Abstract
Tetraspanins function as molecular organizers of multi-protein complexes by assembling primary complexes of a relatively low mass into extensive networks involved in cellular signalling. In this paper, we summarize our studies performed on the tetraspanin D6.1A/CO-029/TM4SF3 expressed by rat carcinoma cells. Primary complexes of D6.1A are almost indistinguishable from complexes isolated with anti-CD9 antibody. Indeed, both tetraspanins directly associate with each other and with a third tetraspanin, CD81. Moreover, FPRP (prostaglandin F2α receptor-regulatory protein)/EWI-F/CD9P-1), an Ig superfamily member that has been described to interact with CD9 and CD81, is also a prominent element in D6.1A complexes. Primary complexes isolated with D6.1A-specific antibody are clearly different from complexes containing the tetraspanin CD151. CD151 is found to interact only with D6.1A if milder conditions, i.e. lysis with LubrolWX instead of Brij96, are applied to disrupt cellular membranes. CD151 probably mediates the interaction of D6.1A primary complexes with α3β1 integrin. In addition, two other molecules were identified to be part of D6.1A complexes at this higher level of association: type II phosphatidylinositol 4-kinase and EpCAM, an epithelial marker protein overexpressed by many carcinomas. The characterization of the D6.1A core complex and additional more indirect interactions will help to elucidate the role in tumour progression and metastasis attributed to D6.1A.
Keywords: carcinoma cell, CD9, D6.1A, prostaglandin F2α receptor-regulatory protein (FPRP), raft, tetraspanin-enriched microdomain (TEM)
Abbreviations: DRM, detergent-resistant membrane; DTSP, dithiobis(succinimidyl propionate); ER, endoplasmic reticulum; FCS, fetal calf serum; FPRP, prostaglandin F2α receptor-regulatory protein; GPI, glycosylphosphatidylinositol; HRP, horseradish peroxidase; MβCD, methyl-β-cyclodextrin; PI4K, phosphatidylinositol 4-kinase; TEM, tetraspanin-enriched microdomain; TfR, transferrin receptor
INTRODUCTION
The tetraspanin D6.1A/CO-029/TM4SF3 was originally described as a tumour-associated antigen expressed by several human carcinoma and astrocytoma cell lines [1]. Higher expression of CO-029 on the metastatic colon carcinoma cell line SW620 when compared with SW480 cells derived from the primary tumour of the same patient may indicate a role for CO-029 in tumour progression [2]. Sato and co-workers [3] found that D6.1A expression increased in a dedifferentiated rat hepatoma cell line and suggested a function for it related to cell proliferation and differentiation. Increased expression of D6.1A was also demonstrated for hepatocellular carcinoma. Here, a possible relevance to haematogenous intrahepatic metastasis was postulated [4,5]. Finally, a mechanistic basis for pro-metastatic functions of D6.1A has also been hypothesized. In these experiments, a weakly metastasizing pancreatic carcinoma cell line was transfected with D6.1A and was found to evoke a severe consumption coagulopathy when grown in rats [6]. Furthermore, the interaction with platelets and leucocytes was suggested to provide tumour cells with a better chance to survive the hostile environment they encounter during metastatic spread [7].
Clearly, a better understanding of the functions of D6.1A on a molecular level is required. According to a widely accepted model, tetraspanins act as adaptors that assemble a multitude of proteins into complexes attached to specific signal transducing molecules [8]. Identification of interaction partners is therefore indispensable for an understanding of the functions conferred by D6.1A. Complicating the situation, various levels of interaction can be distinguished between the tetraspanins and putative partners based on disruption with detergents of increasing stringency [9]. First of all, primary interactions are observed that are direct and occur at high stoichiometry (type I). Some of these associations take place early during biosynthesis, probably in the ER (endoplasmic reticulum). Primary tetraspanin interactions are usually analysed in co-immunoprecipitation experiments using detergents such as Triton X-100, Brij96/Brij97 or digitonin [10,11]. An example of a well-defined primary interaction is the association of the tetraspanin CD151 with α3β1 integrin [12] or the oligomerization of the tetraspanins ROM-1 (rod outer segment membrane protein 1) and peripherin/RDS (retinal degeneration slow) [13].
Secondly, there are interactions that result from the tendency of tetraspanins to interact with each other during the formation of tetraspanin networks (type II). Association of the tetraspanin CD151 with other tetraspanins represents one example of this type of association [14]. These interactions are supposed to occur later during biosynthesis (Golgi apparatus or post-Golgi apparatus) and might be facilitated by palmitoylation of the tetraspanins [15]. The detergent Brij96 is currently used to analyse such complexes; however, milder detergents such as CHAPS might be more suitable because they allow a higher recovery of proteins that are not part of the core complex of tightly associated primary interaction partners.
Finally, a third level of association comprises proteins that are only found in tetraspanin complexes when very mild detergents such as Brij98/Brij99, Brij58 or Brij35 are used for membrane disruption (type III). Signal transducing molecules such as protein kinase C [16] and type II PI4K (phosphatidylinositol 4-kinase) [17] are only detected in tetraspanin complexes under these conditions. These same mild detergent conditions also allow tetraspanin complexes to be recovered from the lipid-rich fractions of isopycnic sucrose gradients [18]. This observation, coupled with reports showing the association of tetraspanin CD9 with ganglioside GM3 [19] and other studies demonstrating the covalent crosslinking of tetraspanins with a photoactivatable radioactive derivative of cholesterol [20], suggest the hypothetical linking of higher-order tetraspanin complexes with lipid rafts. The postulated tetraspanin complexes, in conjunction with these micro-domains of the membrane enriched with long saturated acyl chains and cholesterol, could together serve as signalling platforms [21]. However, other features of tetraspanin complexes suggest critical differences from lipid rafts and require tetraspanin complexes to initiate their own type of microdomains (summarized in [9]).
In the present study, we attempted to characterize the complexes containing the tetraspanin D6.1A at these different levels of association. Our results indicate that D6.1A is a part of a core complex consisting of the tetraspanins CD9 and CD81 and the Ig super-family member FPRP (prostaglandin F2α receptor-regulatory protein; type I interactions). Other D6.1A interactions, e.g. with α3β1 integrin, probably result from a secondary interaction mediated through CD151 [22]. The epithelial marker protein EpCAM and PI4K were identified in D6.1A complexes under mild detergent conditions.
MATERIALS AND METHODS
Cell lines and antibodies
The following rat carcinoma cell lines were used: the pancreatic adenocarcinoma line BSp73ASML [23], the colon carcinoma line PROb [24] and the bladder carcinoma line 804G [25]. Cells were cultivated in RPMI 1640 supplemented with 5% (v/v) FCS (fetal calf serum) and antibiotics.
The monoclonal antibodies used were anti-D6.1A (D6.1), anti-EpCAM (D5.7), anti-C4.4A (C4.4) and anti-α6β4 integrin (B5.5). Generation and characterization of these antibodies have been described in [26–28]. Other antibodies used were anti-β1 integrin (Ha2/5; Pharmingen, San Diego, CA, U.S.A.), anti-α2 integrin (Ha1/29; Pharmingen), anti-α3 integrin (Ralph3.1; Developmental Studies Hybridoma Bank) [29], anti-TfR (OX26; TfR stands for transferrin receptor), anti-Myc (MycL-9E10; both from the European Collection of Animal Cell Culture), anti-caveolin (C13630; Pharmingen), anti-CD9 (RPM.7; Pharmingen), anti-CD81 (TAPA-1; Pharmingen) and anti-hamster IgM (G1888-2; Pharmingen). Hybridomas were adapted to growth in RPMI 1640 supplemented with 10% FCS and antibiotics. For the detection of FPRP, we used a polyclonal antibody raised against rat FPRP [30]. For the detection of rat CD151, a serum was generated by injecting rabbits with a glutathione S-transferase fusion protein comprising amino acids 114–220 of the large extracellular domain of rat CD151 (GenBank® accession number NM_022523). Specificity of the serum was verified by blotting 6×Myc-tagged full-length versions of CD151 and D6.1A (control), which were isolated by immunoprecipitation with anti-Myc antibody from lysates of transfected cells.
Cloning and transfection
Rat CD151 cDNA was amplified with specific primers, by PCR, from a previously described cDNA library [6] and cloned into pcDNA3.1(−) (Invitrogen). A point mutation (Thr887→Cys) changing valine at position 235 to an alanine, identified after sequencing and comparison with the published sequence, was repaired by a second PCR. A 6×Myc tag was added to the N-terminus of CD151 by cloning the 6×Myc tag amplified from CS2+MT [31] into pcDNA3 (Invitrogen). The cDNA for CD151 was cloned in frame with this tag. Detailed information about primers and restriction sites will be given on request.
PROb cells were transfected with ExGen500 (MBI Fermentas, Munich, Germany) according to the manufacturer's instructions and selected in a medium containing 1 mg/ml G418. Clones were derived by limiting dilution and culture in the presence of feeder cells.
