Abstract
The widely held view that SLN (sarcolipin) would be the natural inhibitor of SERCA1 (sarcoplasmic/endoplasmic-reticulum Ca2+-ATPase 1), and PLB (phospholamban) its counterpart for SERCA2 inhibition is oversimplified and partially wrong. The expression of SLN and PLB mRNA and protein relative to SERCA1 or SERCA2 was assessed in ventricle, atrium, soleus and EDL (extensor digitorum longus) of mouse, rat, rabbit and pig. SLN protein levels were quantified by means of Western blotting using what appears to be the first successfully generated antibody directed against SLN. Our data confirm the co-expression of PLB and SERCA2a in cardiac muscle and the very low levels (in pig and rabbit) or the absence (in rat and mouse) of PLB protein in the slow skeletal muscle. In larger animals, the SLN mRNA and protein expression in the soleus and EDL correlates with SERCA1a expression, but, in rodents, SLN mRNA and protein show the highest abundance in the atria, which are devoid of SERCA1. In the rodent atria, SLN could therefore potentially interact with PLB and SERCA2a. No SLN was found in the ventricles of the different species studied, and there was no compensatory SLN up-regulation for the loss of PLB in PLB−/− mouse. In addition, we found that SLN expression was down-regulated at the mRNA and protein level in the atria of hypertrophic hearts of SERCA2b/b mice. These data suggest that superinhibition of SERCA by PLB-SLN complexes could occur in the atria of the smaller rodents, but not in those of larger animals.
Keywords: atrium, cardiomyopathy, phospholamban, sarcolipin, sarcoplasmic/endoplasmic-reticulum Ca2+-ATPase (SERCA), skeletal muscle
Abbreviations: EDL, extensor digitorum longus; NF-SLN, sarcolipin tagged N-terminally with a FLAG epitope; PLB, phospholamban; RPA, RNase-protection assay; RT, reverse transcription; SERCA, sarcoplasmic/endoplasmic-reticulum Ca2+-ATPase; SLN, sarcolipin; WT, wild-type
INTRODUCTION
PLB (phospholamban) and SLN (sarcolipin) are two small homologous intrinsic membrane proteins (respectively comprising 52 and 31 amino acid residues) which are confined to the endoplasmic reticulum or the sarcoplasmic reticulum. Both proteins can inhibit the SERCA (sarcoplasmic/endoplasmic-reticulum Ca2+-ATPase) activity by lowering the apparent Ca2+ affinity of the pump [1]. When both PLB and SLN are co-expressed in vitro, their inhibitory effect becomes synergistic, leading to superinhibition of the pump. This can be partially attributed to the ability of SLN to disrupt PLB pentamers, leading to an increase in the amount of PLB monomers, the inhibitory form of PLB [2]. Also, recent modelling studies suggest that PLB and SLN could bind side-by-side in a lateral groove of SERCA1 forming a more stable complex than in the absence of SLN [3]. This tighter binding would explain part of the superinhibition of SERCA [3].
Studies, mainly on rabbits, have led to the widely accepted view that PLB is expressed predominantly in cardiac and slow-twitch skeletal muscle, where it regulates the SERCA2a isoform of calcium pumps, whereas, in fast-twitch muscle, SLN represents the corresponding regulator of SERCA1 [4]. However, several recent observations suggest that this picture is over-simplified. Minamisawa et al. [5] showed recently that SLN was expressed at the mRNA level in the mouse atria in the absence of significant SERCA1 levels. SLN and PLB can interact in vitro with SERCA1 and SERCA2a, affecting their apparent Ca2+ affinity to the same extent [2]. Moreover, the synergistic action of SLN and PLB on SERCA activity and the recent modelling studies suggest a physiological role for SLN and PLB co-expression. However, only fragmentary data are available on the relative co-expression of SLN and PLB mRNA in various muscles, and there is an almost complete lack of such information at the protein level. This information is necessary in order to assess the full physiological implications of potential synergistic interaction of both regulators. This prompted us to specifically look out for potential co-expression of SLN and PLB in vivo. Therefore a more general study of the relative expression of SLN and PLB in relation to SERCA in different muscles of small and large animals was undertaken.
