, the nematode Caenorhabditis elegans has 284 family members, and the fly Drosophila melanogaster just 21 [2]. What evolutionary forces have shaped the content of the human nuclear receptor family? This is an important question, not only for understanding the general question of how the functions of large gene families evolve, but also because more than 10% of commonly prescribed pharmaceuticals for human disease have nuclear receptors as targets, and understanding the differences in nuclear gene content and function between humans and model organisms may yield information that is critical for the development of novel drugs that target nuclear receptors [1].
One of the seven nuclear receptor subfamilies, the NR5A family, has two members in the human and fly genomes, and just one in C. elegans, while some other subfamilies have several human members for each fly orthologue [1]. NR5A is also called Ftz-F1, because it was first identified as the transcription factor that activates fushi tarazu (ftz) in Drosophila. The ftz-f1 gene has been cloned from Xenopus as well as teleost fish. In mammals, the Ftz-F1 genes can be divided into two subgroups, NR5A1 (SF-1/Ad4BP) and NR5A2 (LRH-1/FTF).
NR5A1 is a transcription factor that controls the expression of many genes, such as steroidogenic cytochrome P450s and gonadotropins. NR5A1 is expressed in the adrenal and gonadal primordia, the ventromedial hypothalamic nucleus, and the developing pituitary primordium. Nr5a1-knockout mice fail to form the adrenal glands and gonads. Thus NR5A1 is essential for adrenal and gonadal development, and sexual differentiation [3].
NR5A2 is expressed more broadly than NR5A1. In the liver, it activates genes that are involved in cholesterol metabolism and bile acid synthesis. NR5A2 is also expressed in the pancreas, ovary, intestine, colon, the adrenal gland and pre-adipocytes for a number of different functions in endocrine and metabolic regulation [4].
A major model for the origin of human paralogues, such as NR5A1 and NR5A2, is that many arose during large-scale genome-amplification events, perhaps two whole-genome-duplication events, that occurred around the origin of vertebrate developmental innovations [5–7]. If the nuclear receptor genes arose by two rounds of whole-genome duplication, then four genes might be expected in the human genome, like the NOTCH genes and HOX clusters, rather than the two for the NR5A subfamily. To begin to understand the relationships of nuclear receptor genes to ancient genome duplications, we cloned a new member of the NR5A nuclear receptor gene family from zebrafish. In the present study, we show by phylogenetic analysis, comparative genomic analysis and by gene expression studies that the zebrafish lineage retains an NR5A gene that is lacking in the human genome. This gene arose in the peri-vertebrate-origin genome-amplification events. In addition, the zebrafish genome has duplicate copies of one of the original vertebrate paralogues that arose in a genome-duplication event specific for ray-fin fish [8–11].
MATERIALS AND METHODS
cDNA cloning and sequence analysis
Total RNA was extracted from zebrafish testis using TRIzol® reagent (Sigma, St. Louis, MO, U.S.A.). To obtain full-length ff1d cDNA, 5′- and 3′-RACE (rapid amplification of 5′- and 3′-cDNA ends) was performed with testis total RNA using the SMART™ cDNA amplification kit (Clontech laboratories, Palo Alto, CA, U.S.A.). For 3′-RACE, the fragments were amplified with the following primers: 5′FTZF1 (5′-TGCGGATAGGATGCGAGGAGGCCGCAAC-3′) for the first PCR and R0 (5′-CATGTATAAGCGAGACCGGGCGTT-3′) for the second PCR. For 5′-RACE, the fragments were amplified with following primers: F01 (5′-CGGCCATGTGGCACATGAGGCCAA-3′) for the first PCR, and F1 (5′-CGGGATGGACTACAGTTATGATGCGGAC-3′) for the second PCR. The RACE products were subcloned and sequenced. The nucleotide sequence was deposited into GenBank® under accession number NM_212834.
Phylogenetic analysis
DNA sequence alignment and homology analysis were performed using SeqWeb 2.1. All phylogenetic analyses were conducted on amino acid sequences. Ftz-F1 and NR6A genes from different animals were imported into CLUSTAL X, and trees were generated from amino acid sequences by the neighbour-joining method using NJplot (http://pbil.univ-lyon1.fr/software/njplot.html). A CG8676PA sequence from Drosophila melanogaster served as an outgroup to root the tree, and alignments used domains C and E, the DNA-binding and ligand-binding domains, which gave 352 amino acids. A bootstrapping method was used as a measure of the statistical validity of each node in the phylogenetic analysis.
Genetic mapping
The ff1a and ff1c genes were mapped as SSCPs (single-strand conformation polymorphisms) on the MOP (Mother of Pearl) and the HS (Heat-Shock) meiotic mapping panels [12–14]. Strain distribution patterns were analysed using MapManager. The ff1b and ff1d genes were mapped on the LN54 radiation hybrid panel [15], and intercalated into the map from the HS panel on the basis of nearby markers mapped on both panels. LocusLink [16] and ZFIN provided data for analysis of conserved syntenies. Mapping primers were: ff1a: ff1a+1830 F, 5′-TGGGTTTGCGCTGGGTGGACAT-3′ and ff1a-2149 R, 5′-TTTTTGGTGAGGGGTTGGAATAA-3′; ff1b: ff1b+1711 F, 5′-CATAACATCACCAGAGGGGAGTCA-3′ and ff1b-1968 R, 5′-TGTGCCGTCAGCCAATCGTT-3′; ff1c: ff1c+27 F, 5′-TACAAGTTAAAACGGCACATTC-3′ and ff1c-288 R, 5′-ACTCACTGCTGACTGAAATGCT-3′; and ff1d: ff1d+1882 F, 5′-AGAAGTTCGCATCACCCTCCACAT-3′ and ff1d-2233 R, 5′-CCACGCGAATACAGAAACACCAAC-3′.
