Abstract
We report a novel SPAG9 (sperm-associated antigen 9) protein having structural homology with JNK (c-Jun N-terminal kinase)-interacting protein 3. SPAG9, a single copy gene mapped to the human chromosome 17q21.33 syntenic with location of mouse chromosome 11, was earlier shown to be expressed exclusively in testis [Shankar, Mohapatra and Suri (1998) Biochem. Biophys. Res. Commun. 243, 561–565]. The SPAG9 amino acid sequence analysis revealed identity with the JNK-binding domain and predicted coiled-coil, leucine zipper and transmembrane domains. The secondary structure analysis predicted an α-helical structure for SPAG9 that was confirmed by CD spectra. Microsequencing of higher-order aggregates of recombinant SPAG9 by tandem MS confirmed the amino acid sequence and mono atomic mass of 83.9 kDa. Transient expression of SPAG9 and its deletion mutants revealed that both leucine zipper with extended coiled-coil domains and transmembrane domain of SPAG9 were essential for dimerization and proper localization. Studies of MAPK (mitogenactivated protein kinase) interactions demonstrated that SPAG9 interacted with higher binding affinity to JNK3 and JNK2 compared with JNK1. No interaction was observed with p38α or extracellular-signal-regulated kinase pathways. Polyclonal antibodies raised against recombinant SPAG9 recognized native protein in human sperm extracts and localized specifically on the acrosomal compartment of intact human spermatozoa. Acrosome-reacted spermatozoa demonstrated SPAG9 immunofluorescence, indicating its retention on the equatorial segment after the acrosome reaction. Further, anti-SPAG9 antibodies inhibited the binding of human spermatozoa to intact human oocytes as well as to matched hemizona. This is the first report of sperm-associated JNK-binding protein that may have a role in spermatozoa–egg interaction.
Keywords: acrosome, c-Jun N-terminal kinase, JNK-interacting protein (JIP), oocyte, sperm-associated antigen 9, spermatozoa–egg interaction
Abbreviations: ERK, extracellular-signal-regulated kinase; HSE, human sperm extract; HZI, hemizona index; JNK, c-Jun N-terminal kinase; JBD, JNK-binding domain; JIP, JNK-interacting protein; MAPK, mitogen-activated protein kinase; MHZ, matched hemizona; SPAG9, sperm-associated antigen 9; SPAG9ΔLZΔT, SPAG9 without JBD, leucine zipper and transmembrane domain; SPAG9ΔT, SPAG9 without transmembrane domain; SWM, sperm-washing medium
INTRODUCTION
Successful fertilization requires a precise series of cell–cell interactions between gametes [1–5]. The sperm surface is covered by a continuous plasma membrane that is divided into distinct domains in which functional molecules are distributed. These domains represent the compartmentalization of functions such as motility, energy production and spermatozoa–egg interacting regions [6]. Sperm surface molecules on the plasma membrane are involved in the recognition of the zona pellucida protein during the early process of spermatozoa–egg interaction (primary binding) [7]. The anterior acrosome located at the anterior head participates in the acrosome reaction, which is an indispensable event during fertilization [4]. Knowledge of the components of the acrosomal membranes and matrix is essential to understand the biology of acrosomal functions including the fusion of the outer acrosomal membrane with the plasma membrane during the acrosome reaction, the interaction of the inner acrosomal membrane with zona pellucida during penetration of the zona (secondary binding) and tertiary egg binding and fusion. Many details of sperm capacitation, spermatozoa–egg interaction and the roles of constituent proteins of the acrosome in these events are still unknown. Molecules, thus characterized as key players in these events, are candidates for targeting by rational drug design or for inclusion in a spermatozoa-based contraceptive vaccine.
In mammals, many sperm molecules display a restricted distribution that corresponds to various sperm regions and this distribution is probably critical for their function during fertilization [8,9]. A few of the signalling molecules including G proteins, tyrosine kinases, the inositol (1,4,5)-trisphosphate receptor, MAPKs (mitogen-activated protein kinases) [10–12], scaffolding protein JIP1 (JNK-interacting protein 1, where JNK stands for c-Jun N-terminal kinase) [13] have been found in the spermatozoa and oocyte; however, the biochemical networks that connect these molecules and their functions are poorly understood. Studies on islet brain-1/JIP1 in the unfertilized oocyte and the spermatozoa indicated that it might play a role during fertilization and may possibly be linked to the JNK pathways [13]. Recently, ERK (extracellular-signal-regulated kinase), a member of the MAPK family, was shown to be associated with human spermatozoa with a direct or indirect function in sperm capacitation [14].
Our previous findings have demonstrated an exclusive expression of SPAG9 (sperm-associated antigen 9) in haploid round spermatid cells during spermatogenesis in the macaque [15], baboon [16] and human [17]. By homology search in the genetic database, SPAG9 cDNA was found to be a member of Unigene cluster Hs. 129872 encoded by chromosome 17. On the basis of structural homology with JIP3, SPAG9 was earlier defined as JIP3γ scaffolding protein [18] and has been recently classified as JIP4 protein [19]. It was found that SPAG9 is structurally distinct from the previously described JIP1 [20] and JIP2 [21] proteins. The purpose of the study described in this report was to extensively characterize SPAG9 protein and to investigate a possible role of SPAG9 molecule in human spermatozoa–egg interaction.
EXPERIMENTAL
Plasmid construction
Molecular cloning of SPAG9 (GenBank® accession number X91879) was described previously [17]. To generate the prokaryotic expression plasmid DNA, a cDNA encoding a complete open reading frame of SPAG9 (comparable with 111–2410 bp of the published SPAG9, amino acid residues from 1–766) having a JBD (JNK-binding domain), a leucine zipper domain, a coiled-coil domain (coil) and a transmembrane domain was amplified using the forward primer 5′-ATGTCCATAATTATATGGAACATTTA-3′, and reverse primer 5′-TAAGTTGATGACCCATTATTAACCA-3′. The product (which contains an NcoI site at the 5′-end and an XhoI site at the 3′-end of the sense strand) was digested with NcoI and XhoI. The NcoI/XhoI fragment was inserted into NcoI/XhoI-digested pET28b(+) vector (Novagen, Madison, WI, U.S.A.) containing multiple cloning sites and a His6-tag coding sequence to obtain pET28b-SPAG9. The nucleotide sequence was confirmed by automated DNA sequencing.
To generate the mammalian expression plasmid DNA, a cDNA encoding a complete open reading frame of SPAG9 (comparable with 111–2410 bp of the published SPAG9 [17], amino acid residues from 1 to 766); a mutant SPAG9ΔT (SPAG9 without transmembrane domain; comparable with 111–2080 bp of the published human SPAG9, amino acid residues from 1 to 658; 108 amino acids deleted at the C-terminus, which has a predicted transmembrane domain, Figure 1A) and a mutant SPAG9ΔLZΔT (SPAG9 without JBD, leucine zipper and transmembrane domains; comparable with 934–2080 bp of the published human SPAG9, amino acid residues from 276 to 658; 275 amino acid residues deleted at the N-terminus, which has JBD, leucine zipper, a portion of the coiled-coil and also the 108 amino acid residues deleted at the C-terminus having its predicted transmembrane domain, Figure 1A) were amplified by PCR with the following primers: forward 5′-ATGTCCATAATTATATGGAACATTTA-3′ and reverse 5′-TAAGTTGATGACCCATTATTAACCA-3′; forward 5′-ATGTCCATAATTATATGGAACATTTA-3′ and reverse 5′-CTGTCTCTGCTGAGCTGTTGC-3′; forward 5′-ATGCTGTGAAGCAAGCCAAAC-3′ and reverse 5′-CTGTCTCTGCTGAGCTGTTGC-3′ respectively. The products (which contain a BamHI site at the 5′-end and a KpnI site at the 3′-end of the sense strand) were digested with BamHI and KpnI. The BamHI/KpnI fragments were inserted into BamHI/KpnI-digested pcDNA 3.1/Myc-His (Invitrogen, Carlsbad, CA, U.S.A.) vector to obtain pcDNA-SPAG9, -SPAG9ΔT, and -SPAG9ΔLZΔT. The nucleotide sequences were confirmed by automated DNA sequencing.