Immunoprecipitation and Western blotting
Cells were labelled with sulphosuccinimidyl-6-(biotinamido)-hexanoate (Biotin-X-NHS; Calbiochem, Bad Soden, Germany) at 0.2 mg/ml for 30 min at 4 °C, washed three times with cold PBS containing 200 mM glycine and lysed with a buffer containing 25 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, protease inhibitor mixture (Roche, Mannheim, Germany) and 1% detergent [LubrolWX (Promega, Mannheim, Germany), Brij96 (Fluka, St. Gallen, Switzerland) or 1% Triton X-100 (Gerbu, Gaiberg, Germany)]. After incubation for 1 h at 4 °C, the insoluble material was removed by centrifugation at 20000 g for 10 min at 4 °C. For immunoprecipitation, an antibody was added for 2 h, followed by Protein G–Sepharose for another 2 h. When anti-β1 integrin antibody Ha2/5 was used, anti-hamster IgM was added before the addition of Protein G–Sepharose. Immunocomplexes were collected by centrifugation at 15000 g (for 20 s), washed four times with lysis buffer and then analysed by SDS/PAGE under non-reducing conditions, unless stated otherwise. For immunoblotting, proteins resolved by SDS/PAGE were transferred to a nitrocellulose membrane and then incubated either with primary and secondary antibodies coupled to HRP (horseradish peroxidase) or with HRP-coupled Extravidin (Sigma) to detect biotinylated proteins. Blots were visualized by chemiluminescence (Amersham Biosciences).
MS analysis
Approx. 2×109 PROb cells were scraped in PBS and pelleted by centrifugation at 480 g. Cells were resuspended in hypo-osmotic lysis buffer [0.3×PBS containing 5 mM EDTA, 10 mM iodoacetamide, 2 mM PMSF and protease inhibitor mixture (Roche)] and disrupted by five successive cycles of freezing in liquid nitrogen and thawing at 37 °C. The cells still intact were removed by centrifugation at 1000 and 2000 g for 5 min each. From the resulting supernatant, cellular membranes were then isolated by centrifugation at 100000 g for 1 h. Membranes were lysed in 1% Brij96 and the insoluble material was removed by centrifugation at 20000 g for 10 min. The lysate was precleared by incubation with Protein G–Sepharose, and D6.1A complexes were then isolated by adding D6.1 antibody and Protein G–Sepharose. D6.1A-associated material was eluted from the immunoprecipitate by adding RIPA buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS). Proteins were precipitated from the eluate with chloroform/methanol [32] and separated by SDS/PAGE in a 4–20% linear gradient gel. Proteins were stained with colloidal Coomassie Blue, cut out and digested with trypsin.
Analysis of the isolated proteins by MS was performed as described previously [33].
Chemical cross-linking
PROb cells were lysed in 1% Brij96 and either 0.5 mM DTSP [dithiobis(succinimidyl propionate); Pierce] in DMSO or DMSO alone was added to equal aliquots of the lysate. After incubation for 1 h at room temperature (20 °C), 50 mM Tris/HCl (pH 7.4) to quench free DTSP and 1% Triton X-100 (final concentration) to disrupt non-covalent interactions were added for another 15 min. These lysates were used for immunoprecipitation or directly resolved by SDS/PAGE and blotted on to a nitrocellulose membrane for Western blotting.
Immunodepletion
Biotinylated PROb cells were lysed in 1% Brij96 or 1% LubrolWX as described above and divided into equal aliquots. Immunodepletion was performed by adding 4× Protein G–Sepharose alone or an antibody coupled with Protein G–Sepharose specific for the proteins mentioned in the legends to the respective Figures. The first three rounds were for 120 min each; the final round was performed overnight. The immunodepleted lysates were used for immunoprecipitation with antibodies mentioned in the legends to the Figures. Samples were analysed by SDS/PAGE and visualized with Extravidin–HRP.
Pulse–chase immunoprecipitation assay
For metabolic labelling, cells were seeded in 6 cm dishes at 1×106 cells/plate and incubated overnight. On the next day, the medium was removed and cells were washed once with a medium lacking methionine and cysteine. Cells were then incubated in a medium without methionine or cysteine and without FCS for 1–2 h. [35S]Met/Cys (0.5 mCi; Pro-mix in vitro cell-labelling mix; Amersham Biosciences) in a Met/Cys-deficient medium, supplemented with 5% dialysed FCS, was added to the starved cells for 15 min. After the removal of the labelling medium, cells were chased by incubation in complete medium containing 5% FCS and a 25-fold excess of methionine and cysteine for various time periods, washed four times with cold PBS and lysed in 1% Brij96. Finally, immunoprecipitations were performed; proteins were separated by SDS/PAGE and blotted on to a nitrocellulose membrane. Signals were revealed by autoradiography.
Gel-permeation chromatography
Gel-permeation chromatography on Sepharose CL6B and Sepharose CL4B was performed as described previously [18].
PI4K assay
PI4K assays were performed as described in [34]. Briefly, samples from immunoprecipitates isolated with anti-D6.1A, anti-CD9, anti-EpCAM and anti-α6β4 integrin antibody were analysed in a 100 μl final volume containing 0.3% Triton X-100, 50 μM ATP, 10 mM MgCl2, 20 mM Hepes (pH 7.5), 0.2 mg/ml sonicated phosphatidylinositol (Sigma) and 5 μCi of [γ-32P]ATP. Samples were incubated for 10 min at room temperature, before the assay was stopped by adding 5 M HCl (25 μl). The lipids were extracted with 160 μl of chloroform/methanol (1:1, v/v). The organic phase was collected and separated by TLC. TLC plates were exposed to an autoradiography film.
Sucrose gradients
Cells were lysed in either 1% Triton X-100, 1% Brij96 or 1% LubrolWX. Lysates were cleared by centrifugation at 20000 g for 10 min at 4 °C. The supernatant was adjusted to 40% (w/v) sucrose, and material corresponding to approx. 2×107 cells in 2 ml total volume was carefully layered on a 0.4 ml cushion of 50% sucrose in 25 mM Hepes, 150 mM NaCl and 5 mM MgCl2. This 40% layer was overlaid with 0.8 ml of 30% sucrose, 0.8 ml of 20% sucrose and 0.4 ml of 5% sucrose prepared in Hepes/NaCl/MgCl2. Samples were centrifuged at 217000 g for 18 h at 4 °C in a Kontron TST60.4 rotor. Eight fractions of equal volume were collected from the top of the gradient; the pellet was resuspended in an equal volume of SDS/PAGE sample buffer. Aliquots of each fraction were separated by SDS/PAGE and analysed with antibodies specific for D6.1A, EpCAM, caveolin, C4.4A and TfR by Western blotting.
RESULTS
D6.1A complexes resemble CD9 complexes and are distinct from CD151 complexes
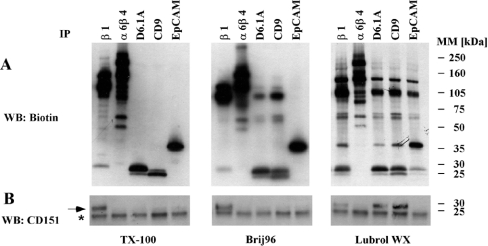
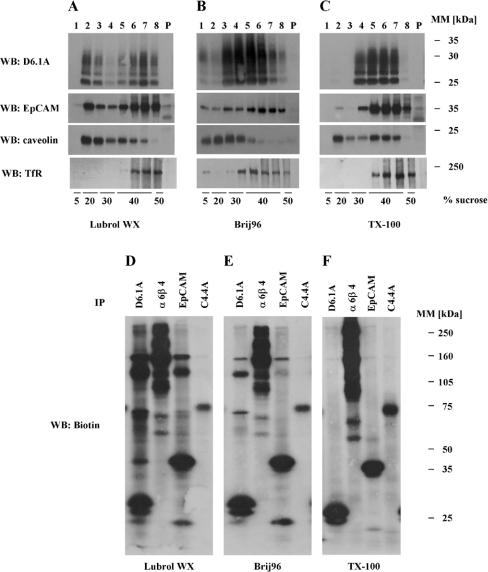
To study D6.1A complexes, we first biotinylated colon carcinoma PROb cells, lysed them under different detergent conditions and looked for associated proteins in immunoisolates of D6.1A. As shown in Figure 1(A), D6.1A does not co-immunoprecipitate any material when Triton X-100 is used for cell lysis. In the presence of Brij96, two bands around 70 kDa and a prominent protein with an apparent molecular mass of approx. 120 kDa are detected. Lowering the stringency further to LubrolWX conditions leads to the isolation of additional co-precipitated proteins; a significantly higher amount of the 120 kDa protein, which corresponds to β1 integrin [6], is present in these D6.1A complexes than under Brij96 conditions. This corresponds to the presence of CD151 in the D6.1A immunoprecipitates. CD151 probably mediates the interaction of α3β1 integrin with other tetraspanins [22], since it is found in D6.1A complexes only in LubrolWX lysates, as shown by immunoblotting with anti-CD151 (Figure 1B). The partial disruption of the interaction of D6.1A with β1 integrin by Brij96 was further corroborated by blotting β1 integrin immunoprecipitates with anti-D6.1A (results not shown). In addition, the LubrolWX detergent allows the isolation of a 35 kDa protein that is probably identical with the epithelial marker protein EpCAM, since an antibody specific for this molecule precipitates a similar pattern of biotinylated proteins; this notion was confirmed by blotting of D6.1A precipitates with an EpCAM-specific antibody and vice versa (results not shown) and by immunodepletion experiments (Figure 6). Under all conditions, the D6.1A coimmunoprecipitation pattern is almost identical with the one found for CD9. Both molecules associate under Brij96 conditions and are separated only if Triton X-100 is used. In contrast, α6β4 integrin, highly expressed in PROb cells, does not co-precipitate either D6.1A or CD9 under any of the conditions analysed. This control proves the specificity of interactions found for D6.1A.