RT (reverse transcription)-PCR
Reverse transcription was performed using the Superscript pre-amplification system (Gibco BRL) with 0.5 μg of oligo(dT)12–18 and 3 μg of each total RNA in a 20 μl reaction mixture as described in [17]. The cDNA product (1 or 2 μl) was used in PCR with ff1a, ff1b, ff1c, ff1d and actin primers for 25–35 cycles at 95 °C for 60 s, 55 °C for 60 s and 72 °C for 60 s. The ff1a primers 5′-GCAGCATCTTCTTCCGGGAACTAAAGG-3′ and 5′-GTACTGTACTCGAGGGCACGTTTGGCGTGCAG-3′ amplified cDNA from nt 1136 to 1674. The ff1b primers 5′-ACACTGCCGTCTGGTTTGTAG-3′ and 5′-GGCAACTGTAACACTACTATGGC-3′ amplified cDNA from nt 1419 to 2063. The ff1c primers 5′-CCCCAACTCCATCACAGAGCTT-3′ and 5′-GCACACATGAGATCATCGCAA-3′ amplified cDNA from nt 842 to 1625. The ff1d primers 5′-GAAAGAAGACGAGGGGAGATGT-3′ and 5′-ACACTCATACGCACTCATACAC-3′ amplified cDNA from nt 1790 to 2321. The actin primers 5′-TCACACCTTCTACAACGAGCTGCG-3′ and 5′-GAAGCTGTAGCCTCTCTCGGTCAG-3′ generated a 340-bp fragment [18]. PCR products were analysed on 1.5% agarose gels.
In situ hybridization
Whole-mount in situ hybridization was performed using digoxigenin-labelled antisense RNA probe and anti-digoxigenin alkaline-phosphatase-conjugated antibody as described previously [19].
RESULTS
Characterization of zebrafish ff1 genes
We have previously isolated three zebrafish ftz-f1 genes, ff1a, ff1b and ff1c [20–22]. In an attempt to understand more about the ftz-f1 gene family, we isolated a fourth cDNA, termed ff1d, from zebrafish testis by using 5′-RACE and 3′-RACE (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/389/bj3890019add.htm). The ff1d cDNA is 2554 bp with a predicted open reading frame of 502 amino acids. In addition, a short ff1d transcript was also obtained using 3′-RACE. The ff1d-short form is 1240 bp in length. It differs from the long form at the 3′-region, probably due to alternative splicing of exons at the 3′-region of the ftz-f1 gene. This shorter transcript encodes a smaller Ff1d protein lacking the C-terminal domain. The C-terminal truncation in Ff1d is due to an early stop codon located 22 amino acids downstream from the point of sequence divergence. The presence of long and short proteins has also been observed in zebrafish Ff1a [21].
Ff1a, Ff1b, Ff1c and Ff1d sequences were aligned (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/389/bj3890019add.htm). These sequences are highly conserved throughout the entire coding region, with Ff1c diverging from the other three. The most conserved region is the DNA-binding domain. The D and E domains have approx. 40–70% sequence identity among these four sequences (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/389/bj3890019add.htm).
Gene phylogenies
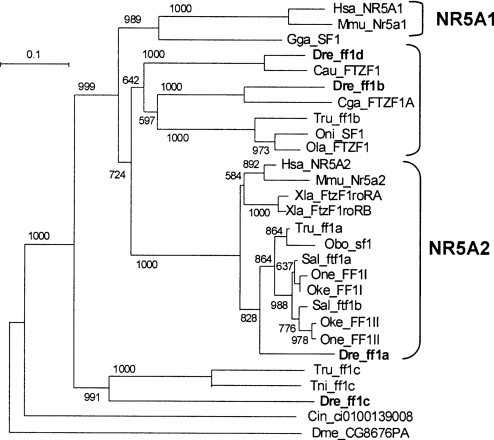
Phylogenetic analysis of ff1 genes can help reveal their origins and relationships to human NR5 genes. Ftz-f1 genes from D. melanogaster and the urochordate Ciona intestinalis served as outgroups to root the tree. The results showed that the tetrapod NR5A1 and NR5A2 genes branched as expected from the known evolutionary relationships of the species (Figure 1). Some of the teleost genes, however, did not fit as neatly into the tree.
Figure 1. Phylogenetic tree of Ftz-F1 (ff1) sequences.
Numbers indicate the number of times the branching was obtained from 1000 bootstrap runs. The marker of 0.1 is the length that corresponds to a 10% sequence difference. Zebrafish ff1b and ff1d can be grouped into the NR5A1 group, and ff1a into the NR5A2 group. Both zebrafish and pufferfish ff1c are segregated as an outgroup. Species abbreviations: Cau, Carassius auratus, goldfish; Cin, Ciona intestinalis, an ascidian; Cga, Clarias gariepinus, North African catfish; Dme, Drosophila melanogaster, fruitfly; Dre, Danio rerio, zebrafish; Gga, Gallus gallus, chicken; Hsa, Homo sapiens, human; Mmu, Mus musculus, mouse; Obo, Odontesthes bonariensis, a percomorph fish; Oke, Oncorhynchus keta, chum salmon; Ola, Oryzias latipes, medaka; One, Oncorhynchus nerka, sockeye salmon; Oni, Oreochromis niloticus, nile tilapia; Sal, Salvelinus alpinus, charr; Tni, Tetraodon nigroviridis, pufferfish; Tru, Takifugu rubripes, pufferfish; Xla, Xenopus laevis, frog. Species_gene name, accession numbers: Cau_FTZF1, AF526537; Cin_FTZF, ci0100139008; Cga_FTZF1A, AY014862; Dme_CG8676PA, NP_476932; Dre_ff1a, NM_131463; Dre_ff1b, AF198086; Dre_ff1c, AF327373; Dre_ff1d, AY212920; Gga_SF1, BAA76713; Hsa_NR5A1, NP_004950; Hsa_NR5A2, NP_003813; Mmu_Nr5a1, NP_620639; Mmu_Nr5a2, NP_109601; Obo_sf1, AY323199; Oke_FF1I, AF242223; Sal_ftf1b, AF468977; Oke_FF1II, AF242224; Ola_FTZF1, AB016834; One_FF1I, AF242225; One_FF1II, AF242226; Oni_SF1, AB060814; Tni_ff1c, CAG12178; Tru_ff1a, scaffold_5225.2457.4; Tru_ff1c, SINFRUP00000152795; Tru_ff1b, SINFRUP00000132871; Sal_ftf1a, AF468978; Xla_FtzF1roRA, AAA18356; Xla_FtzF1roRB, AAA18357.