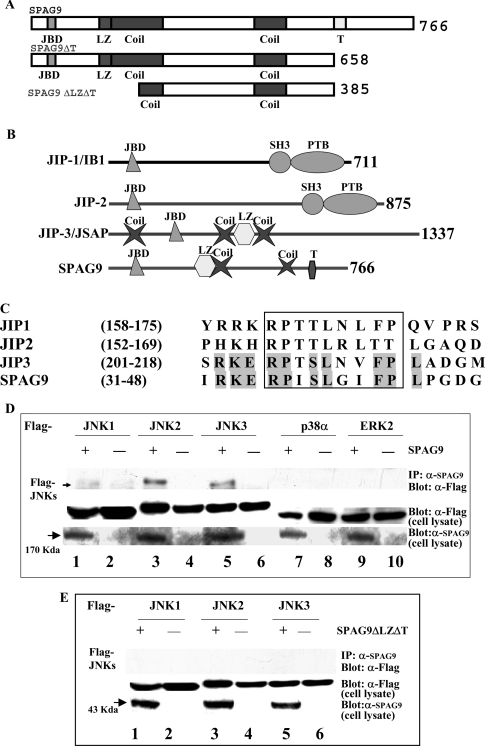
Figure 1. SPAG9 is a new member of the JIP proteins.
(A) Structure of the SPAG9 and deleted mutants are shown schematically. The native SPAG9 is 766 amino acids in length and is shown on the top. The C-terminal deletion mutant SPAG9ΔT and the truncated mutant SPAG9ΔLZΔT are shown below. (B) Schematic illustration of the domain structure of the JIP proteins. JBD, JNK-binding domain; Coil, predicted coiled coil; LZ, leucine zipper; T, predicted transmembrane domain; SH3, Src homology domain 3; PTB, phosphotyrosine binding. (C) Multiple amino acid sequence alignment of SPAG9 and other members of the JNK-binding protein family. The conserved JBD is shown in a box; sequence identity of SPAG9 with JIP3 protein is shown as shaded amino acids. Numbers on the side refer to amino acid positions relative to the first residue. (D) Binding of SPAG9 to mammalian MAPKs. COS-1 cells were co-transfected with 0.5 μg of FLAG–JNK1, FLAG–JNK2, FLAG–JNK3, FLAG–p38α and FLAG–ERK2 along with 1.5 μg of either pcDNA 3.1-Myc-His-empty vector or SPAG9. Cell lysates were immunoprecipitated using anti-SPAG9 antibody and immunoblotted with anti-FLAG monoclonal antibody to show specific interaction of JNKs. Control panels for total cell lysates are shown below. Arrow indicates the position of SPAG9 approx. 170 kDa. (E) Interaction of SPAG9ΔLZΔT and JNKs was investigated as described above which revealed no association. Arrow shows the position of SPAG9ΔLZΔT ∼43 kDa.
The MAPK expression vectors JNK1, JNK2, JNK3, ERK2 and p38α were constructed in pFlag-CMV2 (Kodak, Rochester, NY, U.S.A.) and have been described previously [22]. The expression vectors were a gift from Dr K. Yoshioka (Department of Molecular Pathology, Cancer Research Institute, Kanazawa University, Kanazawa, Japan).
Analyses of protein–protein interactions in vivo
COS-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Life Technologies, Gaithersburg, MD, U.S.A.) in a humidified incubator (5% CO2) at 37 °C. The expression vectors of MAPKs and SPAG9 or SPAG9ΔLZΔT were co-transfected into 5×105 COS-1 cells by Lipofectamine™ procedure (Life Technologies). After 40 h, cells were harvested and lysed in a lysis buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100). Expressions of MAPKs and SPAG9 in cell lysates were probed using respectively anti-FLAG monoclonal antibody Bio-M5 (Sigma) or anti-SPAG9 antibodies raised in rats. For immunoprecipitation, rat anti-SPAG9 antibodies with Protein A–agarose beads (Sigma) were used as described in [23]. The recovered fractions were separated by SDS/PAGE. After electrophoresis, proteins were transferred on to Immobilon-P (Millipore, Bedford, MA, U.S.A.) in a Bio-Rad transblot apparatus at 70 V for 2.5 h at room temperature (22–23 °C). To block non-specific sites, the membrane was treated with 5% (v/v) non-fat milk, 1% BSA and 0.5% Tween 20 in PBS for 45 min at room temperature. Blocked membrane was probed with anti-FLAG monoclonal antibody Bio-M5 (Sigma) in Tris-buffered saline having Tween 20 (0.05%) and visualized with the Amersham enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ, U.S.A.).
Purification of SPAG9 expressed in Escherichia coli and antibody production
Plasmid pET28b-SPAG9 encoding a SPAG9 His6-tagged fusion was transformed in Escherichia coli BL21 (DE3) cells by standard methods. Expression of recombinant His6-tagged SPAG9 in bacterial culture was induced with 1 mM isopropyl β-D-thiogalactopyranoside at 37 °C for 4 h. The recombinant SPAG9 protein was purified using Ni2+-nitrilotriacetate resin (Qiagen, Chatsworth, CA, U.S.A.) according to the manufacturer's instructions. Antibodies to recombinant SPAG9 were raised using alum as an adjuvant in rats and monkeys.
Microsequencing of recombinant protein
The purified protein was subjected to SDS/PAGE (10% polyacrylamide) and visualized by Coomassie Blue staining. The protein band was excised and subjected to LC-tandem MS analysis (W.M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia, VA, U.S.A). The sample was processed as described previously [24]. The spectra resulting from LC-tandem MS were analysed using Sequest (Thermoquest, Palm Beach, FL, U.S.A.) against the non-redundant and expressed sequence tag databases.
CD analysis
The recombinant protein was renatured by stepwise dialysis and was passed through a 0.45 μm filter (Millipore). The far-UV CD spectrum (JASCO-710 spectropolarimeter) of an SPAG9 protein sample (15 μM) in 10 mM Tris/HCl (pH 8.5) was recorded at 25 °C in the wavelength range of 190–250 nm.
Transfection, flow cytometry and immunofluorescence analysis
Plasmid DNA corresponding to three constructs of SPAG9 as described above and pcDNA 3.1 vector alone were purified using Qiagen DNA purification kit (Qiagen) and used for transfection of COS-1 cells by Lipofectamine™ procedure (Life Technologies). For flow cytometric analysis, COS-1 cells were seeded at a density of 3×105 cells/well in a six-well tissue culture plate 18 h before transfection. The cells were trypsinized [0.5% trypsin (Sigma) and 0.2% EDTA], 24 h after transfection with each of the three constructs of SPAG9 and pcDNA 3.1 vector alone, washed twice with PBS, and fixed with 0.4% (w/v) paraformaldehyde in PBS followed by all washings and incubations with rat anti-SPAG9 antibodies followed by goat anti-rat IgG–FITC conjugate (Jackson ImmunoResearch, West Grove, PA, U.S.A.). After the final wash, cells were resuspended in PBS and samples were run on an Elite ESP flow cytometer (Coulter Electronics, Hialeh, FL, U.S.A.) and data analysed using WinMDI (version 2.8) software. Cells stained with just secondary antibody were used to account for the background fluorescence. Flow cytometric analysis was performed as described previously [25]. Cell surface and intracellular localization of SPAG9 and deleted mutant proteins in COS-1 transfectants was examined by fluorescence immunostaining by indirect immunofluorescence microscopy. For staining of cell surface SPAG9 protein, media were removed and cells were incubated with rat anti-SPAG9 antibodies for 2–4 h at 37 °C followed by goat anti-rat IgG–FITC conjugate (Jackson ImmunoResearch). For evaluation of intracellular SPAG9 protein localization, cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Igepal (Sigma) [26] and processed for immunostaining as described above. The stained cells were observed and photographed with ECLIPSE, E 400 Nikon microscope (Nikon, Fukok, Japan).
Gel electrophoresis and immunoblotting
SDS/PAGE was performed by the method of Laemmli [27]. Denatured polyacrylamide gels (10%, v/v) under reducing conditions were used for analysing the cell lysate, culture medium, E. coli expressed proteins and HSE (human sperm extract). The protein solution was diluted with one volume of sample buffer [62 mM Tris/HCl, pH 6.8, 2% (w/v) SDS, 5% (w/v) 2-mercaptoethanol, 10% (v/v) glycerol]. The samples were heated in boiling water for 5 min and loaded on to a 10% polyacrylamide gel. After electrophoresis, proteins were transferred on to nitrocellulose membranes in a Bio-Rad transblot apparatus at 70 V for 2.5 h at room temperature. To block non-specific sites, nitrocellulose paper was treated with 5% non-fat milk, 1% BSA and 0.5% Tween 20 in PBS for 45 min at room temperature. The blocked membrane was probed with primary antibody (anti-SPAG9 antibody raised against recombinant hSPAG9 in rat) and subsequently with goat anti-rat IgG horseradish peroxidase (Jackson ImmunoResearch) as secondary antibody. The blot was developed with 0.05% 3,3′-diaminobenzidene (Sigma). Specificity of anti-SPAG9 antibodies for native SPAG9 in HSE was determined by competition experiment, which included recombinant SPAG9 (15 μg/ml) in the incubation with the primary antibody in immunoblotting procedure. To check for the presence of secreted SPAG9, cell culture supernatants were trichloroacetic acid precipitated. Immunoblotting analysis was performed as described above. Urea/PAGE was used to analyse the recombinant protein. The sample was diluted with one-half volume of sample buffer having 8 M urea, 240 mM Tris/glycine, 0.5 M EDTA, 0.1 M dithiothreitol. The sample was boiled for 3 min, centrifuged at 1250 g for 5 min and loaded on to the gel containing 8 M urea [7% (v/v) resolving gel and 3.5% (v/v) stacking gel]. The gel was run in 240 mM Tris/glycine buffer at 290 V and stained with Coomassie Blue.