Figure 1. D6.1A complexes resemble CD9 complexes, but are distinct from CD151 complexes.
(A) PROb cells were surface-labelled with biotin and then lysed in a buffer containing 1% Triton X-100, 1% Brij96 or 1% LubrolWX. Lysates were cleared by centrifugation at 20000 g for 10 min. Equal portions of the lysates were immunoprecipitated with antibodies directed to the indicated antigens. Isolated material was separated by SDS/PAGE on a 4–20% linear gradient gel, transferred to nitrocellulose and visualized by Extravidin–HRP, followed by chemiluminescence. (B) The membrane depicted in (A) was then incubated with an antiserum recognizing the EC2 domain of rat CD151, and bands were visualized by incubation with anti-rabbit IgG–HRP. A band corresponding to CD151 is marked with an arrow; the asterisk indicates an unspecific band occurring in all lanes.
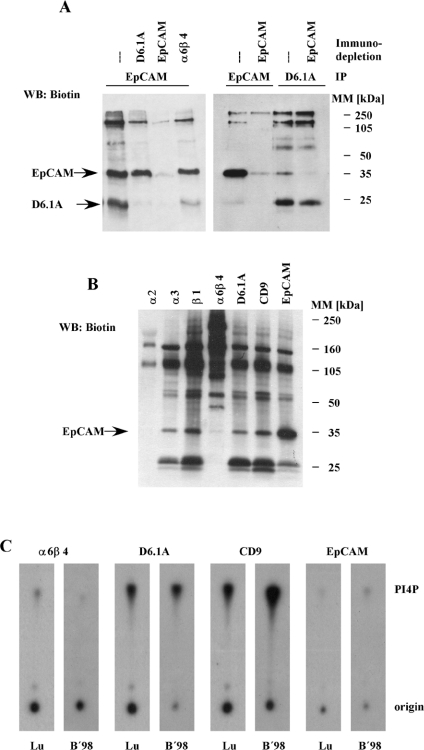
Figure 6. EpCAM is a low stoichiometric partner in D6.1A complexes.
(A) Biotinylated PROb cells were lysed in 1% LubrolWX and immunodepletion was performed in four successive rounds with the indicated antibodies coupled with Protein G–Sepharose or Protein G–Sepharose alone. Equal aliquots of these lysates were subjected to immunoprecipitation with anti-D6.1A and anti-EpCAM antibodies. Precipitates were separated by SDS/PAGE (12% polyacrylamide) and then the biotinylated material was transferred to a nitrocellulose membrane and detected with Extravidin–HRP. (B) Immunoprecipitations with anti-α2 integrin, anti-α3 integrin, anti-α6β4 integrin, anti-D6.1A, anti-CD9 and anti-EpCAM antibodies were performed on 1% LubrolWX lysates of PROb cells and the biotinylated proteins were detected as described for Figure 1(A). (C) BSp73ASML cells were lysed in 1% LubrolWX (Lu) or 1% Brij98 (B'98) and immunoprecipitations were performed with anti-α6β4, anti-D6.1A, anti-CD9 and anti-EpCAM. In vitro PI4K assays were performed on the samples as described in the Materials and methods section. Phosphorylated lipids were separated by TLC and revealed by autoradiography.
In apparent contradiction to published findings [35], we could not detect CD151 in α6β4 immunoprecipitates (Figure 1B). However, when other cell lines were tested, i.e. the pancreatic adenocarcinoma cell line BSp73ASML and the bladder carcinoma line 804G, CD151 was clearly present in α6β4 complexes isolated under Brij96 conditions. Lack of association therefore is peculiar to PROb cells (C. Claas, unpublished work). Association of D6.1A with EpCAM under mild detergent conditions could also be detected in BSp73ASML cells; however, for 804G cells, the expression of EpCAM is too low to allow us to demonstrate this association. All interactions resistant to stringent lysis conditions were the same in all the three cell lines tested.
D6.1A directly associates with CD9, CD81 and FPRP
Complexes isolated in the presence of Brij96 probably depend on direct protein–protein interactions. Identifying the elements that are part of such complexes is key to understanding the functions conferred by the respective tetraspanins. Therefore we isolated D6.1A complexes from Brij96 lysates of PROb cells on a large scale, separated them by SDS/PAGE and analysed the Coomassie Blue-stained bands by MS. Then, 18 peptides were identified that matched the sequence of FPRP, a member of a recently identified subfamily of Ig proteins. FPRP and the related EWI-2 were previously found to interact with tetraspanins CD9 and CD81 [36–39].
Peptides matching sequences from other proteins were found as well. They were disregarded because the corresponding proteins are commonly found contaminants in MS analyses (e.g. heat-shock proteins and keratins) or because their association with D6.1A could not be demonstrated in subsequent co-immunoprecipitation experiments (claudin and rab14).
In contrast, the presence of FPRP in D6.1A complexes was verified by blotting proteins isolated from Brij96 lysates with anti-D6.1A using an anti-serum raised against rat FPRP (Figure 2A). The difference in the published apparent molecular mass of approx. 130 kDa from our protein running at approx. 120 kDa was probably caused by a generally lower level of glycosylation of proteins seen in PROb compared with BSp73ASML and 804G cells [26].
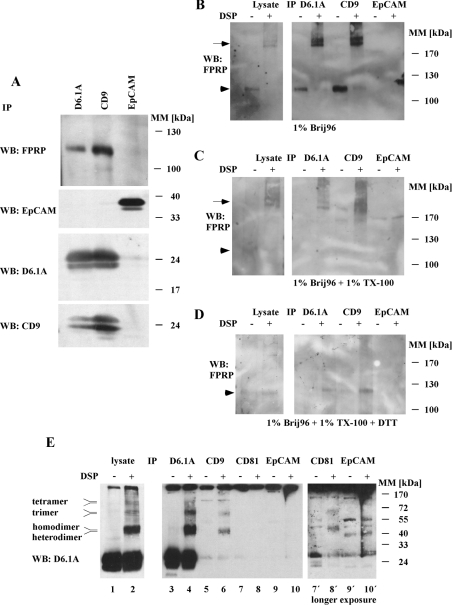
Figure 2. D6.1A directly associates with CD9, CD81 and FPRP.
(A) PROb cells were lysed in 1% Brij96 and immunoprecipitations (IP) were performed with anti-D6.1A, anti-CD9 and anti-EpCAM antibodies. After separation of the isolated complexes by SDS/PAGE and transfer to a nitrocellulose membrane, Western blotting (WB) was performed with anti-FPRP, anti-EpCAM, anti-D6.1A and anti-CD9. (B) Brij96 lysates of PROb cells were treated with 0.5 mM of the reducible cross-linker DTSP (‘DSP’) in DMSO or with DMSO only for 1 h at room temperature, and immunoprecipitations were performed with D6.1A-, CD9- and EpCAM-specific antibodies. Samples were separated by SDS/PAGE (6% polyacrylamide), transferred to nitrocellulose and immunoblotted with anti-FPRP. In parallel, an aliquot of DTSP-treated lysate and control lysate was analysed. The arrow indicates cross-linked material containing FPRP at a mass well beyond 200 kDa; the arrowhead indicates the position of non-cross-linked FPRP at approx. 120 kDa. (C) Same as in (B), but lysates were incubated with 1% Triton X-100 after cross-linking with DTSP to remove any non-cross-linked proteins from the precipitated material. (D) Same as in (C), but samples were separated under reducing conditions (100 mM dithiothreitol) to separate the cross-linked proteins before SDS/PAGE and blotting. (E) PROb cells were lysed in 1% Brij96 and immunoprecipitations were performed with antibodies as in (A–D) and, additionally, with anti-CD81 (lanes 3–10). In parallel, the lysate was analysed in the same way (lanes 1 and 2). Complexes were separated on an 11% gel and immunoblotted with anti-D6.1A. Right panel: lanes 7′, 8′, 9′ and 10′ show a longer exposure of lanes 7–10.
On the basis of results of published work, FPRP is also contained in CD9 complexes. In a control precipitation, it can be shown that FPRP is absent from material collected with an antibody specific for abundantly expressed EpCAM.