One set of teleost ff1 genes, including zebrafish ff1a, was closely related to tetrapod NR5A2 genes with a high bootstrap value (1000/1000). The tree shows that orthologues of zebrafish ff1a exist in the percomorph pufferfish Takifugu rubripes and Odontesthes bonariensis, and several salmonids, and their topology does not contradict the phylogenetic relationships of these species [23]. Each of the three salmonids Oncorhynchus nerka, Oncorhynchus keta and Salvelinus alpinus, has two orthologues of zebrafish ff1a, and they are nested as expected by the species relationship. These results are consistent with a genome duplication event in the salmonid lineage after it diverged from the zebrafish and percomorph lineages [24,25].
A second set of teleost ff1 genes clusters with human NR5A2, but with lower bootstrap values (724/1000). The lower bootstrap values of this set, in contrast with the ff1a set, makes their evolutionary relationships to tetrapod NR5A1 and NR5A2 less clear. Within this set, however, there are two clades, one related to ff1b and the other to ff1d, suggesting that they may come from a gene-duplication event that occurred after the divergence of teleost and tetrapod lineages. Within the ff1b clade, the teleost ff1 genes branch according to their species phylogenies.
The third set of teleost ff1 genes include zebrafish ff1c and a pufferfish gene, and these branch with high bootstrap support (991/1000) as outgroups to the other tetrapod and teleost NR5-related genes. This position is consistent with the interpretation that ff1c genes arose in a gene-duplication event before the divergence of tetrapod and teleost lineages, but that the tetrapod orthologue has since been lost.
Genome context and conserved syntenies
While the phylogenetic analysis showed that ff1a is an orthologue of NR5A2, it left questions about the evolutionary relationships of the other three zebrafish ff1 genes. To discover the chromosomal location of zebrafish Ftz-f1 genes, we performed genetic mapping experiments. We found that zebrafish ff1a, ff1b, ff1c and ff1d mapped to LG22_40.9 cM, LG8_139.1 cM, LG3_80.3 cM and LG21_36.7 cM (see Supplementary Figure S4 at http://www.BiochemJ.org/bj/389/bj3890019add.htm, and Table 1) respectively on the HS meiotic mapping panel [14]. In humans, NR5A paralogues are syntenic with NOTCH paralogues, with NR5A1 at 9q33 and NOTCH1 at 9q34.3, and NR5A2 at 1q32.1 and NOTCH2 at 1p13-p11. Likewise, zebrafish ff1 genes are syntenic with notch paralogues, with ff1b and notch2 on LG8, ff1d and notch1a on LG21, and ff1c and notch3 on LG3 [12,14]. The only zebrafish ff1 gene not known to be syntenic with a notch paralogue is ff1a on LG22, which, however, has conserved syntenies with at least five other loci residing in Hsa1q32.1, syntenic with NOTCH2. Thus conserved long-range syntenies suggest orthologies of ff1a and ff1d with NR5A2 and NR5A1 respectively.
Table 1. Chromosomal localization of human and zebrafish NR5A and NOTCH genes.
Mapping information is listed with linkage group number (LG), map location (cM) and/or mapping panel for zebrafish, and with chromosome number and locus in humans. The accession numbers in the NCBI UniGene database for both species and in the ZFIN database for zebrafish are also listed.
| Zebrafish (Danio rerio) | Human (Homo sapiens) | ||||
|---|---|---|---|---|---|
| Name | Map | ZFIN #/UniGene # | Name | Map | UniGene # |
| notch1a | LG21; 26.10cM (HS) | ZDB-GENE-990415-173/Dr.11847 | NOTCH1 | 9q34.3 | Hs.311559 |
| notch1b | LG5; 232.00cM (MOP) | ZDB-GENE-990415-183/Dr.11846 | NOTCH2 | 1p13-p11 | Hs.8121 |
| notch2 | LG8; 64.90cM (MOP) | ZDB-GENE-000329-4/Dr.604 | NOTCH3 | 19p13.2-p13.1 | Hs.8546 |
| notch3 | LG 3; 146.10cM (MOP) | ZDB-GENE-000329-5/Dr.17815 | NOTCH4 | 6q21.3 | Hs.436100 |
| nr5a1a; ff1b | LG8; 139.1cM | ZDB-GENE-010504-1/Dr.11909 | NR5A1 | 9q33 | Hs.57037 |
| nr5a1b; ff1d | LG21; 36.7cM | ZDB-GENE-040702-6/Dr.30454 | – | ||
| nr5a2; ff1a | LG22; 40.9cM | ZDB-GENE-990415-79/Dr.6870 | NR5A2 | 1q32.1 | Hs.183123 |
| nr5a5; ff1c | LG3; 80.3cM | –/Dr.11921 | – | ||
To understand further the evolutionary relationships of the ff1 genes, we investigated ff1 genes in the genome sequencing databases of zebrafish and fugu (http://fugu.hgmp.mrc.ac.uk/blast/, http://www.sanger.ac.uk/Projects/D_rerio/, http://134.174.23.160/compGenomics/). The ff1a genes of zebrafish and fugu are present on contigs ctg10168.1 (transcript ENSDART00000008771) and Scaffold_5225 (transcript SINFRUT00000152923) respectively. There are no other transcripts present on these contigs, and so these do not provide evidence regarding the evolution of these genes, but the phylogenetic analysis and conserved long-range syntenies are clear that ff1a is an orthologue of human NR5A2.