Indirect immunofluorescence assay
Semen samples were collected from healthy donors and liquefied for 30 min at 37 °C. The spermatozoa were washed twice in Bigger–Whitten–Whittingham medium and centrifuged at 800 g for 10 min. The motile spermatozoa were collected by standard swim-up technique as described in [25]. Briefly, 20×106 spermatozoa fixed in methanol were incubated for 2 h at room temperature with (a) rat anti-hSPAG9 antibodies; (b) with preimmune serum and (c) with neutralized serum [neutralization experiment was performed by including recombinant hSPAG9 (15 μg/ml) in the incubation with the primary antibody] all at the same dilution of 1:500. Samples were washed and incubated with 1:2500 dilution of goat anti-rat IgG–FITC conjugate (Jackson ImmunoResearch) for 1 h at room temperature. After washing, the spermatozoa samples were observed under Nikon microscope with epifluorescence.
Immunoelectron microscopy
Human spermatozoa samples were fixed and prepared for immunoelectron microscopy as described previously [25]. Sections were treated with preimmune serum/neutralized serum [neutralization experiment was performed by including recombinant SPAG9 (15 μg/ml) in the incubation with the primary antibody] or anti-SPAG9 antibodies, washed and then incubated with rabbit anti-rat IgG labelled with 10 nm gold particles (Polysciences, IL, U.S.A.). These sections were observed with a JEOL×100C electron microscope.
Human zona pellucida binding assay using intact oocytes
This assay and hemizona assay were performed according to the guidelines of the Japan Society of Obstetrics and Gynecology. In addition, all experiments using human oocytes and spermatozoa were performed under the patient's informed consent. Human oocytes that had failed to fertilize in standard in vitro fertilization–embryo transfer treatment because of severe male infertility factor were stored at −196 °C in SWM (sperm-washing medium; Irvine Scientific, Santa Ana, CA, U.S.A.) supplemented with 1.5 M propandiol, 0.1 M sucrose and 10% (v/v) cord serum after informed consent from donors was obtained. Before freezing, it was confirmed that all the oocytes had extruded the first polar body, but not the second one, suggesting that they were in metaphase II stage but not fertilized. Preparation of spermatozoa for these experiments was isolated from semen samples obtained from fertile healthy donors. After liquefaction, semen was centrifuged and washed twice in SWM containing 5.0 mg/ml human serum albumin. The motile spermatozoa were collected by a standard swim-up technique and resuspended in SWM (2×107 spermatozoa/ml). Cryopreserved/thawed human oocytes were washed with PBS containing 0.1% BSA to remove the cryoprotectants. Human oocytes were processed as described previously [28]. Rat serum and monkey serum IgG were isolated using Nab™ Protein G spin chromatography kit (Pierce, Rockford, IL, U.S.A.) according to the manufacturer's instructions. The protein concentration was measured by reading absorbance at 280 nm. Binding assays were performed using swim-up spermatozoa pretreated with 100 μg/ml of IgG from rat or monkey anti-SPAG9 antibodies (serum dilution equivalent to 1:6400) for 15 min at 37 °C in 5% CO2 in air. Preimmune IgG (100 μg/ml) was used as negative control. After spermatozoa/antibody incubation, oocytes were added directly to the spermatozoa suspension and the gametes were co-incubated for 3 h at 37 °C in 5% CO2 in air. After gamete coincubation, the oocytes were washed with fresh Biggers–Whitten–Whittingham medium supplemented with 3 mg/ml BSA to remove the loosely bound spermatozoa to the zona pellucida by repeated pipetting. More than 50% of the spermatozoa were freely swimming at the end of the experiment. The number of spermatozoa tightly bound to the zona pellucida was counted using inverted microscope equipped with Hoffman modulation (TE 300, Nikon).
Human hemizona binding assay
The hemizona assay was performed as described previously [28]. Briefly, a human spermatozoa suspension pretreated with IgG from preimmune serum or with IgG from anti-SPAG9 antibodies was introduced in MHZ (matched hemizona). After co-incubation, an HZI (hemizona index) was calculated by counting the number of spermatozoa adhering to the MHZ in test group divided by number of spermatozoa adhering in control MHZ under a phase-contrast microscope [HZI=(number of spermatozoa adhering in test serum/number of bound spermatozoa in control serum)×100].
Chromosomal mapping
Chromosome localization was performed using SPAG9-cDNA labelled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) by nick translation as described previously [29].
Southern hybridization
Genomic DNA was isolated from human blood with QIAmp DNA blood maxiprep kit (Qiagen). Each 10 μg DNA was digested with various restriction enzymes, electrophoresed on a 1% agarose gel and transferred on to Hybond-N nylon membranes (Amersham Biosciences). The blots were analysed using radiolabelled [α-32P]-dCTP (3000 Ci/mmol; Amersham Biosciences) probe of the 5′-end of SPAG9 EcoRI/Hind III fragment (691 bp), full-length SPAG9 cDNA (2523 bp) and 3′-end of SPAG9 fragment (320 bp cloned in SacI/XhoI).
RESULTS
Domain structure of SPAG9
Running the algorithms provided in Simple modular architecture tool on the ExPASy server revealed several features: (1) a characteristic leucine zipper motif; (2) an extended coiled-coil domain (coil) and (3) a transmembrane domain (Figure 1A). Structural homology analysis with JIP proteins showed the presence of signature domains of JIP1, JIP2 and JIP3 (Figure 1B). Recent GenBank® database search indicated two entries describing sequence homology to SPAG9. A cDNA (GenBank® accession number AB028989) encodes a predicted protein that has similarity to the C-terminal region and may represent a fragment of human JIP3. A second partial human cDNA (GenBank® accession number AB011088) also encodes a protein with similarity to the C-terminus of SPAG9 but this protein is the product of a distinct gene [30] and may be a related member of the JIP3 group (JIP3β). GenBank® database searches also showed 100% homology with human protein highly expressed in testis [31], 94% homology with macaque SPAG9 [15], 92.2% with baboon SPAG9 [16], 50% with Mus musculus JIP3/JNK/JSAP1 (JSAP1 stands for stress-activated protein kinase-associated protein 1) [21], 36% with Drosophila melanogaster-Jun/SAPK (stress-activated protein kinase) Sunday driver 1 [32] and 30% with Caenorhabditis elegans coiled-coil protein [33]. The sequence homology suggests that the SPAG9 gene is conserved through evolution.
SPAG9 specifically interacts with JNK and not with other mammalian MAPKs
The amino acid sequence analysis of SPAG9 revealed the primary sequence identity with JBD of JIP. Alignment of the JBD of JIP1, JIP2, JIP3 and SPAG9 exhibited significant sequence identity (Figure 1C). Therefore it was of interest to determine whether SPAG9 could also interact with MAPKs in a similar manner. The binding specificity of SPAG9 with various MAPKs was performed in co-transfection experiments (Figure 1D). We transiently expressed Myc-His-tagged full-length SPAG9 or SPAG9ΔLZΔT with FLAG epitope-tagged JNK1, JNK2, JNK3, ERK2 or p38α in COS-1 cells. The His-tagged proteins were recovered from the cell extracts by affinity binding with anti-SPAG9 antibodies and precipitates were examined for the presence of the MAPKs by immunoblotting with an anti-FLAG antibody. JNK1, JNK2 and JNK3 interacted with SPAG9, although no or very low binding of p38α or ERK2 was observed. JNK3 and JNK2 showed higher binding affinity to SPAG9 compared with JNK1. However, the SPAG9ΔLZΔT construct lacking JBD did not show any interaction with the JNK pathway (Figure 1E), suggesting that the JBD of SPAG9 is involved in JNK interaction.