Addition of the chemical cross-linker DTSP, which reacts with free amino groups of proteins, to Brij96 lysate, followed by subsequent immunoprecipitation of D6.1A and blotting of the isolated complex with anti-FPRP, reveals that the interaction between the two molecules is based on direct association. Close to 100% of total FPRP can be cross-linked with complexes of >200 kDa in mass since the free 120 kDa form present in the control-treated lysate is completely absent from the lysate treated with DTSP (Figure 2B). Addition of Triton X-100 to cross-linked lysates to disrupt non-covalent interactions does not change the mass of the complex (Figure 2C). Reduction of the complexes isolated from Triton X-100-treated lysates with dithiothreitol finally allows the detection of the free FPRP running at 120 kDa again, proving that FPRP is indeed present in the high molecular mass complex (Figure 2D). Corresponding results were obtained if a CD9-specific antibody was used for precipitation. In contrast, there are no cross-linked bands containing FPRP present in complexes precipitated with anti-EpCAM antibody.
The mass of the cross-linked complex in D6.1A and CD9 immunoisolates as well as in total lysates is inconsistent with an FPRP association with tetraspanins in the ratio 1:1 on PROb cells. Thus complexes containing FPRP appear to differ from complexes isolated from A431 cells, where this type of complex is quite prominent [37]. This phenomenon can be explained by the presence of D6.1A in homo- and heterooligomers in PROb cells as demonstrated in Figure 2(E). Lysates prepared with 1% Brij96 were treated first with DTSP or carrier only and then with 1% Triton X-100, as explained for Figure 2(D). Complete lysates and immunoprecipitations performed with antibodies recognizing D6.1A, CD9 and CD81 and with a control antibody directed against EpCAM were separated by non-reducing SDS/PAGE and blotted with anti-D6.1A antibody. In the lysate treated with DTSP, bands appear that are consistent in mass with dimers, trimers and tetramers containing D6.1A. Furthermore, these oligomers exist in doublets of bands, suggesting the presence of D6.1A homo- and heteromers. Indeed, immunoprecipitation with specific antibodies demonstrated that D6.1A directly associates with CD9 and CD81. We hypothesize that the cross-linked products running slightly above adducts with CD9 or CD81 seen only in total lysate and immunoprecipitates with anti-D6.1A (see Figure 2E, lanes 2 and 4), represent complexes richer in D6.1A and D6.1A homodimers. Association of tetraspanins in an oligomeric state with FPRP could account for the FPRP complexes of >200 kDa found in the cross-linking experiments depicted in Figures 2(B)–2(D).
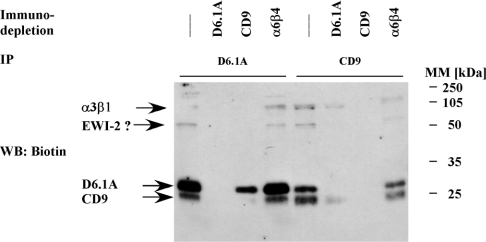
Association of CD9 and CD81 with FPRP and the related EWI-2 has been characterized as highly stoichiometric [36,39]. An estimate of the stoichiometry of interaction between D6.1A and CD9 comes from immunodepletion experiments summarized in Figure 3. Brij96 lysates of surface-biotinylated PROb cells were subjected to four rounds of immunodepletion with anti-D6.1A, anti-CD9, anti-α6β4 or no antibody. Immunoprecipitations were subsequently performed on these lysates with D6.1A- or CD9-specific antibody. Complexes separated by SDS/PAGE were then checked for the presence of biotinylated material by blotting with Extravidin–HRP. Immunodepletion of nearly all the D6.1A leads to a substantial decrease of surface-associated CD9, by approx. 50% as determined by semiquantitative densitometry. When performed in the other order, approx. 70% of the surface-associated D6.1A is removed when the lysate was completely cleared of CD9. Immunodepletion performed with an antibody raised against highly expressed α6β4 integrin did not cause a significant decrease in the recovery of D6.1A and CD9. Effects were less pronounced when we examined for total D6.1A in CD9-depleted lysates or the other way round, indicating that considerable amounts of non-associated tetraspanins are present on intracellular membranes (results not shown).
Figure 3. Stoichiometry of the association of D6.1A with CD9.
PROb cells were surface-biotinylated and lysed in 1% Brij96. The lysate was divided into four equal aliquots and depleted with Protein G–Sepharose alone or with Protein G–Sepharose coupled with a monoclonal antibody specific for D6.1A, CD9 and α6β4 integrin respectively. After four successive rounds of immunodepletion, immunoprecipitations were performed with anti-D6.1A or anti-CD9 on the lysates. Complexes were separated by SDS/PAGE (12% polyacrylamide) and the biotinylated proteins were revealed by blotting with Extravidin–HRP. The question mark indicates that the identity of the 70 kDa protein as EWI-2 has yet to be verified, although work published by other groups has suggested that this protein could be EWI-2 (see the Discussion).
Hemler and co-workers [36] found that CD81 complexes isolated from 293 cells under Brij96 conditions and subjected to gel filtration were clearly included within a CL6B column, indicating that the complexes were significantly smaller than 4×106 Da. Moreover, they showed that complexes of CD81 with CD9 and FPRP could be separated from complexes containing CD81, CD9 and a protein probably identical with EWI-2. In an analogous experiment performed on PROb cells, we also found that complexes of D6.1A, CD9 and FPRP are separated within the column (Figure 4).
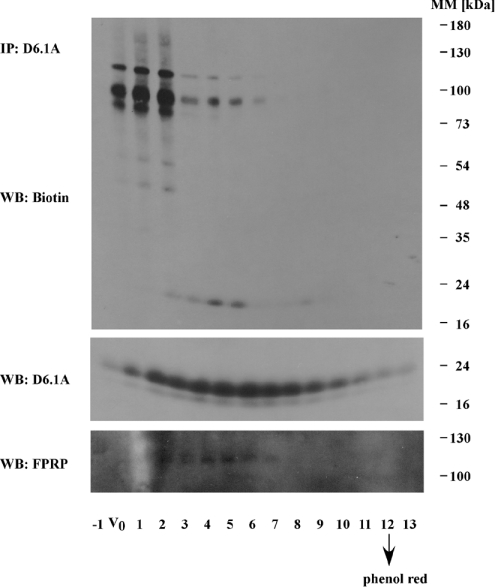
Figure 4. Analysis of D6.1A complexes by gel filtration on a Sepharose CL6B column.
Brij96 lysate from approx. 2×107 biotinylated PROb cells was fractioned on a Sepharose CL6B column (30 cm×0.8 cm). Fractions were collected from the leading edge of the Blue Dextran (approximately the V0) to Phenol Red (indicating the small molecule elution point). Aliquots of each fraction were immunoprecipitated with anti-D6.1A antibody and immunoblotted for biotinylated material with Extravidin–HRP. α3 and β1 integrins are found mainly in fractions V0, 1 and 2. The elution profile of total D6.1A- and D6.1A-associated FPRP was also analysed by immunoblotting with the respective antibodies.
It also appears that the associated integrin can be efficiently separated from the core complex of directly interacting D6.1A, CD9 and FPRP by Sepharose CL6B gel filtration. Integrin-containing complexes are eluted close to the approximate void volume of the column and contain only very small amounts of D6.1A. All of these results further support the conclusions suggested by the co-immunoprecipitations seen in Figure 1; association of D6.1A (and CD9) with integrin is based on a different type of interaction than with the core complex.
Association of D6.1A with FPRP during biosynthesis
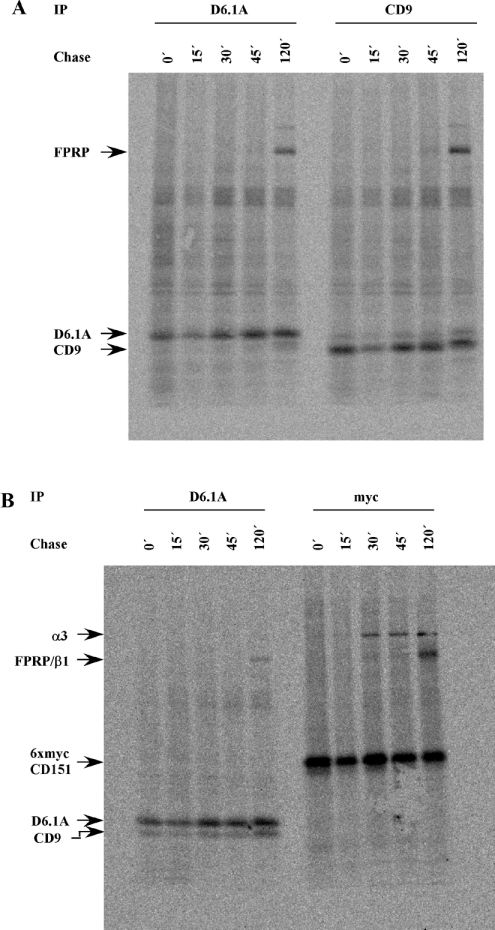
To address the question of whether D6.1A and CD9 show differences in their kinetics of association with FPRP, we performed pulse–chase experiments with 35S-labelled Met/Cys. To detect early events in tetraspanin biosynthesis, PROb cells were labelled for only 15 min before unlabelled chase medium was added to the cells. Figure 5(A) shows that both D6.1A- and CD9-specific antibodies precipitated FPRP within 2 h after the cells had been metabolically labelled. When time points between 45 and 120 min were analysed, we could detect FPRP in association with D6.1A within 60 min after labelling (results not shown). Since the association of CD151 with α3β1 integrin was known to follow a much faster time course in MDA-MB-231 cells [14], we also examined the association of CD151 with integrin in PROb cells. A rat CD151-specific antibody suitable for immunoprecipitation is not available; therefore we cloned the cDNA for rat CD151 and attached a 6×Myc tag to the N-terminus of the molecule. A clone of PROb cells stably expressing this construct was used for the experiment depicted in Figure 5(B). In agreement with the results mentioned above, association of CD151 with integrin seems to occur significantly earlier in biosynthesis than the interaction of D6.1A and CD9 with FPRP. In fact, the interaction between CD151 and α3β1 is the only one proven so far to occur already in the ER [9]. Attempts to evaluate at what step during biosynthesis FPRP associates with D6.1A and CD9 did not give unambiguous results. Although immunoprecipitations of D6.1A and CD9 from cells treated with Brefeldin A, a reagent blocking the translocation of proteins from the ER to the Golgi apparatus [40], led to the detection of proteins still associated with both tetraspanins, none of these proteins could be reprecipitated with FPRP-specific serum (results not shown). However, the FPRP antisera used are of relatively low avidity and, hence, a conclusion from this experiment could not be drawn.