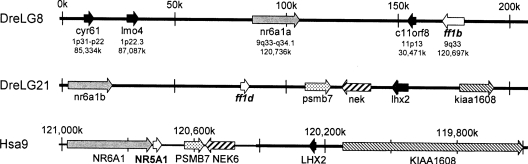
The phylogenetic analysis suggested that the ff1b gene of zebrafish is orthologous to fugu predicted protein SINFRUP-00000132871 on scaffold_7529, although the bootstrap value was not high (597/1000). In tBLASTn analysis, zebrafish ff1b hit fugu SINFRUP00000132871 better than ff1d did (e-score of 3.3e-47 compared with 1.1e-38 respectively), consistent with the phylogenetic analysis. Fugu ff1b was the only predicted protein on its scaffold, but the zebrafish ff1b gene is on connoting ctg12124 with four other genes. In tBLASTn analysis, these other genes hit strongly the human genes CYR61 (e-score 1e-73, 1p31-p22, 1_85334k), LMO4 (e-score 2e-70, 1p22.3, 1_87087k), NR6A1 (e-core 0.0, 9q33-q34.1, 9_120736k), and C11orf8 (e-score 1e-69, 11p13, 11_30471k) (Figure 2). As Figure 2 shows, NR6A1 is the nearest gene to NR5A1 in the human genome, with only 39 kb of non-transcribed DNA between them in the human genome, and NR6A1 and NR5A1 are transcribed in the same direction. Thus the nearest-neighbour test strongly supports the orthology of ff1b to NR5A1. As Figure 2 shows, however, the zebrafish orthologue of human C11orf8 lies between ff1b and nr6a1a. Furthermore, the orientation of ff1b and nr6a1a are convergent, rather than in the same direction as in humans. This arrangement is to be expected if the original gene order was …→nr6a1a→ff1b→…c11orf8→…, and then an inversion occurred which made the new order …→nr6a1a→…←c11orf8← ff1b←. The orientation may also be due to problems with the genome assembly.
Figure 2. Comparison of human chromosome Hsa9 and zebrafish chromosomes DreLG8 and DreLG21.
The gene segments are shown as thick lines and gene orientations are indicated by arrows. Zebrafish ff1b and ff1d, and human NR5A1 are all located next to NR6A1. Besides, ff1d and NR5A1 are in the same gene orientation syntenic with several similar genes.
The phylogenetic and tBLASTn analysis suggest that ff1d is a duplicate of the ff1b gene. This conclusion is also supported by the genomic analysis for zebrafish (an orthologue of ff1d was not found in the current fugu genomic database). The orthologues of five human genes in addition to ff1d are on the zebrafish contig ctg11365.1, and all are contiguous in human as well (Figure 2). These genes include KIAA1608 (e-score 0.0, 9q34.11, 9_119595k), LHX2 (e-score 6e-70, 9_120230k), NEK6 (e-score 2e-24, 9q33.3-q34.11, 9_120472k), PSMB7 (e-score e-121, 9q34.11-q34.12, 9_120569k), and NR6A1 (e-score 5e-47, 9q33-q34.1 9_120734k). The gene order and transcription direction of this set of six genes has not changed in the 450 million years since the divergence of zebrafish and human lineages. Thus this genomic arrangement very strongly supports both the assignment of ff1d as a zebrafish orthologue of human NR5A1, and the conclusion that ff1d and ff1b are gene duplicates because of their linkage to nr6a1a and nr6a1b, which the phylogeny suggests are themselves duplicates (see Supplementary Figure S5 at http://www.BiochemJ.org/bj/389/bj3890019add.htm).
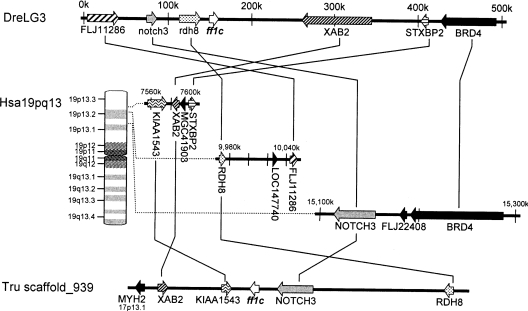
The phylogeny (Figure 1) showed that ff1c of both zebrafish and fugu fell as an outgroup to the NR5A1 and NR5A2 clades. Because ff1c is found in fugu and zebrafish, whose lineages separated early in the teleost radiation, it is probably found in most teleosts, which constitute half of vertebrate species. The absence of this gene in tetrapods raises the question of why ff1c has been retained for hundreds of millions of years in ray-fin fish, but lost in the lobe-fin lineage leading to humans. The genomic analysis of the regions surrounding ff1c in both zebrafish (contig ctg30266) and fugu (scaffold_939) shows orthology of this chromosome segment to a portion of Hsa19p13 near NOTCH3 (Figure 3). Several groups have noted that the four human NOTCH paralogues anchor four paralogous chromosome segments including parts of chromosomes 1, 6, 9 and 19 [26,27] that probably arose by two successive rounds of duplication, probably during two whole-genome-duplication events hypothesized to have occurred at about the origin of vertebrates [7]. Our genomic analysis suggests that the parent of this group of paralogous chromosome segments contained an ff1 gene, and that after the two rounds of duplication, there were four ff1 genes each syntenic with a NOTCH gene. Subsequently, the ff1 gene syntenic with NOTCH4 was lost before the divergence of human and teleost lineages, and the ff1c gene linked to NOTCH3 was eventually lost in the human lineage, but was retained in the teleost lineage. Thus there is no orthologue of ff1c in human. Comparison of gene orders and transcription directions between zebrafish, pufferfish and human show that several inversions have occurred in this otherwise rather well conserved region of the genome (Figure 3).
Figure 3. Comparison of human, zebrafish and fugu chromosome segments surrounding the ff1c gene.