Chromosomal localization of the SPAG9 gene and gene structure
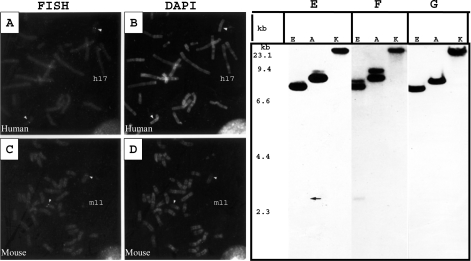
The fluorescence in situ hybridization analysis showed double consistent fluorescent signal on human chromosome 17 at q21.33 and on mouse chromosome 11 at band C (Figures 2A and 2C). Southern hybridization studies revealed similar sizes and pattern of hybridizing bands when probed with full-length SPAG9, 5′- and 3′-ends of SPAG9 cDNA suggesting that human genome contains a single copy of SPAG9 gene (Figures 2E–2G). A Blast search revealed the complete SPAG9 gene sequence on human chromosome 17 (GenBank® accession number AC005920). The gene sequence analysis further revealed that the SPAG9 gene has 19 exons and the positions of 19 exons were mapped based on cDNA sequence with consensus splice sites. Sequence analysis of the intron–exon junctions (see Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890073add.htm) revealed that all the splice sites observed the GT-AG paradigm. The exon sequence length varied from 39 to 333 bp. A homology search on the NCBI database revealed that a nucleotide sequence of SPAG9 was assigned in the UniGene cluster Hs. 129872. This gene cluster is shown to be encoded by chromosome 17q21, which supports our chromosomal localization.
Figure 2. Chromosomal localization of the SPAG9 gene.
Mapping of SPAG9 in human and mouse chromosome by fluorescence in situ hybridization. Human and mouse metaphase spread hybridized with SPAG9 cDNA. The signals are visible on both copies of chromosomes. (A) Arrow heads; human chromosome 17 (h17). (C) Arrow heads; mouse 11C (m11). (B, D) DAPI (4,6-diamidino-2-phenylindole) banding of human (arrows h17) and mouse chromosomes (arrows m11). (E–G) Southern blotting of SPAG9 gene after digestion with different restriction endonucleases and probed with different SPAG9 cDNA probes. (E) The 5′-end of SPAG9 cDNA, (F) full-length SPAG9 cDNA and (G) the 3′-end SPAG9 cDNA. Identical band numbers, pattern and the sizes indicate that the human genome contains a single copy SPAG9 gene (arrow indicates the location of faint band). The markers on the left represent a ladder λDNA/HindIII fragments (Gibco BRL, Gaithersburg, MD, U.S.A.). Abbreviations: E, EcoRV; A, ApaI; K, KpnI.
Characterization of the recombinant SPAG9
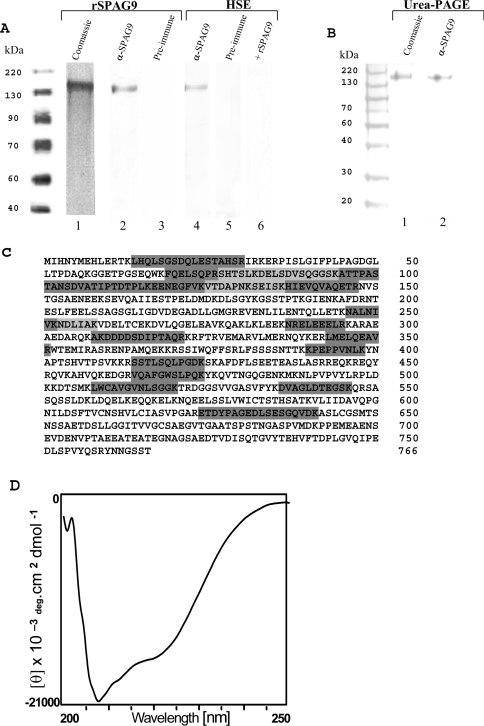
Affinity-purified recombinant SPAG9 expressed in E. coli revealed a major band at approx. 170 kDa in an SDS/PAGE (10% polyacrylamide) under reduced conditions (Figure 3A, lane 1). Since the deduced molecular mass of SPAG9 is 83.9 kDa, the approx. 170 kDa molecular mass is apparently due to the dimerization of the protein. To find if the protein would run as a monomer in the presence of urea, we performed a urea/PAGE. However, as shown in Figure 3(B, lane 1), the protein showed an apparent mobility of 170 kDa even in urea/PAGE. In immunoblot analysis, the antibodies to recombinant SPAG9 specifically recognized the recombinant protein, showing a single band of approx. 170 kDa (Figure 3A, lane 2, Figure 3B, lane 2 urea/PAGE). The preimmune serum did not recognize the recombinant SPAG9 (Figure 3A, lane 3). In the Triton X-100 solubilized HSE, the anti-SPAG9 antibodies recognized native protein of approx. 170 kDa in the Western-blot procedure (Figure 3A, lane 4). The specific band was not recognized by the preimmune serum (Figure 3A, lane 5). Competition experiment was performed by including recombinant SPAG9 in the incubation with the primary antibody, which resulted in loss of immunoreactivity with native SPAG9 in HSE (Figure 3A, lane 6). The affinity-purified SPAG9 (Figure 3A, lane 1) was subjected to MS analysis. The monoisotopic mass of 83.788 kDa is very close to the deduced value of 83.9 kDa for SPAG9. Furthermore, MS sequence analysis of 17 different internal peptide regions encompassing 214 amino acids (Figure 3B) reconfirmed the protein sequence reported earlier (NCBI accession number gi/4504525/CAA, 83.9 kDa, pI 4.94, SPAG9). Proper folding and secondary structure of SPAG9 protein was investigated by the CD spectrum analysis. The CD spectra exhibited minima at 220 and 208 nm, which is characteristic of an α-helical confirmation (Figure 3C). The helical content calculated from CD spectra was found to be 40.2%, which was in agreement with the theoretical predictions of 41.38% [34,35].
Figure 3. Characterization of SPAG9 protein.
Western-blot analysis of rSPAG9 and HSE using denatured SDS/PAGE gel under reducing conditions. (A) Affinity-purified rSPAG9 stained with Coomassie Brilliant Blue is shown in lane 1. Western blotting of rSPAG9 (lane 2) and HSE (lane 4) show a specific band of approx. 170 kDa. Preimmune control revealed no reactivity (lanes 3 and 5). Specificity of HSE is shown in lane 6 by including recombinant SPAG9 (15 μg/ml) in the incubation with primary antibody, which resulted in loss of immunoreactivity with native SPAG9 in HSE proteins. (B) Urea/PAGE of rSPAG9. Coomassie Blue staining (lane 1) and Western blotting of rSPAG9 (lane 2) show a specific band of approx. 170 kDa. Prestained protein molecular-mass standards (BENCHMARK, Gibco BRL). (C) Primary sequencing of SPAG9 showing the peptide stretches (light and dark grey shaded) that matched with published SPAG9 (EMBL Nucleotide Sequence Database accession no. X91879) protein identified by MS. (D) CD of the purified and refolded recombinant SPAG9 protein exhibited minima at 220 and 208 nm.
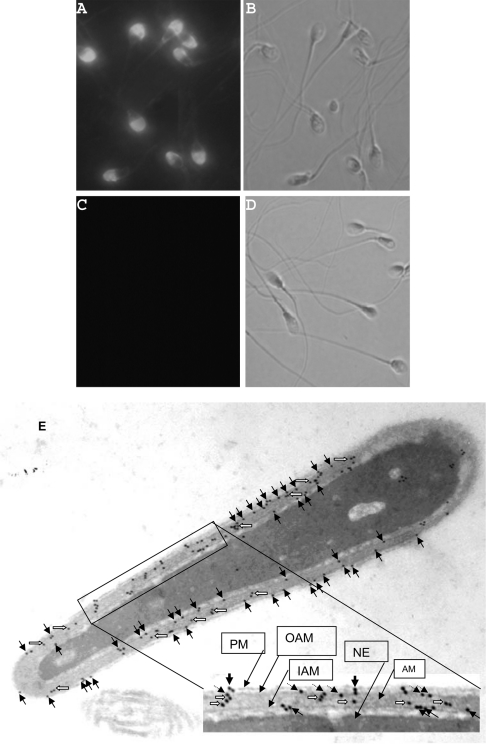
Localization of SPAG9 in human spermatozoa
Immunofluorescence studies on fixed human swim-up spermatozoa with anti-SPAG9 antibodies revealed a selective localization of SPAG9 to the acrosomal compartment of the sperm head (Figure 4A). The preimmune or neutralized serum failed to show any reactivity with any region of the spermatozoa (Figure 4C). A direct evidence of the site of localization was provided at ultra structural level by electron microscopy. The studies on intact spermatozoa demonstrated that the SPAG9 protein was associated with the plasma membrane, the outer and inner acrosomal membranes, although some of the gold particles were also seen in the acrosome matrix (Figure 4E). In all instances, the preimmune serum showed background levels of immunoreactivity (results not shown).