Figure 5. Association of D6.1A and CD9 with FPRP during biosynthesis.
PROb cells (A) or PROb cells expressing 6×mycCD151 (B) were starved for 2 h and then labelled with 0.5 mCi of [35S]Met/Cys in 1 ml of RPMI 1640 containing 5% dialysed FCS for 15 min at 37 °C (zero time=0 min, 0′). Cells were subsequently incubated in complete medium containing a 25-fold excess of unlabelled methionine and cysteine for various time periods. At the indicated time points, cells were put on ice and lysed with a buffer containing 1% Brij96. Immunoprecipitations were performed with monoclonal antibodies recognizing D6.1A, CD9 or a Myc tag attached to CD151. Samples were separated on a 5–15% linear gradient gel, transferred to nitrocellulose and exposed to a phosphoimager screen for 3 days. The radiolabelled band at 120 kDa found in D6.1A and CD9 immunoprecipitates is FPRP. This was verified by reprecipitation with FPRP-specific serum after elution of the associated protein from D6.1A and CD9 complexes using RIPA buffer (results not shown).
Interactions of D6.1A retained under mild detergent conditions
It is well established that certain associations in tetraspanin complexes are not seen if relatively stringent detergents like Brij96 or Triton X-100 are used. These so-called secondary and tertiary interactions might depend on tetraspanin–tetraspanin interactions or on incomplete solubilization of raft-like subdomains [9]. As shown in Figure 1, this also pertains to D6.1A complexes. When cells were lysed in 1% LubrolWX (a detergent having a hydrophilic to lipophilic balance of 14.9, compared with 13.1 for Brij96, and therefore predicted to be less efficient in disrupting the membranes), integrin, CD151 and EpCAM are found to be associated with D6.1A in addition to the core complex of FPRP and CD9. Integrin and CD151 are already known to be part of secondary tetraspanin complexes [22], so we concentrated on the characterization of the interaction with EpCAM. To estimate the stoichiometry of EpCAM in D6.1A complexes, immunodepletion experiments were performed as described above. Apparently, only a small amount of the total EpCAM expressed by PROb cells can be precipitated with a D6.1A-specific antibody, since recovery of D6.1A from EpCAM-depleted cells is virtually unaltered (Figure 6A). In the reverse experiment, namely immunodepletion of D6.1A and immunoprecipitation of EpCAM from the cleared lysate, no apparent change in levels of EpCAM was observed either.
To check whether EpCAM might associate with β1 integrins rather than with tetraspanin complexes, co-immunoprecipitations were performed with anti-α2β1 integrin and anti-α3β1 antibody, and the associated biotinylated material was detected by blotting with Extravidin–HRP. Whereas α3β1 is a prominent element in tetraspanin complexes through its tight interaction with CD151 [11], α2β1 integrin is rarely found associated with tetraspanins [41]. The association pattern of EpCAM is that of a true tetraspanin-associated protein. It is present at background levels in α2β1 precipitates, but is seen as one of the pools of protein isolated together with α3β1 (Figure 6B).
Finally, another protein frequently found in tetraspanin complexes under mild detergent conditions, type II PI4K [17], was studied for its presence in D6.1A complexes. A D6.1A-specific antibody precipitates comparable amounts of PI4K activity as found in anti-CD9 precipitates (Figure 6C). In contrast and again underscoring the low stoichiometry of interaction with D6.1A, EpCAM is associated with little PI4K activity, not significantly above the levels precipitated with an anti-α6β4 antibody.
EpCAM is present in DRM (detergent-resistant membrane), but its interaction with tetraspanins does not depend on incomplete solubilization
It has been emphasized by several groups that TEMs (tetraspanin-enriched microdomains) share the characteristics of membrane subdomains commonly referred to as ‘rafts’ (summarized in [9]). An easy method indicative of raft association for a given protein is to check for its presence in ‘light’, lipid-rich fractions of sucrose density gradients. For tetraspanins, it has been shown that flotation to these fractions depends on the presence of cholesterol, since removal of cholesterol by treatment of cells with MβCD (methyl-β-cyclodextrin) leads to their confinement in the bottom fractions of such gradients [18]. On the other hand, TEMs differ from ‘conventional’ rafts by their solubility in Triton X-100. Only milder detergents render TEMs intact so that their buoyancy is preserved [18]. EpCAM possesses features very similar to those found for tetraspanins (Figures 7A–7C). In sucrose gradients run with lysates of BSp73ASML cells, both D6.1A and EpCAM localize to DRM fractions only when lysates were prepared with LubrolWX or Brij96, whereas they are completely soluble when Triton X-100 was used as the detergent. In contrast, caveolin, a marker for conventional Triton X-100-insoluble rafts [42], localizes to the DRM fractions under all detergent conditions. Blotting for the non-raft marker TfR [42] demonstrates that the gradients depicted efficiently separated completely solubilized material from DRM fractions. The fact that DRM association of D6.1A and EpCAM is found after Brij96 solubilization conditions (see Figures 1B and 7E) excludes incomplete solubilization as the basis for interaction of D6.1A with EpCAM.
Figure 7. Association of D6.1A and EpCAM with DRM under various detergent conditions.
(A–C) BSp73ASML cells were lysed in a buffer containing 1% LubrolWX (A), 1% Brij96 (B) or 1% Triton X-100 (C). Lysates were cleared by centrifugation at 20000 g for 10 min and the supernatants corresponding to approx. 2×107 cells were adjusted to 40% sucrose. The sucrose-containing supernatant was layered on to a cushion of 50% sucrose and successively overlaid with a buffer containing 30, 20 and 5% sucrose respectively. Gradients thus formed were centrifuged at 217000 g for 18 h in a Kontron TST60.4 rotor. Eight fractions of equal volume were removed with a pipette from the top of each gradient and the pellet was resuspended in Laemmli sample buffer. Equal volumes of each fraction and the pellet were resolved by SDS/PAGE, transferred to nitrocellulose and immunoblotted with antibodies directed to the indicated antigens. (D–F) PROb cells were surface-biotinylated and lysed in a buffer containing 1% LubrolWX (D), 1% Brij96 (E) or 1% Triton X-100 (F). Immunoprecipitations were performed with anti-D6.1A, anti-α6β4, anti-EpCAM and anti-C4.4A antibodies. Precipitates were separated on a 5–15% linear gradient gel and biotinylated proteins were revealed as described for Figure 1(A).
In addition, neither EpCAM nor D6.1A under any detergent condition was found to be associated with a marker molecule of conventional rafts, GPI (glycosylphosphatidylinositol)-anchored C4.4A (Figure 7D–7F). The same observations account for immunoprecipitations performed with anti-caveolin (results not shown).
D6.1A complexes isolated from LubrolWX lysates are separated within a Sepharose CL4B column
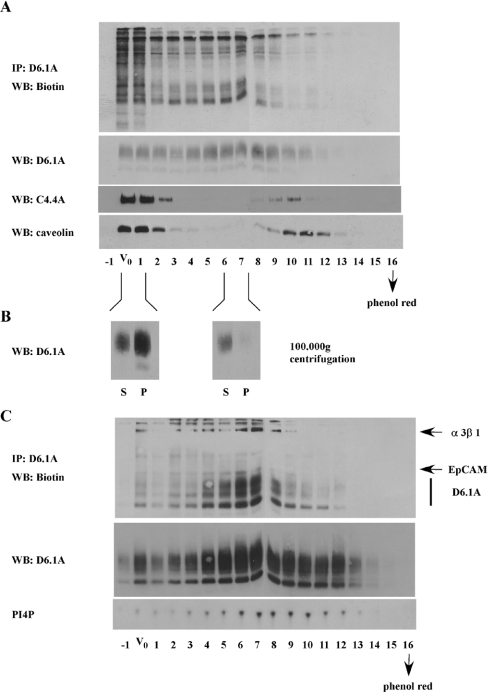
Analysis by gel filtration on Sepharose CL4B of D6.1A complexes isolated under LubrolWX conditions corroborated the results described in the previous section. It has been described that complexes of tetraspanin CD63 and αLβ2 integrin isolated under mild detergent conditions and analysed by gel filtration were found to occur in incompletely solubilized membrane subdomains of human neutrophils and therefore were excluded from a Sepharose CL4B column [43]. As demonstrated in Figures 8(A) and 8(B), D6.1A complexes from 804G cells are eluted with the void volume as well as with markers of smaller mass. In contrast, a large amount of C4.4A and caveolin is excluded from the column; only much smaller amounts can be detected in a region centred on fractions 10 and 11. In contrast, D6.1A is eluted in a region centred on fractions 5–8. We hypothesize that fractions 10 and 11 contain small amounts of completely solubilized caveolin and C4.4A. Centrifugation at 100000 g (for 1 h) and blotting of the super-natant and pellet of material pooled from the void volume and from fractions 6 and 7 reveal that CL4B gel filtration provides an efficient way of separating completely solubilized tetraspanin complexes from material that still contains membrane lipids. Very little of the D6.1A from fractions 6 and 7 is pelleted at 100000 g for 1 h (i.e. is vesicular), whereas large amounts of D6.1A are pelleted from fractions taken at the void volume.