Recent studies have shown that SLN mRNA is present in the heart, notably where PLB is abundantly expressed, and suggested that changes in SLN expression might alter cardiac function in several cardiomyopathies. Indeed, SLN mRNA was found in human [6], mouse [5] and rat atria [7], but not in the ventricle. Furthermore, it was recently shown that SLN mRNA is down-regulated in atria of patients with chronic atrial fibrillation [8] and in atria of activated H-Ras transgenic mice [5]. Thus altered SLN expression may contribute to different Ca2+ handling in cardiac disease. However, comparing SLN mRNA levels may only give a partial, possibly even misleading, picture. Several studies have shown that the expression of PLB must be strongly controlled at the protein level [9–10]. A similar post-transcriptional control of SLN expression might exist. Therefore the present study included both mRNA and protein assays.
EXPERIMENTAL
Animals
All animal experimentation was performed in accordance with approved protocols of the local ethics committee and conforms to the Guide for the Care and Use of Laboratory Animals of the National Academy of Sciences (http://www.nap.edu/readingroom/books/labrats/). The animals used in the present study were: adult mice (strain 129sV/Swiss 50/50, 12 weeks old), adult rats (Wistar, 15 weeks old), young adult rabbits (White New Zealand, 3 months old, 3.5 kg) and young domestic pigs (27–32 kg; Seghers GenTech, Lebbeke, Belgium).
RT (reverse transcription) PCR
Total RNA was isolated from EDL (extensor digitorum longus) (fast-twitch skeletal muscle), soleus (slow-twitch skeletal muscle), atria and ventricles from mouse, rat, rabbit and pig using the TRIzol® method (Invitrogen). RNA was treated with DNase I to avoid genomic DNA contamination. cDNA was prepared from 1 μg of RNA in a final volume of 20 μl according to the manufacturer's protocol (Thermoscript RT-PCR kit; Invitrogen). The amount of cDNA mixture for PCR was adjusted to obtain equal levels of SERCA for each tissue. Negative controls without reverse transcriptase were tested for each sample. Primers were designed for SLN, PLB and SERCA1/2 of mouse, rat, rabbit and pig (Supplementary Table A at http://www.BiochemJ.org/bj/389/bj3890151add.htm). The number of cycles was chosen to fall in the exponential phase of amplification. The SERCA primers were designed to co-amplify SERCA1 and SERCA2 with the same efficiency. SERCA1 and SERCA2 each contain a different unique restriction site in the amplified product (Supplementary Table B at http://www.BiochemJ.org/bj/389/bj3890151add.htm). Hence, the relative contribution of both isoforms could be assessed after restriction analysis of the PCR product.
RPA (RNase-protection assay)
To check mRNA levels in different mouse models, we generated a 111-nucleotide SLN RNA probe which we used for RPA. Therefore we cloned the SLN coding region in a pGEMTeasy vector and linearized it with SpeI. T7 RNA polymerase was added with α-32P-radiolabelled UTP to create an antisense probe. A standard RPA protocol was used as described previously [9].
Production of anti-SLN antibodies
A peptide (MERSTQELFINFC) corresponding to the 12-residue-long cytosolic part of mouse/rat SLN with an additional cysteine added to its C-terminus was conjugated to keyhole-limpet haemocyanin. Rabbits were immunized with this antigen, and the serum obtained was designated SLN78. For affinity purification, the immunizing peptide was coupled to AF-Amino Toyopearl 650 M. The bound antibodies were eluted from the affinity gel with 100 mM glycine (pH 2.5). The affinity-purified antibodies were designated SLNAP78. Both antibody production and affinity purification occurred at Eurogentec (Seraing, Belgium).
Western-blot analysis
Isolated tissue from EDL, soleus, atria and ventricles of mouse, rat, rabbit and pig were homogenized in a buffer containing 10 mM imidazole (pH 7.0), 300 mM sucrose and Complete™ protease inhibitor cocktail (Roche). For mouse atria, soleus and EDL, tissue was pooled from six animals. Protein concentration was determined using the BCA (bicinchoninic acid) kit (Pierce). Amounts of homogenates were adjusted to obtain equal SERCA loading for each tissue. Samples were separated on 4–12% Bis-Tris gradient gels (NuPAGE™ electrophoresis system; Invitrogen) and blotted on to PVDF membranes (0.22 μm; Bio-Rad) according to the manufacturer's protocol. Blots were probed with a polyclonal antiserum for SLN78 (1:500), an affinity-purified polyclonal anti-SLN antibody (SLNAP78, 1:200), a monoclonal anti-PLB antibody (mA1, 1:20000, Upstate Biotechnology) and with a home-made pan-specific polyclonal anti-SERCA antibody TRY2 (1:2000) [11]. Detection was performed with alkaline-phosphatase-coupled secondary antibodies, using the Vistra enhanced chemifluorescence system (Amersham Biosciences).