Figure 4. Localization of SPAG9 in human spermatozoa.
Epifluorescent photomicrograph showing the immunofluorescence pattern of the human spermatozoa. (A) Rat anti-SPAG9 antibody reacted strongly with the acrosomal compartment of sperm head. (C) Preimmune or neutralized serum did not react with any of the region of the spermatozoa. The phase-contrast pictures of (A) and (C) are (B) and (D) respectively (original magnification was ×630). (E) Immunoelectron microscopy of human spermatozoa. Longitudinal section of sperm head revealed immunogold labelling of SPAG9 over the acrosomal compartment. The arrows indicate gold particles associated with the plasma membrane (solid arrows), outer acrosomal membrane (dotted arrows), inner acrosomal membrane (solid arrows) and acrosomal matrix (open arrows). PM, plasma membrane; OAM, outer acrosomal membrane; IAM, inner acrosomal membrane; AM, acrosomal matrix (original magnification was ×45000).
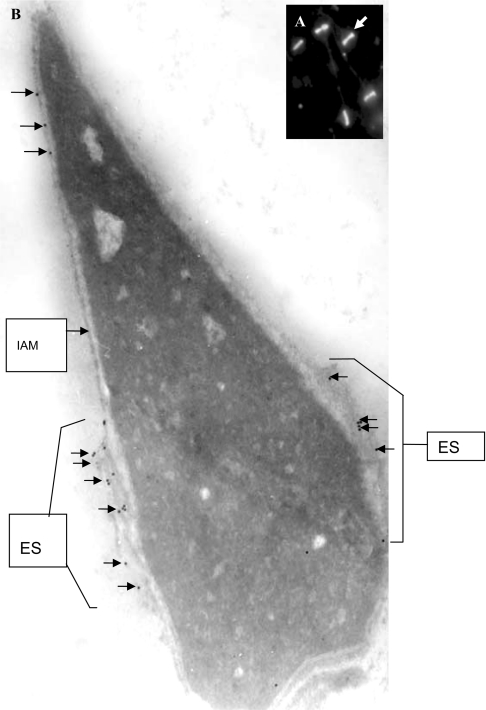
In immunofluorescence studies on human spermatozoa after stimulation with calcium ionophore A23187, which induced the acrosome reaction in a high percentage of live spermatozoa, anti-SPAG9 antibodies stained mainly on the equatorial segment of the central region of the sperm head (Figure 5A). Localization of the equatorial region was further confirmed at ultrastructural level by electron microscopy, which revealed that the SPAG9 protein was mainly localized at the equatorial region of the acrosomal compartment, although a few gold particles were also seen on the inner acrosome membrane (Figure 5B). In all instances, the preimmune serum showed background levels of immunoreactivity (results not shown).
Figure 5. Immunolocalization of SPAG9 in acrosome-reacted spermatozoa.
Immunolocalization staining showing equatorial localization of SPAG9 in the human sperm head. (A) Indirect immunofluorescence light microscopic image (arrows, original magnification was ×630). (B) Electron micrograph (longitudinal section) (arrows, original magnification was ×67000). Few gold particles are also shown to be associated with the inner acrosomal membrane (arrows). IAM, inner acrosomal membrane; ES, equatorial segment.
Expression of SPAG9 protein in mammalian cells
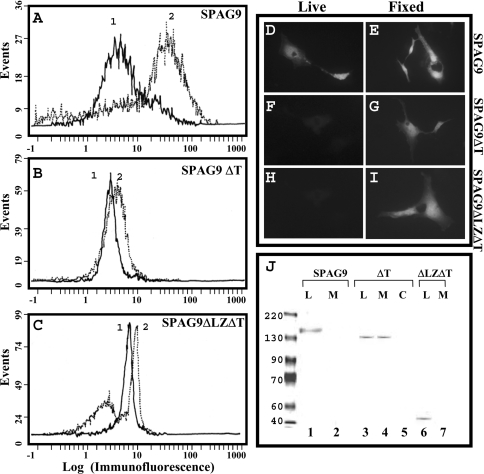
We investigated the role of the predicted leucine zipper and transmembrane domain of SPAG9 by transfection studies in COS-1 cells. Fluorescence-activated cell-sorter analysis of COS-1 cells transfected with SPAG9 having leucine zipper and transmembrane domain revealed a surface localization for SPAG9 (Figure 6A). Peak 2 shows displacement of fluorescence on the x-axis representing surface binding of anti-SPAG9 antibodies when compared with peak 1 in vector alone (Figure 6A). However, no or very low surface distribution shown as displacement of fluorescence on x-axis for both SPAG9ΔT and SPAG9ΔLZΔT was observed (Figures 6B and 6C, reactivity with anti-SPAG9 antibodies for surface localization, peak 1: pcDNA vector alone; peak 2: pcDNA-SPAG9ΔT or SPAG9ΔLZΔT), suggesting that leucine zipper and transmembrane domains are required for secretion and proper anchoring. In addition, the surface and cellular location of the expressed SPAG9 and its deleted mutants were determined employing indirect immunofluorscence microscopy (Figures 6D–6I). Approx. 2 days post-transfection, COS-1 cells were stained with rat anti-SPAG9 antibodies. When live cells were stained (no fixation/permeabilization step), cell surface localization of SPAG9 in COS-1 transfectants was detected (Figure 6D), whereas there was no detectable cell surface staining in COS-1 SPAG9ΔT (Figure 6F) or COS-1 SPAG9ΔLZΔT (Figure 6H) transfectants, suggesting a role for the leucine zipper and transmembrane domain in surface localization. There was no visible staining of COS-1 cells transfected with vector alone (results not shown). For intracellular localization, COS-1 transfectants were fixed, permeabilized and immunostained with rat anti-SPAG9 antibodies. As expected, cytoplasmic localization of SPAG9 protein in COS-1 cells transfected with SPAG9 (Figure 6E), SPAG9ΔT (Figure 6G) and SPAG9ΔLZΔT (Figure 6H) was observed. No staining was detected with pcDNA vector alone (results not shown). Immunoblotting analysis further confirmed the pattern of expression in all three constructs (Figure 6J). In cells transfected with SPAG9 (Figure 6J, lane 1) and SPAG9ΔLZΔT (Figure 6J, lane 6), the protein was detected only in the cell lysates and not in the medium. However, in cells transfected with SPAG9ΔT (Figure 6J, lanes 3, 4), the protein was detected in the medium as well as in the cell lysates suggesting the potential role for the leucine zipper in directing the protein to the surface and transmembrane domain for anchoring the protein. In addition, the deduced amino acid sequence of SPAG9 predicted a molecular mass of 83.9 kDa, whereas immunoblotting analysis revealed that it runs at an anomalous position corresponding to approx. 170 kDa. However, SPAG9ΔLZΔT, which lacks JBD, leucine zipper and the transmembrane domain revealed immunoreactivity with a protein band of approx. 43 kDa in immunoblotting, which is in agreement with the deduced mass of 42.5 kDa. In addition, SPAG9ΔT, which lacks a transmembrane domain revealed immunoreactivity with a protein band of apparent molecular mass of approx. 140 kDa in immunoblotting as against the deduced mass of 72.7 kDa. These deletion experiments indicated that the N-terminal region (residues 1–275) of SPAG9 was involved in the anomalous migration of SPAG9/SPAG9ΔT in SDS/PAGE.
Figure 6. Flow cytometric, immunoflourescence microscopy and immunoblot analysis of SPAG9 and its deletion mutants in COS-1 cells.