The zebrafish chromosome segment DreLG3 is shown at the top, and the human chromosome segments homologous with the DreLG3 are shown below. A fugu gene segment corresponding to the region is shown at the bottom. NOTCH3 and/or ff1c, RDH8, XAB2 and STXBP2 can be found in linked loci in zebrafish, fugu and human.
Expression of ff1 genes during development
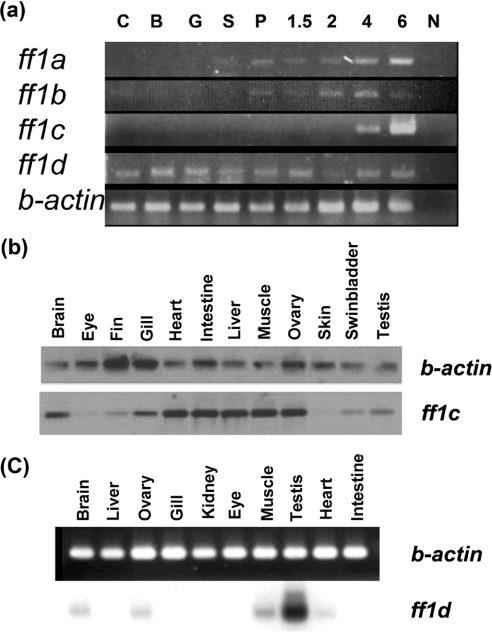
Gene functions are determined partly by their expression patterns. To understand further the relationships among different ff1 family members and to provide insights into gene functions, we carried out RT-PCR analysis to learn their expression patterns during development. Expression of ff1a and ff1b was first detected at about the segmentation stage, and continued through development (Figure 4a). The ff1c gene, however, was silent during most of the embryonic stages, and was turned on at the larval stage at approx. 4 days post-fertilization. The ff1d transcripts were present from fertilization throughout all stages of development (Figure 4a). Thus these genes were differentially expressed during development, indicating that they may have different functions.
Figure 4. Expression patterns of (a) ff1 genes during early development, and (b) ff1c and (c) ff1d in different adult tissues detected by RT-PCR.
RNA was isolated from different stages or from adult tissues, and primers for specific ff1 genes were used for RT-PCR. The abbreviations for the developmental stages are as follows: C, cleavage; B, blastula, G, gastrula, S, segmentation, P, pharyngula. 1.5, 2, 4 and 6 represent the number of days post-fertilization. N, negative control. Expressions of ff1a and ff1b genes begin during segmentation and become more evident during pharyngula. The ff1c gene is expressed starting at 4 days post-fertilization. Transcripts of ff1d start maternally at the cleavage stage and persist throughout all examined time points. Actin expression was used as a control.
The differential expression patterns of these four ftz-f1 genes were also exemplified in the adult tissues. Earlier work already showed expression of ff1a and ff1b in a variety of different tissues [20,21,28–30]. In the present study, we show that ff1c transcripts are detected in many adult tissues (Figure 4b). Thus, although ff1c is silent during the embryonic stage, after it is turned on at the larval stage around day 4, it is expressed in many adult tissues, including intestine and liver, like NR5A2, and gonads, like NR5A1. The ff1d transcripts were present in fewer adult tissues, mainly in the testis, but lower levels were also found in ovary, brain, muscle and heart (Figure 4c).
To examine expression patterns among ff1 orthologues in more detail, we performed whole-mount in situ hybridization with zebrafish embryos (Figure 5). Transcripts of ff1a were apparent in the hypothalamus and trigeminal ganglion in the head region, and in the spinal neurons, somites and endodermal cells in the trunk (Figures 5A and 5E), as reported previously [20]. This expression pattern is similar to that of mammalian Nr5a2 [4]. Expression of ff1b is more restricted than ff1a, mainly in the hypothalamus in the head and head kidney primordia in the trunk (Figures 5B and 5F), as reported previously [31]. This expression pattern is very similar to that of mouse Nr5a1 [32], supporting further the orthology of ff1b with NR5A1. The ff1c transcripts were not detected in zebrafish embryos at 26 h post-fertilization (Figures 5C and 5G), consistent with the results obtained by RT-PCR. The area of ff1d expression is quite broad, mainly in anterior and dorsal neural tissues in the head and trunk (Figures 5D and 5H). Thus the difference in the expression patterns of ff1b and ff1d indicate that they may have very different functions during embryogenesis. The partitioning of ancestral subfunctions between duplicated genes such as this is expected from the duplication, degeneration, complementation hypothesis for the evolution of gene duplicates [33].
Figure 5. Expression patterns of ftz-f1 homologues in zebrafish head (A–D) and trunk (E–H) regions 26 h post-fertilization.
In the head, both ff1a (A) and ff1b (B) are detected in the hypothalamus (hy), but the sizes of expression domains are different. In addition, ff1a is expressed in trigeminal ganglion (tg). In contrast with ff1a and ff1b, ff1c (C) expression is not detected at this stage, and ff1d (D) is ubiquitously expressed. In the trunk, ff1a (E) is detected in spinal neurons (sn), somites (sm) and endoderm. Different from wide distribution of ff1a, ff1b (F) is detected only in head kidney primordia (hk). The ff1c gene is not expressed in the trunk (G), and ff1d is ubiquitous expressed in the dorsal part of the trunk (H).
DISCUSSION
In the present paper, we describe the characterization of a new zebrafish nr5a gene and genetic mapping experiments that showed that each of the four zebrafish nr5a genes is located on a different chromosome. While teleosts have four nr5a genes, mammals have just two. Phylogenetic analyses showed that the NR5A family can be divided into the NR5A1 and NR5A2 subgroups [34]. Our studies of zebrafish show that these two groups arose before the divergence of teleost and mammalian lineages, probably in genome-amplification events at the base of vertebrate evolution. Zebrafish ff1a and human NR5A2 have highly conserved sequences, and this conservation of sequence suggests the conservation of function maintained since the teleost/tetrapod divergence 450 million years ago. Zebrafish ff1b and ff1d probably arose by the duplication of an ancestral NR5A1 gene in a genome duplication event that punctuated ray-fin fish evolution [8–12,35]. These genes have the distinctive signs of having resulted from a whole-genome-duplication event, in that they are present on paralogous chromosomes and are sisters in a phylogeny compared with the genome of a related species that did not undergo whole-genome duplication [8–12]. And ff1c probably arose in the genome-amplification events at the base of vertebrate evolution and was lost in the human lineage.