Preparation of SERCA, SLN and PLB standards
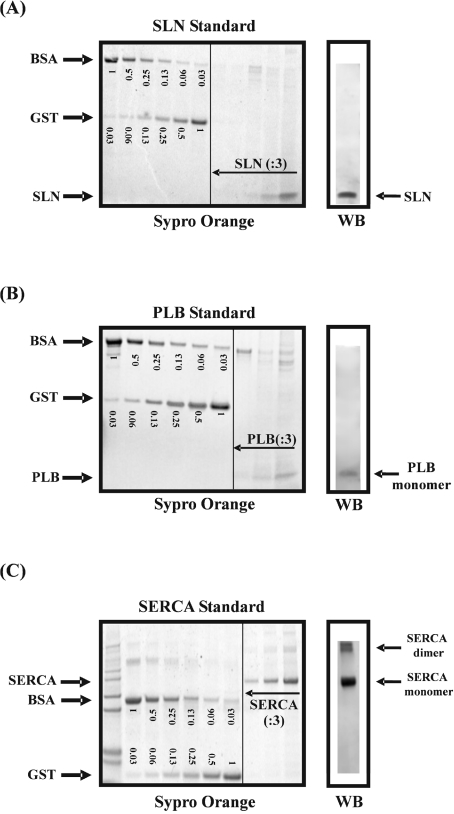
SERCA was enriched from rabbit EDL. Homogenates were fractionated at 4 °C for 20 min at 1000 g, 20 min at 15000 g and 30 min at 200000 g. Microsomes were treated further with 0.6 M KCl and 100 mM Na2CO3 (pH 11). SLN was purified from pig EDL by the procedure of Ohnoki and Martonosi [12]. Briefly, a microsomal fraction was prepared which was treated further with 9 vol. of methanol. Acidified methanol/chloroform (2:1, with 10 mM HCl) was added to the residue and 5 vol. of cold diethyl ether were used to precipitate the crude proteolipid fraction. Finally, SLN was extracted with and stored in n-butanol at −80 °C. The identity and purity of the extract was confirmed by SDS electrophoresis and N-terminal amino acid sequence analysis (Edman degradation), which was performed on a Procise 492 microsequencer (Applied Biosystems) running in pulsed liquid mode. The sample was loaded on to a polybrene-coated glass fibre disc, previously pre-cycled four times. To obtain a protein standard for PLB, PLB was extracted from rabbit ventricle according to the butanol extraction method of Ohnoki and Martonosi [12], as described for the purification of SLN. We observed that PLB co-extracted with SLN from tissue that contains both proteins. The purity of the PLB extract was analysed on a polyacrylamide gel, stained with Sypro Orange (Invitrogen). The amounts of SERCA, PLB and SLN were compared with a known amount of BSA and GST (glutathione S-transferase) on a gel stained with Sypro Orange. The fluorescent signal was visualized with a Storm840 FluorImager (Amersham Biosciences).
Statistics
Data shown are means±S.E.M. Statistical analysis was performed using Student's t test or by ANOVA followed by Bonferroni's t test. P<0.05 is considered to be significant.