(A–C) Flow cytometric analysis of SPAG9, SPAG9ΔT and SPAG9ΔLZΔT constructs. Peak 1, pcDNA 3.1 vector control. Peak 2 in each construct reveals displacement of fluorescence on the x-axis representing surface binding of anti-SPAG9 antibodies showing surface location of SPAG9. Note that only SPAG9 transfected cells revealed surface localization (A). (D–I) Fluorescence microscopy of cells transfected with SPAG9, SPAG9ΔT and SPAG9ΔLZΔT constructs showing surface localization of SPAG9 in live and fixed (permeablized) cells (original magnification was ×500). Only SPAG9 transfected cells revealed surface localization (D). (J) Immunoblot analysis of SPAG9 and its mutants (SPAG9ΔT, SPAG9ΔLZΔT) in cell lysates (L) and culture medium (M). Note the secretion of SPAG9ΔT in the medium fraction. Lane 5, pcDNA vector cell lysate control (C). It may be noted that the SPAG9 protein (lane 1) and SPAG9ΔT (lanes 3 and 4) move with anamolous mobility corresponding to an apparent molecular mass of approx. 170 and 140 kDa respectively. However, SPAG9ΔLZΔT (lane 6), the deleted mutant lacking JBD, leucine zipper and the transmembrane domain, runs according to its deduced molecular mass of approx. 43 kDa. Prestained protein molecular-mass standards (BENCHMARK, Gibco BRL) were used.
Role of SPAG9 in human spermatozoa–oocyte interaction
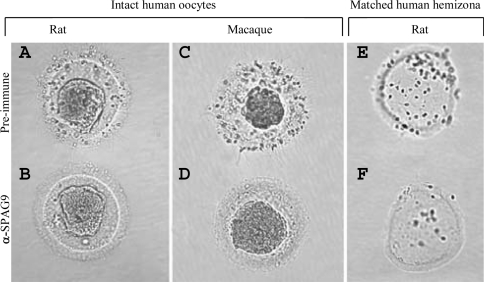
Inhibition of spermatozoa attachment and tight binding with intact oocytes was observed in spermatozoa treated with IgG from anti-SPAG9 antibodies from rat (100% inhibition) and macaque (98% inhibition) [Figures 7B and 7D, Table 1(i)], whereas spermatozoa incubated with IgG from preimmune serum did not inhibit the spermatozoa attachment with zona pellucida [Figures 7A and 7C, Table 1(i)]. In MHZ assays, spermatozoa treated with IgG from anti-SPAG9 antibody from rat (90% inhibition) and macaque (88% inhibition) also revealed decrease in spermatozoa attachment and tight binding [Figure 7F, Table 1(ii)] as compared with spermatozoa treated with IgG from preimmune serum [Figure 7E, Table 1(ii)].
Figure 7. Role of SPAG9 in human spermatozoa binding to human zona pellucida.
Intact human oocytes were co-incubated with spermatozoa preincubated with (A) IgG from rat preimmune serum, (B) IgG from rat anti-SPAG9 antibody, (C) IgG from the macaque preimmune serum and (D) IgG from macaque anti-SPAG9 antibody. Note the inhibition in binding of spermatozoa to oocytes (B, D). (E, F) Spermatozoa binding in matched human hemizona. Note the inhibition of binding of spermatozoa in MHZ by rat anti-SPAG9 antibody (F).
Table 1. Effects of rat and monkey antibodies (IgG) against SPAG9 on human spermatozoa binding to intact oocytes (i) and in hemizona (ii).
DISCUSSION
In mammals, fertilization is completed by the direct interaction of spermatozoa and eggs, a process mediated primarily by gamete surface proteins. A two-step model for the spermatozoa–egg interaction has been proposed. The first step is based on plasma membrane interactions, the second step on introduction of spermatozoa factor(s) into the egg cytoplasm [36]. On the basis of the model, it is probable that both physiological processes may be involved in fertilization. On following the acrosome reaction further, the equatorial region is known to come directly in contact with the egg, leading to the later events of egg penetration. Our findings indicated that the SPAG9 acrosomal molecule is not only restricted to acrosomal compartment but also persists in the equatorial segment of the post-acrosome reaction. Such maturation-associated modifications are considered to be important for spermatozoa–egg interaction.
We have identified a spermatozoa-specific SPAG9 protein, classified as JIP4 [19] which is structurally related to JIP3. The members of JIP3 group include the JIP3a and JIP3b proteins, which appear to represent the protein products of alternatively spliced transcripts derived from the JIP3 gene. The other JIP3 group of proteins includes JIP3β [18], which is also structurally related to JIP3 [17,30]. Both JIP3 and JIP3β [30] are expressed in the brain, whereas SPAG9 (JIP4) was shown to be expressed in the testis [15–17,31]. Although many JIP proteins are related in structure and/or function, the genes that encode these proteins are not closely linked in the genome. The human JIP1 and JIP2, which are related in function, are located on the human chromosome 11(11p 11.2-p12, 12 exons) and human chromosome 22 (22q13, 12 exons) respectively [37,38], whereas the human JIP3 gene is located on the human chromosome 16 (16p13.3, 30 exons). In contrast, the SPAG9 gene maps on to human chromosome 17 (17q21.33) and contains 19 exons, which has synteny with mouse chromosome 11-band C.
Co-transfection studies in COS-1 cells indicated that SPAG9 exhibits different binding affinities with a variety of mammalian MAPKs. Thus SPAG9 co-precipitated with JNK1, JNK2 and JNK3 but not with ERK2 or p38α MAPKs. JNK3 and JNK2 showed higher binding affinity to SPAG9 compared with JNK1. These binding interactions are similar to those of JIP1, JIP2 and JIP3, which also bind JNK [18,21,22]. Furthermore, a SPAG9 mutant lacking JBD failed to interact with the JNK pathway. These results are in agreement with recent reports of JIP3/JSAP1 [22] that demonstrated a direct interaction between the JNK binding region and the JNK pathway. Since SPAG9 and JIP3 exhibited identical amino acid sequences in the JNK binding region, it is not surprising that both these proteins showed similar binding interactions with JNK [22].
Experimental evidences have shown that JIP proteins can form oligomeric complexes, which may be dimers or higher order aggregates. It is probable that the coiled-coil nature of JIP proteins makes them assemble as multimeric complexes for specific functions [23]. Sequence analysis of SPAG9 showed two coiled-coil structures with an embedded leucine zipper motif. The leucine residues have a special function in leucine zipper dimerization and form the interface between the two α-helices in a coiled-coil structure. Far-UV CD spectrometry analysis supported this observation and revealed a predominant α-helical structure of SPAG9 (40.2%) that is in agreement with predictions from secondary structure analysis (41.38%) [34,35]. Our deletion analysis indicated that the N-terminal region (residues 1–275) of SPAG9, which included an extended coiled-coil and leucine zipper domain, was sufficient for aggregation of the SPAG9 molecules (Figure 6J). Further, we showed that both the expressed recombinant SPAG9 and native SPAG9 in HSE form aggregates, as they migrate with an apparent molecular mass of approx. 170 kDa on SDS/PAGE. It may be further noted that the apparent mobility of this protein remains invariant at approx. 170 kDa under both reducing and non-reducing conditions in SDS/PAGE (10% polyacrylamide). Anomalous retardation seen in SDS/PAGE is probably either due to extreme anomaly in the shape of the monomer or a consequence of the strong aggregation tendency of the monomers under the condition of electrophoresis. It appears that the anomalous migration of SPAG9 may not be dependent on disulphide bonds but may rather have its origins in strong hydrophobic interactions including the leucine zipper, the transmembrane domain and the coiled coils, which are the hallmarks of this protein. This is corroborated by the fact that SPAG9ΔLZΔT, which lacks JBD, leucine zipper and transmembrane domains runs on SDS/PAGE (10% polyacrylamide) gel true to its molecular mass. The anomalous mobility persists even in 8 M urea gel, where it shows an apparent mobility of approx. 170 kDa. The observation of a dominant band at 170 kDa in PAGE and the observation of a single peak at approx. 84 kDa in mass spectral analysis suggests that aggregation may be solvent-driven and may not occur under the MS conditions. The high-order aggregates of SPAG9 may be significant, since similar complexes have been detected for other members of the family of scaffold proteins, including JIP1 [20], JIP2 [21] and JIP3 [22]. Thus it is quite probable that the SPAG9 may be involved in the JNK pathway.