Based on our phylogenetic analysis, chromosome mapping and comparative genomics, we assign the zebrafish ff1 genes the following names: ff1a becomes nr5a2, ff1b becomes nr5a1a, ff1c becomes nr5a5 and ff1d becomes nr5a1b (Table 1). We previously called the ff1b gene nr5a4 [4,21], but the experiments reported in the present paper make, for the first time, the historical past of the genes evident, allowing us to assign more biologically meaningful names.
The assignment of ff1a to nr5a2 is very clear because data from phylogenetic analysis, chromosomal locations and expression patterns all agree that ff1a in zebrafish and NR5A2 in human are derived from a single gene in their last common ancestor. The assignment of ff1b and ff1d to nr5a1 is somewhat more difficult. Our phylogenetic analysis gave ambiguous results, indicating that ff1b could be more similar to NR5A2. The ff1b gene resides in LG8 in a region resembling both Hsa1 and Hsa9, where NR5A1 and NR5A2 reside respectively. Yet ff1b and ff1d genes appear as gene duplicates from phylogenetic analyses. A particularly telling finding is that both genes are adjacent to duplicate copies of nr6a1, whose human counterpart NR6A1 is next to NR5A1 on Hsa9. In addition, many ff1d neighbouring genes are also syntenic to NR5A1 on human Hsa9. Furthermore, our gene expression data and functional analysis also show that ff1b is most similar to mammalian Nr5a1. Expression of ff1b in the head kidney primordia and hypothalamus (Figure 5) is similar to that of mammalian Nr5a1. The function of ff1b in the development of zebrafish interrenal primordia is also similar to that of mouse Nr5a1 in the adrenal gland [29,36]. Thus, in addition to sequence homology, gene structure and chromosomal locations, the conserved expression and function between zebrafish ff1b and mouse Nr5a1 support further the notion that ff1b can indeed be classified as nr5a1a.
It has been suggested that the partitioning of ancestral subfunctions between gene duplicates can permit evolutionary changes in tissue-specific functional domains of encoded proteins [33,37], and that these tissue-specific subfunctions could provide targets for drugs that might be more tissue-specific and thus avoid some undesirable side effects [38]. Future work may reveal whether the duplicated ff1b and ff1d genes might contribute to enhanced design of drugs that target NR5A genes.
We demonstrate that the tandem arrangement of NR5A1 and NR6A1 that is found in the human lineage is shared by zebrafish, suggesting its origin by ancient tandem duplication. We propose here a model that can explain the evolutionary relationships of all these genes (Figure 6). The existence of many duplicated chromosome segments in the human genome has suggested that the vertebrate lineage experienced two rounds of whole-genome duplication, the ‘2R hypothesis’ [35,39], although some authors consider that the results could equally be explained by segmental duplication [40,41].
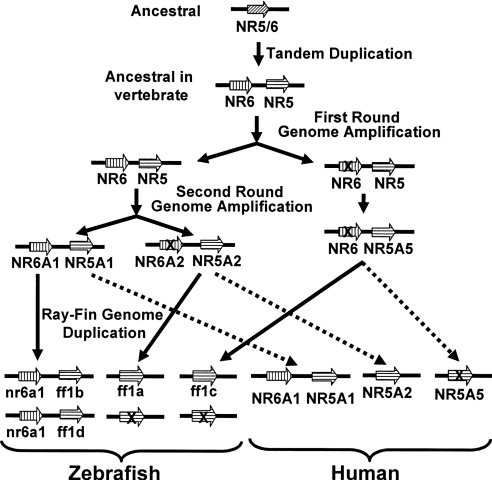
Figure 6. Model of NR5A/NR6A gene evolution.
The ancestral gene segment contains one NR5 gene. After tandem duplication, this gene became the ancestor of NR5 and NR6 in vertebrates. Two more rounds of gene amplification ensue, resulting in four sets of the gene segment. Some of the genes were subsequently lost in the lobe-fin lineage, leading to the current human genome with one NR6A1 and three NR5A genes. In the ray-fin lineage, the genome was duplicated further, followed by loss of chromosomal segments, forming zebrafish chromosomes with two nr6a1 and four ff1 genes.
Whether vertebrate genome evolves through the whole-genome-duplication or segmental-duplication event, evidence shows that the chromosome segment containing NOTCH and many other genes was duplicated in the vertebrate lineage, in the ‘1/6/9/19p paralogon’ [27,42]. The NR5A gene has just two paralogues in the human genome, with no copy currently on the Hsa19 paralogous chromosome segment. The genetic mapping and phylogenetic analysis reported in the present paper are consistent with the hypothesis that ff1c is the descendent of an NR5A gene present in the last common ancestor of human and zebrafish that has since become lost in the lineage leading to humans. Understanding the functions of the zebrafish copy of the NR5A gene missing from the human genome may be informative about evolutionary differences between members of the ray-fin fish lineage, such as zebrafish, and members of the lobe-fin fish lineage, such as human.
This phenomenon – the retention of an ancient paralogue in the fish lineage and its loss in the human lineage – has been recognized only recently. For example, an EVX paralogue lies adjacent to the mammalian HOXA and HOXD clusters, but none exists adjacent to the mammalian HOXB or HOXC clusters [43]. The chromosome segment that duplicated to give rise to the four mammalian HOX clusters undoubtedly had an EVX gene adjacent to the group-13 gene [44–46]. Because eve1 exists at this location in the zebrafish hoxba cluster [9], it must have been present adjacent to the HOXB cluster in the last common ancestor of zebrafish and human, but subsequently became lost in the human lineage. The same situation most likely happened with ff1c. We propose that many teleost genes that are termed ‘novel’ may be found, after subsequent genomic and phylogenetic analysis, to have origins like ff1c and eve1.