Figure 8. Analysis of D6.1A complexes isolated from LubrolWX lysates, by gel filtration on a Sepharose CL4B column.
(A) Surface-biotinylated 804G cells (∼2×107) were lysed in 1% LubrolWX and fractionated on a Sepharose CL4B column (50 cm×1 cm). Fractions of approx. 1 ml each were collected and void volume and the small molecule elution point were determined as described for Figure 4. Aliquots of each fraction were immunoprecipitated with anti-D6.1A and the associated biotinylated material was revealed by Extravidin–HRP. The distribution of total D6.1A, C4.4A and caveolin was determined by blotting aliquots of each fraction with the respective antibodies. (B) Fractions corresponding to the void volume (V0+1) and from within the gradient (6+7) found to contain peaks of D6.1A-associated proteins as shown in (A) were pooled and subjected to ultracentrifugation at 100000 g for 2 h in a SW41 rotor. The pellet was washed once with lysate buffer and then solubilized in 1% LubrolWX+0.1% SDS. Equal volumes of supernatant (S) and pellet (P) were precipitated with chloroform/methanol, separated on an 11% gel and immunoblotted with anti-D6.1A. (C) BSp73ASML cells were fractionated on a Sepharose CL4B column as described in (A). Immunoprecipitations with anti-D6.1A were performed on each fraction. Biotinylated material as well as D6.1A were revealed by Extravidin–HRP and immunoblotting with anti-D6.1A. In addition, in vitro PI4K assays were performed on D6.1A precipitates to determine the elution profile of type II PI4K associated with D6.1A.
Since 804G cells express very little EpCAM, we repeated this gel-filtration experiment with BSp73ASML cells to examine the elution profile of EpCAM. As seen in Figure 8(C), EpCAM as well as PI4K can be precipitated with anti-D6.1A from fractions eluting after the void volume. This result reinforces the hypothesis that complexes of D6.1A with EpCAM and PI4K can occur in solubilized membrane subdomains.
DISCUSSION
Primary interactions of D6.1A
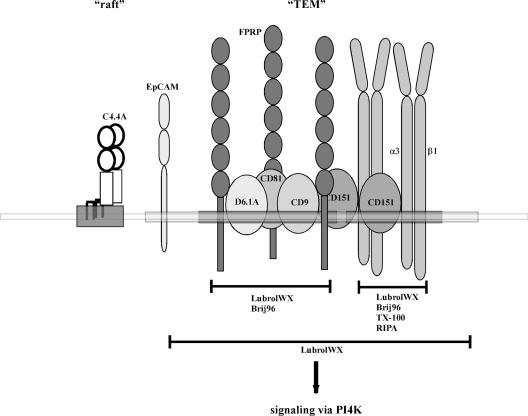
According to a recent hypothesis, the role of tetraspanins is to serve as adaptors tying together a multitude of associated proteins and thereby assemble complexes with specific signalling capabilities [8]. The aim of the present study was to determine the position and function of the tetraspanin D6.1A (CO-029 and TM4SF3) within this network. Figure 9 summarizes the findings with regard to proteins found in D6.1A complexes and characteristics of their interactions.
Figure 9. Various levels of association identified for D6.1A on carcinoma cells.
A schematic representation of the interactions determined for D6.1A and the different levels of association. Results presented in this paper were supplemented with observations published by other laboratories (see the text for details). According to this model, D6.1A is part of a core complex of directly associating proteins comprising, other than D6.1A, the tetraspanins CD9 and CD81 and the Ig family member FPRP (membrane region marked dark grey). This complex is connected to another primary complex consisting of CD151 and α3β1 integrin, probably by interactions among D6.1A, CD9 and CD81 on the one hand and CD151 on the other. In contrast with the interactions within the primary complexes that are stable in Brij96 (D6.1A/CD9/CD81/FPRP) or even RIPA detergent conditions (CD151/α3β1), this latter association is disrupted by Brij96 and withstands LubrolWX detergent only. This is also true for EpCAM, a protein that becomes part of the complex by a so far undefined type of interaction. The whole complex of primary and secondary interaction partners (the corresponding membrane region drawn in light grey) can occur in microdomains (TEM) that share raft-like features (a raft subdomain is represented by a region of the membrane harbouring the GPI-linked protein C4.4A). Nevertheless, TEMs are distinct from rafts as evident, e.g., from the lack of co-precipitation of TEM-associated proteins with raft-associated molecules and vice versa (see Figure 7). Moreover, TEM-associated proteins may get access to defined signalling molecules such as PI4K.
Herein, we establish that FPRP (prostaglandin F2α receptor-regulatory protein; also known as EWI-F or CD9P-1) is a prominent partner directly interacting with D6.1A. This Ig super-family member owes its name to the negative regulatory function it exerts on the prostaglandin F2α receptor, probably by down-regulating the ability of the receptor to bind the ligand [44]. The regulatory function of FPRP extends to other G-protein-coupled receptors as well [45]. In this respect, it is of interest that the presence of a G-protein-coupled receptor, GPR56, could be shown in CD81 complexes [46].
FPRP belongs to a newly defined family of proteins with high structural similarity. Other members of this subfamily besides FPRP include CD101, IGSF3 and EWI-2 proteins [38,39]. FPRP and EWI-2 were found to interact directly with tetraspanins CD9 and CD81 [36–39]. A third tetraspanin, CD82, was reported to associate with EWI-2 [47]. In the present study, we also find an as yet unidentified surface-biotinylated protein with an apparent molecular mass of approx. 70 kDa in D6.1A complexes; approx. 70 kDa is consistent with the mass of EWI-2. Identification of this approx. 70 kDa molecule is in progress.
We also demonstrate here that D6.1A is found associated with CD9 and CD81. Previously, the association of CD81 with FPRP has been demonstrated in the human HT1080 cell line [36]. HT1080 cells express very little CD9 and do not express the D6.1A human homologue CO-029 [10], suggesting that interaction of CD81 with FPRP occurs independent of the other two tetraspanins. CD9 has also been observed to interact directly with FPRP in human A431 cells [37], which express large amounts of both CD9 and CD81 [10]. In that study, formation of cross-linked adducts consistent in mass with one molecule of CD9 and one molecule of FPRP were detected. Thus CD9 is also capable of interacting directly with FPRP independent of other tetraspanins. Cross-linking experiments on PROb cells with the bifunctional reagent DTSP lead to the detection of FPRP-D6.1A and FPRP-CD9 complexes with a stoichiometry different from 1:1, but in accordance with oligomers of D6.1A and CD9 to interact with FPRP, leaving open the possibility that CD9 mediates the interaction between FPRP and D6.1A. Other experiments, performed on HT1080 double-transfectants expressing D6.1A and rat FPRP, indicated that D6.1A does associate with FPRP in the absence of CD9 (results not shown). Furthermore, the expression level of CD81 in the PROb cell line used here is probably too low to account for the large amounts of FPRP in D6.1A complexes, which are comparable with the amounts of FPRP found associated with CD9. Nevertheless, we cannot exclude at the moment that association of D6.1A with FPRP does depend on the presence of either CD9 or CD81. We could demonstrate however that D6.1A does not enter pre-existent complexes of FPRP and CD9. Both tetraspanins show the same time kinetic interaction with FPRP.
Therefore the interactions of D6.1A with tetraspanins CD9 and CD81 and FPRP fulfil criteria formulated for primary interactions within TEMs: they are of high stoichiometry (as shown by immunodepletion), rely on direct protein–protein interaction (shown by cross-linking with DTSP) and are of relatively small mass (complexes are retained within a Sepharose CL6B column).