To investigate the function of the leucine zipper and transmembrane domains for proper localization of SPAG9, we constructed deletion mutants of SPAG9, namely SPAG9ΔLZΔT and SPAG9ΔT and examined the involvement of leucine zipper and transmembrane domain in cellular localization by transfected COS-1 cells (which do not express endogenous JIP1, JIP2 and JIP3). Indirect immunofluorescence studies in live COS-1 cells transfected with SPAG9 demonstrated the surface localization of SPAG9 protein whereas, no surface localization was observed for both SPAG9ΔT and SPAG9ΔLZΔT. This observation was further strengthened by flow cytometric analysis, which revealed a distinct surface localization for SPAG9 and no or very low surface distribution for both SPAG9ΔT and SPAG9ΔLZΔT, supporting the hypothesis that the leucine zipper domain and transmembrane segment are involved in proper localization of SPAG9. However, for the deletion mutant SPAG9ΔT having a leucine zipper but no transmembrane domain, the protein was secreted and was detected in a medium supporting the role of leucine zipper in the secretion of protein. A similar observation was reported in Chinese-hamster ovary cells and in a yeast expression system, using leucine zipper motifs as a replacement of the hydrophobic transmembrane region, which allowed the assembly and secretion of recombinant HLA-DR2 molecules (where HLA stands for human histocompatibility leucocyte antigen) [39]. Although SPAG9 lacks a signal sequence, it was found to be anchored and secreted. In this context, basic fibroblast growth factor-2 [40] and pro-inflammatory cytokine interleukin-1β [41] are also shown to be secreted even though they lack a signal peptide. In recent studies, highly enriched spermatozoa membrane proteins (plasmalemma and inner and outer acrosomal membranes) were separated by a two-dimensional electrophoresis technique and were identified using matrix-assisted laser desorption ionization-MS and peptide mapping. In support of our investigation, this study also found that SPAG9 was associated with enriched spermatozoa membrane proteins [42]. Thus our studies demonstrate that the leucine zipper domain and the transmembrane segment are involved in transport and membrane anchoring respectively. These results are also in agreement with a report that demonstrates a direct interaction between the N-terminal half of JIP3 [32] and suggested that JIP3 is a transmembrane protein that provides a direct link between kinesin and the membrane.
Immunofluorescence and electron microscopic studies of ejaculated spermatozoa showed that SPAG9 is specifically located in the acrosome and hence is an acrosomal molecule. However, indirect immunofluorescence study in acrosome-reacted sperms showed the presence of SPAG9 exclusively in the equatorial region of the sperm head. In addition, SPAG9 was also shown to be associated with the inner acrosomal membrane in acrosome-reacted spermatozoa in the electron microscopic study. A possible explanation for this localization is that during the acrosome reaction, remodelling and rearrangement of spermatozoa protein takes place. It is quite probable that some of the SPAG9 protein seen in the inner acrosomal membrane may be the residue of such a rearrangement process. The human in vitro spermatozoa–egg binding and hemizona binding assays demonstrated that the antibody raised against SPAG9 inhibited binding of the spermatozoa with zona pellucida, which suggested that SPAG9 may have a role in spermatozoa adherence and/or in the subsequent fertilization process. Since human studies involving oocytes for spermatozoa penetration assay have ethical problems, the zona-free hamster egg penetration assays were performed using anti-SPAG9 antibodies. A significant inhibition (96.5%, P<0.0001) of human spermatozoa penetration was observed in zona-free hamster egg assay [46]. Considering the high concentrations of SPAG9 in the anterior acrosome and its equatorial localization, it was not surprising to get such a high inhibition of spermatozoa binding. It is appropriate to add here that an SAMP14 (sperm acrosomal membrane protein 14) [43], which has an equatorial distribution post-acrosome reaction, has also been shown to be involved in spermatozoa–egg interactions.
Although a few potential candidate binding partners for spermatozoa–egg interactions have been described, the precise roles of these molecules have not been established [44,45]. Hence, the identification of such molecules that participate in gamete interactions, particularly those retained after the remodelling events of acrosomal exocytosis, could greatly enhance the understanding of this process. SPAG9 is an attractive addition to this group. Finally, the results obtained in the present study pose the question: what could be the requirement for the regulation of MAP kinase signalling and for the MAP kinase pathway in the haploid germ cell? For much of spermiogenesis, spermatids and spermatozoa are transcriptionally inactive as a consequence of nuclear condensation; yet the SPAG9 protein persists and is incorporated into spermatozoa. As a JIP family member, SPAG9 merits further evaluation as a JNK scaffolding protein in the upstream of MAPKs module that is retained on the equatorial compartment after acrosomal exocytosis. Therefore SPAG9 protein, in addition to mediating adherence, may also be involved in transducing signals during the gamete fusion, which needs to be addressed in future studies.
Online data
Acknowledgments
We thank Professor S. K. Basu, Director, National Institute of Immunology (New Delhi, India) for his constant encouragement of this work. We are especially grateful to Dr N. E. Sherman (Biomolecular Research Facility, University of Virginia Health System, Charlottesville, VA, U.S.A.) for technical assistance with MS. We thank Dr D. Sahal and Dr V. Kumar (International Centre for Genetic Engineering and Biotechnology, New Delhi, India) and Dr R. Pal (National Institute of Immunology) for helpful discussions and a critical reading of this paper. We acknowledge Dr K. Yoshioka for gifts of pFlag-CMV2-JNK1, -JNK2, -JNK3, -ERK2 and p38α expression vectors. This work was supported by grants from the Department of Biotechnology, Government of India, Mellon Foundation and CONRAD, U.S.A., Indo-U.S. Programme on Contraceptive Reproductive Health Research (CRHR).
References
- 1.Evans J. P., Florman H. M. The state of the union: the cell biology of fertilization. Nat. Cell Biol. 2002;4(Suppl.):S57–S63. doi: 10.1038/ncb-nm-fertilityS57. [DOI] [PubMed] [Google Scholar]
- 2.Singson A., Zannoni S., Kadandale P. Molecules that function in the steps of fertilization. Cytokine Growth Factor Rev. 2001;12:299–304. doi: 10.1016/s1359-6101(01)00013-2. [DOI] [PubMed] [Google Scholar]
- 3.Vacquier V. D. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- 4.Yanagimachi R. Mammalian fertilization. In: Knobil E., Neill J. D., editors. The Physiology of Reproduction. New York: Raven Press; 1994. pp. 189–317. [Google Scholar]
- 5.Talbot P., Shur B. D., Myles D. G. Cell adhesion and fertilization: steps in oocyte transport, sperm-zona pellucida interactions, and sperm-egg fusion. Biol. Reprod. 2003;68:1–9. doi: 10.1095/biolreprod.102.007856. [DOI] [PubMed] [Google Scholar]
- 6.Travis A. J., Kopf G. S. The spermatozoon as machine: compartmentalized metabolic and signaling pathways bridge cellular structure and function. In: DeJonge C., Barratt C. L. R., editors. Assisted Reproductive Technology. Cambridge, U.K.: Cambridge University Press; 2002. pp. 26–39. [Google Scholar]
- 7.Wassarman P. M., Litscher E. S. Sperm-egg recognition mechanisms in mammals. Curr. Top. Dev. Biol. 1995;30:1–19. doi: 10.1016/s0070-2153(08)60562-1. [DOI] [PubMed] [Google Scholar]
- 8.Primakoff P., Myles D. G. A map of the guinea pig sperm surface constructed with monoclonal antibodies. Dev. Biol. 1983;98:417–428. doi: 10.1016/0012-1606(83)90371-8. [DOI] [PubMed] [Google Scholar]
- 9.Ramalho-Santos J., Schatten G., Moreno P. Control of membrane fusion during spermiogenesis and the acrosome reaction. Biol. Reprod. 2002;67:1043–1051. doi: 10.1095/biolreprod67.4.1043. [DOI] [PubMed] [Google Scholar]
- 10.Lu Q., Sun Q. Y., Breitbart H., Chen D. Y. Expression and phosphorylation of mitogen-activated protein kinases during spermatogenesis and epididymal sperm maturation in mice. Arch. Androl. 1999;43:55–66. doi: 10.1080/014850199262733. [DOI] [PubMed] [Google Scholar]