RESULTS
Levels of SLN and PLB mRNA normalized to those of SERCA
Mainly because antibodies directed against SLN were previously unavailable, and because the systematic quantification of the SLN proteolipid via butanol extraction is cumbersome, earlier studies on SLN expression only assessed the relative expression levels of its mRNA [5]. In a first series of experiments, we therefore also measured the mRNA levels of SLN and PLB in total RNA extracts from cardiac ventricle, cardiac atrium, slow-twitch soleus and fast-twitch EDL from mice, rats, rabbits and pigs by means of RT-PCR. Since SLN and PLB act as regulators of SERCA, the relative expression levels of SLN and PLB mRNA were normalized to those of SERCA. Hence, the amount of total input RNA in the RT-PCR assay of SLN and PLB was adjusted to yield equal SERCA signals in parallel assays. The obtained SLN/SERCA and PLB/SERCA mRNA ratios were expressed as percentages of the ratio obtained for atria, tissue that contains both SLN and PLB mRNA. According to their fibre-type composition, muscles express SERCA1 or SERCA2, or a mixture of both. Therefore the primers used in the RT-PCR were designed to co-amplify SERCA1 and SERCA2 cDNA with the same efficiency. The relative expression levels of SERCA1 and SERCA2 were then determined by restriction analysis of the amplified DNAs as detailed in Supplementary Table B (http://www.BiochemJ.org/bj/389/bj3890151add.htm).
The results depicted in Figure 1 and Table 1 confirm the general view that SLN mRNA is highly expressed in the fast-twitch EDL of rabbit and, furthermore, now also indicates that this is also the case for the pig EDL. Remarkably, the expression of SLN relative to SERCA is nearly as high in the slow-twitch soleus of rabbit and pig. Surprisingly, in the smaller animals, such as rat and mouse, no SLN mRNA was detected in EDL. Instead, both rodents express small amounts of SLN mRNA in the soleus, but clearly show the highest SLN mRNA levels in the cardiac atria, a tissue that does not express SERCA1. High SLN mRNA expression levels in the murine and rat atria have been reported earlier [5,7]. Furthermore, it is clear that the ventricle does not contain SLN.
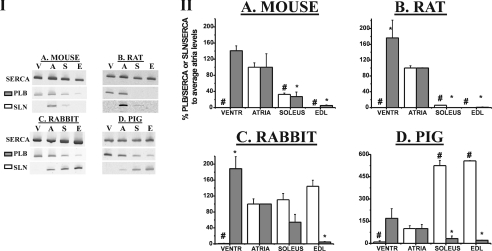
Figure 1. Determination of SLN/SERCA and PLB/SERCA mRNA ratios using RT-PCR.
The amount of cDNA used for amplification was adjusted to yield equal amounts of SERCA. The panels in (I) show representative examples of RT-PCR on cDNA of different muscle types (V, ventricles; A, atria; S, soleus; E, EDL). Four different species, as indicated, were analysed in parallel. The primers and PCR conditions summarized in Supplementary Table A (http://www.BiochemJ.org/bj/389/bj3890151add.htm) were used to determine expression levels of SLN, PLB and SERCA. The panels in (II) summarize the PLB/SERCA (grey) or SLN/SERCA (white) mRNA ratio represented as mean percentage (±S.E.M.) compared with atrial levels. For mouse, tissue was pooled from six different animals to prepare total RNA (n=2). For other species, n=3. #SLN/SERCA mRNA ratio significantly different (P<0.05) compared with atria. *PLB/SERCA mRNA ratio significantly different (P<0.05) compared with atria. VENTR, ventricles.
Table 1. Summary of SERCA, SLN and PLB mRNA expression levels.
Summary of data presented in Figure 1. Relative ratios of SLN/SERCA (a) and PLB/SERCA (b) normalized to the average percentage of atria as determined by RT-PCR. For mouse, tissue was pooled from six different animals to prepare total RNA (n=2). For other species, n=3. Analysis was performed in triplicate. Results are expressed as means±S.E.M. N.D., none detected.
| (a) SLN/SERCA mRNA ratios | ||||
|---|---|---|---|---|
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | N.D.† | N.D.† | N.D.† | 0.5±0.2† |
| Atria | 100.0±10.7 | 100.0±5.9 | 100.0±12.8 | 100.0±19.4 |
| Soleus | 32.7±3.1† | 5.9±0.1† | 110.8±15.6 | 524.2±35.3† |
| EDL | N.D.† | N.D.† | 144.4±15.4 | 556.7±2.1† |
| (b) PLB/SERCA mRNA ratios | ||||
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | 140.8±12.1 | 176.1±35.0* | 188.5±30.2* | 169.1±65.4 |
| Atria | 100.0±33.7 | 100.0±0.7 | 100.0±0.2 | 100±28.0 |
| Soleus | 27.2±11.3* | N.D.* | 54.2±19.8 | 32.2±18.0* |
| EDL | 5.2±1.4* | 0.7±0.5* | 4.5±1.3* | 21.4±1.5* |
† SLN/SERCA mRNA ratio significantly different (P<0.05) compared with atria.