In addition to gene structure and chromosomal locations, gene expression patterns provide clues to the functions and evolutionary relationships of genes. We showed that the ff1a gene is expressed in many of the same organs in which mammalian NR5A2 is expressed [47]. In contrast with ff1a and ff1b, ff1c and ff1d do not have expression patterns similar to mammalian NR5A genes. For ff1c, this is probably because the gene orthologous to ff1c was lost during mammalian evolution. For ff1d, the duplicate of ff1b, regulatory sequences may have been free to change during evolution because of the maintenance of its duplicate gene ff1b, and perhaps by the retention of ff1c.
Many of the NR5A genes have multiple promoters and are differentially spliced. The zebrafish nr5a2 gene has two promoters and is alternatively spliced at the 3′-region, resulting in the formation of four isoforms with identical central protein portions, but different in their N- and C-termini [21]. Both human and mouse NR5A1 genes also have the same structure [48,49]. We find that the zebrafish ff1d gene has two different 3′-sequences (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/389/bj3890019add.htm), which is probably due to alternative splicing, as has been demonstrated for its orthologous genes in the human and mouse lineages.
Our investigation has shown that the zebrafish genome has twice the number of NR5A genes as the human genome, and that the developmental genetic functions of these genes, as reflected by their expression patterns, are different between ray-fin (zebrafish) and lobe-fin (human) lineages. Because these genes regulate important developmental functions, including the development of the gonads and adrenals, and control critical functions in the development of sterol metabolism in liver and other tissues, further investigation of the ‘extra’ genes in teleosts may reveal ray-fin-specific functions that could have been involved in the divergence of ray-fin and lobe-fin fishes.
Online data
Acknowledgments
This work was supported by grants NSC93-2321-B-001-018 from the National Science Council, and AS91IZ2PP from Academia Sinica, Republic of China to B.-c.C., grants R01RR10715 and P01HD22486 from NIH (National Institutes of Health) and IBN-9728587 from NSF (National Science Foundation), U.S.A., to J.P., and R-154-000-076-112 from National University of Singapore, Singapore, to W.K.C.
References
- 1.Maglich J. M., Sluder A. E., Willson T. M., Moore J. T. Beyond the human genome: examples of nuclear receptor analysis in model organisms and potential for drug discovery. Am. J. Pharmacogenomics. 2003;3:345–353. doi: 10.2165/00129785-200303050-00005. [DOI] [PubMed] [Google Scholar]
- 2.Gissendanner C. R., Crossgrove K., Kraus K. A., Maina C. V., Sluder A. E. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Parker K. L., Rice D. A., Lala D. S., Ikeda Y., Luo X., Wong M., Bakke M., Zhao L., Frigeri C., Hanley N. A., et al. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 2002;57:19–36. doi: 10.1210/rp.57.1.19. [DOI] [PubMed] [Google Scholar]
- 4.Fayard E., Auwerx J., Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Wolfe K. H. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 6.Hughes A. L. Phylogenies of developmentally important proteins do not support the hypothesis of two rounds of genome duplication early in vertebrate history. J. Mol. Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- 7.Holland P. W., Garcia-Fernandez J., Williams N. A., Sidow A. Gene duplications and the origins of vertebrate development. Dev. Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 8.Taylor J. S., Braasch I., Frickey T., Meyer A., Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 10.Naruse K., Tanaka M., Mita K., Shima A., Postlethwait J., Mitani H. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 2004;14:820–828. doi: 10.1101/gr.2004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature (London) 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 12.Postlethwait J. H., Yan Y. L., Gates M. A., Horne S., Amores A., Brownlie A., Donovan A., Egan E. S., Force A., Gong Z., et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 13.Kelly P. D., Chu F., Woods I. G., Ngo-Hazelett P., Cardozo T., Huang H., Kimm F., Liao L., Yan Y. L., Zhou Y., et al. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 2000;10:558–567. doi: 10.1101/gr.10.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods I. G., Kelly P. D., Chu F., Ngo-Hazelett P., Yan Y. L., Huang H., Postlethwait J. H., Talbot W. S. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hukriede N. A., Joly L., Tsang M., Miles J., Tellis P., Epstein J. A., Barbazuk W. B., Li F. N., Paw B., Postlethwait J. H., et al. Radiation hybrid mapping of the zebrafish genome. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt K. D., Maglott D. R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo I.-C., Huang C., Chung B.-c. Differential regulation of the CYP11A1 (P450scc) and ferredoxin genes in adrenal and placental cells. DNA Cell Biol. 1993;12:849–860. doi: 10.1089/dna.1993.12.849. [DOI] [PubMed] [Google Scholar]
- 18.Hwang S. P. L., Tsou M. F., Lin Y. C., Liu C. H. The zebrafish BMP4 gene: sequence analysis and expression pattern during embryonic development. DNA Cell Biol. 1997;16:1003–1011. doi: 10.1089/dna.1997.16.1003. [DOI] [PubMed] [Google Scholar]
- 19.Chiang E. F.-L., Pai C.-I., Wyatt M., Yan Y.-L., Postlethwait J., Chung B.-c. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- 20.Lin W., Wang H. W., Sum C., Liu D., Hew C. L., Chung B.-c. Zebrafish ftz-f1 gene has two promoters, is alternatively spliced, and is expressed in digestive organs. Biochem. J. 2000;348:439–446. [PMC free article] [PubMed] [Google Scholar]