Secondary and tertiary interactions of D6.1A
Several pieces of data suggest that the interaction of D6.1A with α3β1 integrin is different from the primary D6.1A interactions described above. First, although integrin is detectable in D6.1A precipitates isolated from Brij96 lysates, significantly more integrin is found when the less harsh LubrolWX detergent is used during cell lysis. In contrast, complexes of D6.1A with FPRP occur in similar quantities under both detergent conditions (results not shown). Secondly, there is very little CD151 seen in D6.1A and CD9 immunoprecipitates from Brij96 lysates. If, instead, CD151 was first overexpressed in PROb cells, then Western blots could identify small amounts of CD151 in D6.1A complexes isolated under Brij96 conditions (results not shown). Since CD151 has been shown to mediate the interaction between α3β1 and the tetraspanin network [22], we believe that our demonstration of integrin in the D6.1A complexes under Brij96 conditions is due to the high sensitivity of our assays detecting biotinylated integrin. Finally, gel filtration of D6.1A complexes on Sepharose CL6B under Brij96 conditions revealed that the integrin associated with D6.1A can be separated from the core complex and is eluted close to the void volume.
The results presented here are also consistent with the previously mentioned model [22] wherein small primary complexes are connected through secondary interactions to larger ‘super-complexes’ (D6.1A–CD9–FPRP complexes and CD151–α3β1 integrin respectively). Results demonstrating that EWI-2 is linked by CD9 and CD81 to α3β1 and that it may regulate certain α3β1-mediated functions [48] are suggestive of secondary tetraspanin complexes integrating signals from different receptors.
Previously, most of the proteins found to associate with the tetraspanins were identified while using detergents such as CHAPS, Brij35, Brij58 or Brij98; all of these detergents are relatively less harsh than Brij96 or Triton X-100. In the present study, we have chosen to use LubrolWX during analysis of interactions of D6.1A with non-core complex-associating partner molecules. LubrolWX has been shown to be suitable for the isolation of microdomains with raft-like features containing the pentaspan protein prominin [49]. In our experiments, LubrolWX was superior to Brij98 in solubilizing α6β4 integrin and GPI-linked C4.4A, even if Brij98 was used at 37 °C. Furthermore, in comparison with CHAPS, the pattern of biotinylated proteins co-immunoprecipitating with D6.1A under LubrolWX conditions was more restricted, but the proteins present in the complex occurred at higher amounts (results not shown). Two of the molecules detectable in our D6.1A complexes only under milder detergent conditions are type II PI4K and EpCAM. EpCAM is an epithelial marker protein overexpressed in many carcinomas [50] and involved in homotypic cell–cell adhesion [51]. Both type II PI4K and EpCAM were also found in comparable amounts in CD9 complexes. Interestingly, we noted recently that immunoprecipitates of CD44 isolated under the same detergent conditions also contained EpCAM, D6.1A and CD9 [52]. Immunodepletion experiments revealed that only a small fraction of the total EpCAM is associated with D6.1A. However, the lack of association of EpCAM with α2β1 integrin, which itself has little tendency to interact with tetraspanins [41], suggests that EpCAM is a true part of the TEMs. In contrast, the CD46 receptor for measles virus, which is also found in tetraspanin complexes, is found in association with α1, α2, α3, α5 and α6 integrins, indicating that its interaction is with β1 integrin rather than specifically with the tetraspanins [53]. In experiments described here, the relatively low levels of CD151 association may be due to the limited sensitivity of the antisera used or to the low abundance of CD151. In contrast, when PROb cells overexpressing CD151 were used for immunoprecipitation under LubrolWX conditions, CD151 was readily detected in EpCAM precipitates (results not shown). At present, it is unknown whether the interaction of EpCAM with TEMs represents a secondary interaction according to the criteria put forward by Hemler [9]. It is clearly possible that the association of EpCAM with TEMs is mediated by another as yet unidentified tetraspanin. Analysis of the proteins associated with EpCAM under more stringent detergent conditions and by MS might answer this question. The functional relevance of the association of EpCAM with TEMs is also as yet unidentified. EpCAM is reported to mediate homophilic adhesion of cells [51] and to play a role in proliferation [54]. It is feasible to assume a positive effect on these functions by clustering in TEMs. Both cell–cell adhesion and proliferation have also been shown to be influenced by tetraspanins [55,56].
The interaction of D6.1A and EpCAM does not rely on incomplete solubilization or intact raft domains; rather it can withstand treatment with the cholesterol-removing agent MβCD (results not shown). Furthermore, use of Brij96 conditions disrupts the interaction between D6.1A and EpCAM while leaving DRM intact. Finally, complexes of D6.1A and EpCAM can be isolated by gel filtration on a Sepharose CL4B column; these separated complexes are resistant to centrifugation at 100000 g and thus are not of a vesicular nature. Although it seems that raft-like subdomains are not the basis of the TEM architecture, this concept of subdomains with characteristic lipid composition existing in biological membranes may still be relevant for understanding the functions of TEMs. Two observations are of particular interest in this respect. First, it has been reported that the localization of the LubrolWX-insoluble protein prominin to microvilli of Madin–Darby canine kidney cells depends on the presence of cholesterol [49]. Secondly, in hepatic cells, distinct trafficking pathways are used by proteins that are insoluble in LubrolWX but soluble in Triton X-100, as well as by the proteins that are insoluble in both detergents. Cholesterol depletion leads to mistargeting of proteins that are LubrolWX-insoluble but Triton X-100-soluble [57].
Lastly, D6.1A is the only protein in the complex also containing CD9, CD81 and FPRP that encodes a putative late endosomal targeting sequence (YCQI). Type II PI4K has been reported to be involved in regulating the intracellular traffic originating from the trans-Golgi compartment [58]. Therefore future work on tetraspanin function should include an examination of intracellular trafficking.
In summary, results presented here provide evidence that D6.1A is one of the primary elements in complexes containing the protein FPRP together with tetraspanins CD9 and CD81 in carcinoma cells. This core complex associates with α3β1 integrin, CD151, PI4K and EpCAM at a higher level of integration. Future work will concentrate on the elucidation of the functions specifically conferred by D6.1A, with the primary focus on a possible role for D6.1A in regulating the intracellular localization of the associated complexes.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft grant no. ZO 40/8 (to M.Z. and C.C.) and by a grant from the Tumorzentrum Heidelberg/Mannheim (to M.Z.).
References
- 1.Szala S., Kasai Y., Steplewski Z., Rodeck U., Koprowski H., Linnenbach A. J. Molecular cloning of cDNA for the human tumor-associated antigen CO-029 and identification of related transmembrane antigens. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6833–6837. doi: 10.1073/pnas.87.17.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huerta S., Harris D. M., Jazirehi A., Bonavida B., Elashoff D., Livingston E. H., Heber D. Gene expression profile of metastatic colon cancer cells resistant to cisplatin-induced apoptosis. Int. J. Oncol. 2003;22:663–670. [PubMed] [Google Scholar]
- 3.Tanaka F., Hori N., Sato K. Identification of differentially expressed genes in rat hepatoma cell lines using subtraction and microarray. J. Biochem. (Tokyo) 2002;131:39–44. doi: 10.1093/oxfordjournals.jbchem.a003075. [DOI] [PubMed] [Google Scholar]
- 4.Kanetaka K., Sakamoto M., Yamamoto Y., Yamasaki S., Lanza F., Kanematsu T., Hirohashi S. Overexpression of tetraspanin CO-029 in hepatocellular carcinoma. J. Hepatol. 2001;35:637–642. doi: 10.1016/s0168-8278(01)00183-0. [DOI] [PubMed] [Google Scholar]
- 5.Kanetaka K., Sakamoto M., Yamamoto Y., Takamura M., Kanematsu T., Hirohashi S. Possible involvement of tetraspanin CO-029 in hematogenous intrahepatic metastasis of liver cancer cells. J. Gastroenterol. Hepatol. 2003;18:1309–1314. doi: 10.1046/j.1440-1746.2003.03182.x. [DOI] [PubMed] [Google Scholar]
- 6.Claas C., Seiter S., Claas A., Savelyeva L., Schwab M., Zoller M. Association between the rat homologue of CO-029, a metastasis-associated tetraspanin molecule and consumption coagulopathy. J. Cell Biol. 1998;141:267–280. doi: 10.1083/jcb.141.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bick R. L. Coagulation abnormalities in malignancy: a review. Semin. Thromb. Hemost. 1992;18:353–372. doi: 10.1055/s-2007-1002575. [DOI] [PubMed] [Google Scholar]
- 8.Boucheix C., Rubinstein E. Tetraspanins. Cell. Mol. Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemler M. E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 10.Serru V., Le Naour F., Billard M., Azorsa D. O., Lanza F., Boucheix C., Rubinstein E. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem. J. 1999;340:103–111. [PMC free article] [PubMed] [Google Scholar]
- 11.Yauch R. L., Kazarov A. R., Desai B., Lee R. T., Hemler M. E. Direct extracellular contact between integrin alpha(3)beta(1) and TM4SF protein CD151. J. Biol. Chem. 2000;275:9230–9238. doi: 10.1074/jbc.275.13.9230. [DOI] [PubMed] [Google Scholar]