* PLB/SERCA mRNA ratio significantly different (P<0.05) compared with atria.
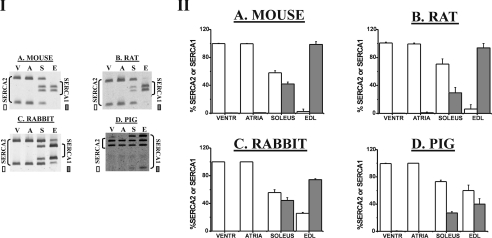
The PLB/SERCA mRNA ratio of all four animal species is the highest in ventricle and the lowest in EDL. For rabbit and pig, this corresponds roughly to a complementary expression pattern as compared with that of SLN mRNA (Figure 1 and Table 1). Figure 2 and Table 2 show the percentages of SERCA2 and SERCA1 mRNA compared with total SERCA mRNA expression levels for the different animal species. Conforming to the general expectation, with the notable exception of the pig, SERCA1 is most highly expressed in fast-twitch EDL of all species. In the slow-twitch soleus SERCA2 mRNA prevails in all four species.
Figure 2. Determination of SERCA isoforms using ratio PCR.
Mouse (A), rat (B), rabbit (C) and pig (D) were analysed in parallel. The amount of cDNA used for RT was adjusted to yield equal amounts of total SERCA. Following PCR, restriction digestion (see enzymes in Supplementary Table A at http://www.BiochemJ.org/bj/389/bj3890151add.htm) was performed to discriminate between SERCA1 and SERCA2. The panels in (I) show representative examples of ratio PCRs on cDNA of different muscle types (V, ventricles; A, atria; S, soleus; E, EDL). The panels in (II) summarize the data expressed as the means (±S.E.M.) of percentage SERCA1/total SERCA (grey) or percentage SERCA2/total SERCA (white) mRNA ratios. Mouse tissue was pooled from six different animals to prepare total RNA (n=2). For other species, n=3. VENTR, ventricles.
Table 2. Summary of ratio PCR of different SERCA isoforms.
Summary of data presented in Figure 2. Results are the percentage SERCA2 (a) and percentage SERCA1 (b) of total SERCA (SERCAtot) mRNA levels per tissue per species. For mouse, tissue was pooled from six different animals to prepare total RNA (n=2). For other species, n=3. Analysis was performed in triplicate. Results are expressed as means±S.E.M. N.D., none detected.
| (a) Percentage SERCA2/SERCAtot | ||||
|---|---|---|---|---|
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | 99.8±0.4 | 99.8±1.3 | 100.0 | 99.5±0.4 |
| Atria | 99.7±0.6 | 99.2±1.2 | 100.0 | 100.0 |
| Soleus | 58.0±2.8 | 70.5±7.6 | 55.7±4.1 | 73.1±2.0 |
| EDL | 2.5±2.4 | 6.3±4.5 | 25.7±1.3 | 60.0±8.1 |
| (b) Percentage SERCA1/SERCAtot | ||||
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | 0.3±0.3 | 0.2±0.2 | N.D. | 0.5±0.4 |
| Atria | 0.3±0.2 | 0.8±0.7 | N.D. | N.D. |
| Soleus | 42.0±2.8 | 29.5±7.6 | 44.3±4.1 | 26.9±2.0 |
| EDL | 98.5±4.0 | 93.7±6.3 | 74.3±1.3 | 40.0±8.1 |
Characterization of the anti-SLN antibody
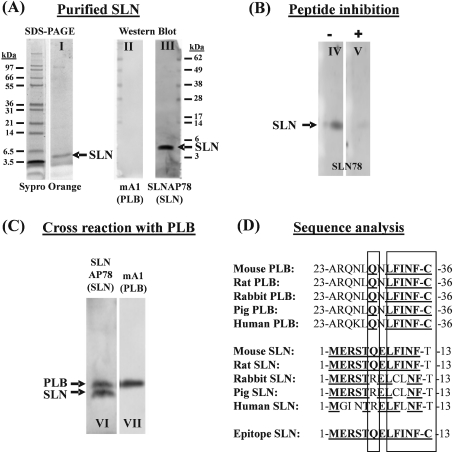