- 11.Luconi M., Barni T., Vannelli G. B., Krausz C., Marra F., Benedetti P. A., Evangelista V., Francavilla S., Properzi G., Forti G., Baldi E. Extracellular signal-regulated kinases modulate capacitation of human spermatozoa. Biol. Reprod. 1998;58:1476–1489. doi: 10.1095/biolreprod58.6.1476. [DOI] [PubMed] [Google Scholar]
- 12.Wadewitz A. G., Winer M. A., Wolgemuth D. J. Developmental and cell lineage specificity of raf family gene expression in mouse testis. Oncogene. 1993;8:1055–1062. [PubMed] [Google Scholar]
- 13.Thompson T. A., Haefliger J. A., Senn A., Tawadros T., Magara F., Ledermann B., Nicod P., Waeber G. Islet-Brain1/JNK-interacting protein-1 is required for early embryogenesis in mice. J. Biol. Chem. 2001;276:27745–27748. doi: 10.1074/jbc.C100222200. [DOI] [PubMed] [Google Scholar]
- 14.De Lamirande E., Gagnon C. The extracellular signal-regulated kinase (ERK) pathway is involved in human sperm function and modulated by the super oxide. Mol. Hum. Reprod. 2002;8:124–135. doi: 10.1093/molehr/8.2.124. [DOI] [PubMed] [Google Scholar]
- 15.Jagadish N., Rana R., Selvi R., Mishra D., Shankar S., Mohapatra B., Suri A. Molecular cloning and characterization of a haploid germ cell specific sperm associated antigen 9 (SPAG9) from the macaque. Mol. Reprod. Dev. 2005;71:58–66. doi: 10.1002/mrd.20245. [DOI] [PubMed] [Google Scholar]
- 16.Shankar S., Mohapatra B., Verma S., Selvi R., Jagadish N., Suri A. Isolation and characterization of a haploid germ cell specific sperm associated antigen 9 (SPAG9) from the baboon. Mol. Reprod. Dev. 2004;69:186–193. doi: 10.1002/mrd.20164. [DOI] [PubMed] [Google Scholar]
- 17.Shankar S., Mohapatra B., Suri A. Cloning of a novel human testis mRNA specifically expressed in testicular haploid germ cells, having unique palindromic sequences and encoding a leucine zipper dimerization motif. Biochem. Biophys. Res. Commun. 1998;243:561–565. doi: 10.1006/bbrc.1997.7943. [DOI] [PubMed] [Google Scholar]
- 18.Kelkar N., Gupta S., Dickens M., Davis R. J. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol. Cell. Biol. 2000;20:1030–1043. doi: 10.1128/mcb.20.3.1030-1043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison D. K., Davis R. J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 20.Whitmarsh A. J., Cavanagh J., Tournier C., Yasuda J., Davis R. J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 21.Yasuda J., Whitmarsh A. J., Cavanagh J., Sharma M., Davis R. J. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito M., Yoshioka K., Akechi M., Yamashita S., Takamatsu N., Sugiyama K., Hibi M., Nakabeppu Y., Shiba T., Yamamoto K. I. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a scaffold factor in the JNK signaling pathway. Mol. Cell. Biol. 1999;19:7539–7548. doi: 10.1128/mcb.19.11.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison L. T., Wang Q. T., Studzinski G. T. Butyrate-induced G2/M block in Caco-2 colon cancer cells is associated with decreased p34cdc2 activity. Proc. Soc. Exp. Biol. Med. 1999;222:150–156. doi: 10.1046/j.1525-1373.1999.d01-125.x. [DOI] [PubMed] [Google Scholar]
- 24.Flickinger C. J., Rao J., Bush L. A., Sherman N., Oko R., Herr J. C. Outer dense fiber proteins are dominant post-obstruction autoantigens in adult Lewis rats. Biol. Reprod. 2001;64:1451–1459. doi: 10.1095/biolreprod64.5.1451. [DOI] [PubMed] [Google Scholar]
- 25.Suri A., Chabbra S., Upadhyay S. Identification of human sperm antigen recognized by serum of an immunoinfertile woman: a candidate for immunocontraception. Am. J. Reprod. Immunol. 1996;36:317–326. doi: 10.1111/j.1600-0897.1996.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 26.Sui G., Soohoo C., Affar E., Gay F., Shi Y., Forrester W. C., Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Burkman L. J., Coddington C., Franken D. R., Kruger T. F., Rosenwaks Z., Hodgen G. D. The hemizona assay: development of a diagnostic test for the binding of human spermatozoa to the human zona pellucida to predict fertilization potential. Fertil. Steril. 1988;49:688–697. [PubMed] [Google Scholar]
- 29.Okumara K., Matsushima Y., Matsumura K., Nakamura K., Taguchi H., Kitagawa Y. Mapping of human DNA-binding Nuclear Protein (NP220) to chromosome band 2P13.1-P13.2 and its relation to matrin 3. Biosci. Biotechnol. Biochem. 1998;62:1640–1642. doi: 10.1271/bbb.62.1640. [DOI] [PubMed] [Google Scholar]
- 30.Suyama M., Nagase T., Ohara O. HUGE: a database for human large proteins identified by Kazusa cDNA sequencing project. Nucleic Acid Res. 1999;27:338–339. doi: 10.1093/nar/27.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuoka H., Ihn H., Medsger T. A., Hirakata M., Kawakami Y., Ikeda Y., Kuwana M. A novel protein highly expressed in testis is overexpressed in systemic sclerosis fibroblasts and targeted by autoantibodies. J. Immunol. 2003;171:6883–6890. doi: 10.4049/jimmunol.171.12.6883. [DOI] [PubMed] [Google Scholar]
- 32.Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., Gindhart J. G., Goldstein L. S. B. Kinesin-dependent axonal transport is mediated by the Sunday Driver (SYD) protein. Cell (Cambridge, Mass.) 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 33.Byrd D. T., Kawasaki M., Walcoff M., Hisamoto N., Matsumoto K., Jin Y. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron. 2001;32:787–800. doi: 10.1016/s0896-6273(01)00532-3. [DOI] [PubMed] [Google Scholar]
- 34.Sreerama N., Woody R. W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 35.Geourjon C., Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 36.Parrington J., Swann K., Shevchenko V. I., Sesay A. K., Lal F. A. Calcium oscillations in mammalian eggs triggered by a soluble sperm protein. Nature (London) 1996;379:364–368. doi: 10.1038/379364a0. [DOI] [PubMed] [Google Scholar]
- 37.Girardin S. E., Yaniv M. A direct interaction between JNK1 and CrkII is critical for Rac1-induced JNK activation. EMBO J. 2001;20:3437–3446. doi: 10.1093/emboj/20.13.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mooser V., Maillard A., Bonny C., Steinmann M., Shaw P., Yarnall D. P., Burns D. K., Schorderet D. F., Nicod P., Waeber G. Genomic organization, fine-mapping, and expression of the human islet-brain 1 (IB1)/c-Jun-amino-terminal kinase interacting protein-1 (JIP-1) gene. Genomics. 1999;55:202–208. doi: 10.1006/geno.1998.5641. [DOI] [PubMed] [Google Scholar]
- 39.Kalandadze A., Galleno M., Foncerrada L., Strominger J. L., Wucherpfennig K. W. Expression of recombinant HLA-DR2 molecules. J. Biol. Chem. 1996;271:20156–20162. doi: 10.1074/jbc.271.33.20156. [DOI] [PubMed] [Google Scholar]
- 40.Engling A., Backhaus R., Stegmayer C., Zehe C., Seelenmeyer C., Kehlenbach A., Schwappach B. Biosynthetic FGF-2 is targeted to non-lipid raft microdomains following translocation to the extracellular surface of CHO cells. J. Cell Sci. 2002;115:3619–3631. doi: 10.1242/jcs.00036. [DOI] [PubMed] [Google Scholar]
- 41.MacKenzie A., Wilson H. L., Kiss-Toth E., Dower S. K., North R. A., Suprenant A. Rapid secretion of interleukin-1β by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 42.Bohring C., Krause E., Habermann B., Krause W. Isolation and identification of sperm membrane antigens recognized by antisperm antibodies, and their possible role in immunological infertility disease. Mol. Hum. Repod. 2001;7:113–118. doi: 10.1093/molehr/7.2.113. [DOI] [PubMed] [Google Scholar]
- 43.Shetty J., Wolkowicz M. J., Digilio L. C., Klotz K. L., Jayes F. L., Diekman A. B., Westbrook V. A., Farris E. M., Hao Z., Coonrod S. A., et al. SAMP14, a novel, acrosomal membrane-associated, glycosylphosphatidylinositol-anchored member of the Ly-6/Urokinase type plasminogen activator receptor superfamily with a role in sperm-egg interaction. J. Biol. Chem. 2003;278:30506–30515. doi: 10.1074/jbc.M301713200. [DOI] [PubMed] [Google Scholar]
- 44.Mcleskey S. B., Dowds C., Carballada R., White R. R., Saling P. M. Molecules involved in mammalian sperm-egg interaction. Int. Rev. Cytol. 1998;177:57–113. doi: 10.1016/s0074-7696(08)62231-7. [DOI] [PubMed] [Google Scholar]
- 45.Suri A. Sperm specific proteins – potential candidate molecules for fertility control. Reprod. Biol. Endocrinol. 2004;2:10–15. doi: 10.1186/1477-7827-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagadish N., Rana R., Mishra D., Garg M., Selvi R., Suri A. Vaccine. 2005. Characterization of immune response in mice to plasmid DNA encoding human sperm associated antigen 9 (SPAG9) in the press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.