- 12.Yauch R. L., Berditchevski F., Harler M. B., Reichner J., Hemler M. E. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell. 1998;9:2751–2765. doi: 10.1091/mbc.9.10.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loewen C. J., Molday R. S. Disulfide-mediated oligomerization of Peripherin/Rds and Rom-1 in photoreceptor disk membranes. Implications for photoreceptor outer segment morphogenesis and degeneration. J. Biol. Chem. 2000;275:5370–5378. doi: 10.1074/jbc.275.8.5370. [DOI] [PubMed] [Google Scholar]
- 14.Berditchevski F., Gilbert E., Griffiths M. R., Fitter S., Ashman L., Jenner S. J. Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 2001;276:41165–41174. doi: 10.1074/jbc.M104041200. [DOI] [PubMed] [Google Scholar]
- 15.Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X. A., Bontrager A. L., Hemler M. E. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem. 2001;276:25005–25013. doi: 10.1074/jbc.M102156200. [DOI] [PubMed] [Google Scholar]
- 17.Yauch R. L., Hemler M. E. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4-kinase. Biochem. J. 2000;351:629–637. [PMC free article] [PubMed] [Google Scholar]
- 18.Claas C., Stipp C. S., Hemler M. E. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 2001;276:7974–7984. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami Y., Kawakami K., Steelant W. F., Ono M., Baek R. C., Handa K., Withers D. A., Hakomori S. Tetraspanin CD9 is a ‘proteolipid,’ and its interaction with alpha 3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin-5-dependent cell motility. J. Biol. Chem. 2002;277:34349–34358. doi: 10.1074/jbc.M200771200. [DOI] [PubMed] [Google Scholar]
- 20.Charrin S., Manie S., Thiele C., Billard M., Gerlier D., Boucheix C., Rubinstein E. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 2003;33:2479–2489. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- 21.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 22.Charrin S., Manie S., Billard M., Ashman L., Gerlier D., Boucheix C., Rubinstein E. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 2003;304:107–112. doi: 10.1016/s0006-291x(03)00545-x. [DOI] [PubMed] [Google Scholar]
- 23.Zoller M., Matzku S., Goerttler K. High incidence of spontaneous transplantable tumours in BDX rats. Br. J. Cancer. 1978;37:61–66. doi: 10.1038/bjc.1978.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reisser D., Olsson N. O., Martin F. In vivo and in vitro reactivity of rat spleen cells against regressor and progressor colon-cancer cell variants. Int. J. Cancer. 1993;53:651–656. doi: 10.1002/ijc.2910530421. [DOI] [PubMed] [Google Scholar]
- 25.Riddelle K. S., Green K. J., Jones J. C. Formation of hemidesmosomes in vitro by a transformed rat bladder cell line. J. Cell Biol. 1991;112:159–168. doi: 10.1083/jcb.112.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claas C., Herrmann K., Matzku S., Moller P., Zoller M. Developmentally regulated expression of metastasis-associated antigens in the rat. Cell Growth Differ. 1996;7:663–678. [PubMed] [Google Scholar]
- 27.Matzku S., Wenzel A., Liu S., Zoller M. Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res. 1989;49:1294–1299. [PubMed] [Google Scholar]
- 28.Herlevsen M., Schmidt D. S., Miyazaki K., Zoller M. The association of the tetraspanin D6.1A with the alpha6beta4 integrin supports cell motility and liver metastasis formation. J. Cell Sci. 2003;116:4373–4390. doi: 10.1242/jcs.00760. [DOI] [PubMed] [Google Scholar]
- 29.DeFreitas M. F., Yoshida C. K., Frazier W. A., Mendrick D. L., Kypta R. M., Reichardt L. F. Identification of integrin alpha 3 beta 1 as a neuronal thrombospondin receptor mediating neurite outgrowth. Neuron. 1995;15:333–343. doi: 10.1016/0896-6273(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 30.Orlicky D. J., Nordeen S. K. Cloning, sequencing and proposed structure for a prostaglandin F2 alpha receptor regulatory protein. Prostaglandins Leukot. Essent. Fatty Acids. 1996;55:261–268. doi: 10.1016/s0952-3278(96)90007-1. [DOI] [PubMed] [Google Scholar]
- 31.Rupp R. A., Snider L., Weintraub H. Xenopus embryos regulate the nuclear localization of XmyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 32.Wessel D., Flugge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn J., Gotting C., Schnolzer M., Kempf T., Brinkmann T., Kleesiek K. First isolation of human UDP-D-xylose: proteoglycan core protein beta-D-xylosyltransferase secreted from cultured JAR choriocarcinoma cells. J. Biol. Chem. 2001;276:4940–4947. doi: 10.1074/jbc.M005111200. [DOI] [PubMed] [Google Scholar]
- 34.Wong K., Meyers R., Cantley L. C. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- 35.Sterk L. M., Geuijen C. A., Oomen L. C., Calafat J., Janssen H., Sonnenberg A. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin alpha6beta4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 2000;149:969–982. doi: 10.1083/jcb.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stipp C. S., Orlicky D., Hemler M. E. FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 2001;276:4853–4862. doi: 10.1074/jbc.M009859200. [DOI] [PubMed] [Google Scholar]
- 37.Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- 38.Clark K. L., Zeng Z., Langford A. L., Bowen S. M., Todd S. C. PGRL is a major CD81-associated protein on lymphocytes and distinguishes a new family of cell surface proteins. J. Immunol. 2001;167:5115–5121. doi: 10.4049/jimmunol.167.9.5115. [DOI] [PubMed] [Google Scholar]
- 39.Stipp C. S., Kolesnikova T. V., Hemler M. E. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 2001;276:40545–40554. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- 40.Ktistakis N. T., Linder M. E., Roth M. G. Action of brefeldin A blocked by activation of a pertussis-toxin-sensitive G protein. Nature (London) 1992;356:344–346. doi: 10.1038/356344a0. [DOI] [PubMed] [Google Scholar]
- 41.Hemler M. E., Mannion B. A., Berditchevski F. Association of TM4SF proteins with integrins: relevance to cancer. Biochim. Biophys. Acta. 1996;1287:67–71. doi: 10.1016/0304-419x(96)00007-8. [DOI] [PubMed] [Google Scholar]
- 42.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skubitz K. M., Campbell K. D., Skubitz A. P. CD63 associates with CD11/CD18 in large detergent-resistant complexes after translocation to the cell surface in human neutrophils. FEBS Lett. 2000;469:52–56. doi: 10.1016/s0014-5793(00)01240-0. [DOI] [PubMed] [Google Scholar]
- 44.Orlicky D. J. Negative regulatory activity of a prostaglandin F2 alpha receptor associated protein (FPRP). Prostaglandins Leukotrienes Essent. Fatty Acids. 1996;54:247–259. doi: 10.1016/s0952-3278(96)90055-1. [DOI] [PubMed] [Google Scholar]
- 45.Orlicky D. J., Lieber J. G., Morin C. L., Evans R. M. Synthesis and accumulation of a receptor regulatory protein associated with lipid droplet accumulation in 3T3-L1 cells. J. Lipid Res. 1998;39:1152–1161. [PubMed] [Google Scholar]
- 46.Little K. D., Hemler M. E., Stipp C. S. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol. Biol. Cell. 2004;15:2375–2387. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X. A., Lane W. S., Charrin S., Rubinstein E., Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003;63:2665–2674. [PubMed] [Google Scholar]
- 48.Stipp C. S., Kolesnikova T. V., Hemler M. E. EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 2003;163:1167–1177. doi: 10.1083/jcb.200309113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roper K., Corbeil D., Huttner W. B. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 50.Balzar M., Winter M. J., de Boer C. J., Litvinov S. V. The biology of the 17-1A antigen (Ep-CAM) J. Mol. Med. 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- 51.Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., Warnaar S. O. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 1994;125:437–446. doi: 10.1083/jcb.125.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt D. S., Klingbeil P., Schnolzer M., Zoller M. CD44 variant isoforms associate with tetraspanins and EpCAM. Exp. Cell Res. 2004;297:329–347. doi: 10.1016/j.yexcr.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 53.Lozahic S., Christiansen D., Manie S., Gerlier D., Billard M., Boucheix C., Rubinstein E. CD46 (membrane cofactor protein) associates with multiple beta1 integrins and tetraspans. Eur. J. Immunol. 2000;30:900–907. doi: 10.1002/1521-4141(200003)30:3<900::AID-IMMU900>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 54.Schon M. P., Schon M., Klein C. E., Blume U., Bisson S., Orfanos C. E. Carcinoma-associated 38-kD membrane glycoprotein MH 99/KS 1/4 is related to proliferation and age of transformed epithelial cell lines. J. Invest. Dermatol. 1994;102:987–991. doi: 10.1111/1523-1747.ep12384258. [DOI] [PubMed] [Google Scholar]
- 55.Shigeta M., Sanzen N., Ozawa M., Gu J., Hasegawa H., Sekiguchi K. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J. Cell Biol. 2003;163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi W., Fan H., Shum L., Derynck R. The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced EGF receptor activation and cell proliferation. J. Cell Biol. 2000;148:591–602. doi: 10.1083/jcb.148.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slimane T. A., Trugnan G., Van IJzendoorn S. C., Hoekstra D. Raft-mediated trafficking of apical resident proteins occurs in both direct and transcytotic pathways in polarized hepatic cells: role of distinct lipid microdomains. Mol. Biol. Cell. 2003;14:611–624. doi: 10.1091/mbc.E02-08-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell (Cambridge, Mass.) 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]