- 21.Chai C., Chan W. K. Developmental expression of a novel Ftz-F1 homologue, ff1b (NR5A4), in the zebrafish Danio rerio. Mech. Dev. 2000;91:421–426. doi: 10.1016/s0925-4773(99)00312-3. [DOI] [PubMed] [Google Scholar]
- 22.Xia J. M.S. Thesis. Singapore: National University of Singapore; 2003. Characterization of zebrafish ff1c gene. [Google Scholar]
- 23.Nelson J. S. Fishes of the World. New York: John Wiley; 1994. [Google Scholar]
- 24.Allendorf F. W., Utter F. M., May B. P. Gene duplication within the family Salmonidae: II. Detection and determination of the genetic control of duplicate loci through inheritance studies and the examination of populations. In: Markert C. L., editor. Isozymes. New York: Academic Press; 1975. pp. 415–432. [Google Scholar]
- 25.Phillips R., Rab P. Chromosome evolution in the Salmonidae (Pisces): an update. Biol. Rev. Cambridge Philos. Soc. 2001;76:1–25. doi: 10.1017/s1464793100005613. [DOI] [PubMed] [Google Scholar]
- 26.Kasahara M., Nakaya J., Satta Y., Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- 27.Katsanis N., Fitzgibbon J., Fisher E. M. Paralogy mapping: identification of a region in the human MHC triplicated onto human chromosomes 1 and 9 allows the prediction and isolation of novel PBX and NOTCH loci. Genomics. 1996;35:101–108. doi: 10.1006/geno.1996.0328. [DOI] [PubMed] [Google Scholar]
- 28.von Hofsten J., Jones I., Karlsson J., Olsson P. E. Developmental expression patterns of FTZ-F1 homologues in zebrafish (Danio rerio) Gen. Comp. Endocrinol. 2001;121:146–155. doi: 10.1006/gcen.2000.7582. [DOI] [PubMed] [Google Scholar]
- 29.Hsu H. J., Lin G., Chung B. C. Parallel early development of zebrafish interrenal glands and pronephros: differential control by wt1 and ff1b. Development. 2003;130:2107–2116. doi: 10.1242/dev.00427. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y. W., Gao W., Teh H. L., Tan J. H., Chan W. K. Prox1 is a novel coregulator of Ff1b and is involved in the embryonic development of the zebra fish interrenal primordium. Mol. Cell. Biol. 2003;23:7243–7255. doi: 10.1128/MCB.23.20.7243-7255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu H. J., Hsiao P., Kuo M. W., Chung B. C. Expression of zebrafish cyp11a1 as a maternal transcript and in yolk syncytial layer. Gene Expr. Patterns. 2002;2:219–222. doi: 10.1016/s1567-133x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 32.Parker K. L., Schimmer B. P. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 33.Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellinger-Ziegelbauer H., Hihi A. K., Laudet V., Keller H., Wahli W., Dreyer C. FTZ-F1-related orphan receptors in Xenopus laevis: transcriptional regulators differentially expressed during early embryogenesis. Mol. Cell. Biol. 1994;14:2786–2797. doi: 10.1128/mcb.14.4.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer A., Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 36.Chai C., Liu Y., Chan W. K. Ff1b is required for the development of steroidogenic component of the zebrafish interrenal organ. Dev. Biol. 2003;260:226–244. doi: 10.1016/s0012-1606(03)00219-7. [DOI] [PubMed] [Google Scholar]
- 37.Hughes A. L. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. London Ser. B. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 38.Postlethwait J., Amores A., Cresko W., Singer A., Yan Y. L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Gibson T. J., Spring J. Evidence in favour of ancient octaploidy in the vertebrate genome. Biochem. Soc. Trans. 2000;28:259–264. doi: 10.1042/bst0280259. [DOI] [PubMed] [Google Scholar]
- 40.Skrabanek L., Wolfe K. H. Eukaryote genome duplication – where's the evidence? Curr. Opin. Genet. Dev. 1998;8:694–700. doi: 10.1016/s0959-437x(98)80039-7. [DOI] [PubMed] [Google Scholar]
- 41.Hughes A. L., da Silva J., Friedman R. Ancient genome duplications did not structure the human Hox-bearing chromosomes. Genome Res. 2001;11:771–780. doi: 10.1101/gr.160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasahara M. New insights into the genomic organization and origin of the major histocompatibility complex: role of chromosomal (genome) duplication in the emergence of the adaptive immune system. Hereditas. 1997;127:59–65. doi: 10.1111/j.1601-5223.1997.t01-1-00059.x. [DOI] [PubMed] [Google Scholar]
- 43.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 44.Ferrier D. E., Minguillon C., Holland P. W., Garcia-Fernandez J. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol. Dev. 2000;2:284–293. doi: 10.1046/j.1525-142x.2000.00070.x. [DOI] [PubMed] [Google Scholar]
- 45.Minguillon C., Garcia-Fernandez J. Genesis and evolution of the Evx and Mox genes and the extended Hox and ParaHox gene clusters. Genome Biol. 2003;4:R12. doi: 10.1186/gb-2003-4-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard S. L., Holland P. W. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 2000;10:1059–1062. doi: 10.1016/s0960-9822(00)00676-x. [DOI] [PubMed] [Google Scholar]
- 47.Rausa F. M., Galarneau L., Belanger L., Costa R. H. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver, intestine and pancreas. Mech. Dev. 1999;89:185–188. doi: 10.1016/s0925-4773(99)00209-9. [DOI] [PubMed] [Google Scholar]
- 48.Oba K., Yanase T., Nomura M., Morohashi K., Takayanagi R., Nawata H. Structural characterization of human Ad4bp (SF-1) gene. Biochem. Biophys. Res. Commun. 1996;226:261–267. doi: 10.1006/bbrc.1996.1343. [DOI] [PubMed] [Google Scholar]
- 49.Ninomiya Y., Okada M., Kotomura N., Suzuki K., Tsukiyama T., Niwa O. Genomic organization and isoforms of the mouse ELP gene. J. Biochem. (Tokyo) 1995;118:380–389. doi: 10.1093/oxfordjournals.jbchem.a124918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.