We raised a rabbit antiserum (SLN78) against the cytosolic part (i.e. the peptide comprising the first 12 amino acids counting from the N-terminal side) of the rat/mouse SLN. A cysteine residue was added to the C-terminus of the antigen peptide to link it to keyhole-limpet haemocyanin as a carrier protein. These antibodies were used for Western blotting. To convert relative expression levels of SLN, PLB and SERCA into absolute values, SLN, PLB and SERCA proteins were purified and used as an absolute standard. Purified SLN was obtained from pig EDL via the butanol extraction method (Figure 3A, lane I), and its protein sequence was confirmed by direct sequencing (Edman degradation). Notably, on a Western blot, no PLB could be detected in the purified SLN fraction (Figure 3A, lane II). The SLNAP78 antibody recognizes purified SLN on a Western blot, as indicated in lane III of Figure 3(A), and addition of the antigen peptide in solution suppresses the antibody-SLN binding (Figure 3B, lanes IV and V). Unfortunately, the anti-SLN antibody can cross-react with PLB, as indicated in Figure 3(C), lanes VI and VII. The extra cysteine residue on the antigen peptide of SLN causes greater sequence similarity with PLB and might therefore favour this unwanted cross-reaction of the antibody (Figure 3D). Our Western blots of PLB−/− and SERCA2b/b atria depicted in Figure 6(C) indicate further that the anti-SLN antibodies cross-react with PLB. Indeed, the additional band with lower mobility that reacts positively with anti-SLN antibodies is absent in PLB−/− and becomes more pronounced in SERCA2b/b mice that respectively lack or show up-regulation of PLB. Furthermore, when the blots were stained in parallel for PLB (Figure 6C), the expression levels of PLB correlate nicely with the strength of the upper band.
Figure 3. Characterization of the anti-SLN antibody.
(A) Purified SLN (0.1 μg) was visualized with Sypro Orange after SDS/PAGE (lane I). After transfer on to a PVDF membrane, the SLNAP78 antibody recognizes the purified SLN (lane III). No PLB is present in the purified SLN (lane II) as shown by incubation of the blot with an anti-PLB antibody. (B) Addition of antigenic peptide in solution (+) during primary antibody incubation (lane V) can reduce the antibody reaction with purified SLN (−) (lane IV). (C) The SLNAP78 antibody cross-reacts with PLB. Homogenate of mouse atria (70 μg) was transferred to a blot and stained in parallel with the SLNAP78 (lane VI) and anti-PLB antibody (lane VII). PLB (apparent molecular mass 5 kDa) and SLN (apparent molecular mass 4 kDa) were discriminated by size. (D) Comparison of the amino acid sequence of homologous parts of PLB, SLN and the epitope used to develop anti-SLN antibodies. Bold and underlined amino acids are identical with the epitope. As indicated by the two rectangles, the C-terminal part of the PLB sequence shows striking similarity to the corresponding mouse and rat SLN sequence, and to the epitope.
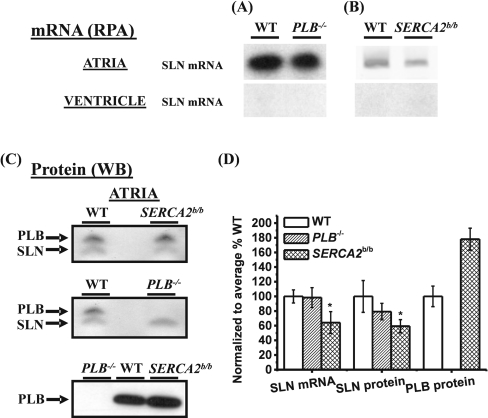
Figure 6. Representative examples of the relative quantification of SLN mRNA and protein expression in atria or ventricle of PLB−/− and SERCA2b/b.
RPA was performed to estimate SLN mRNA levels in atria or ventricle of PLB−/− (A) and SERCA2b/b (B). (C) After Western blotting (WB) of 60 μg homogenates, the SLNAP78 antibody was used to determine relative SLN protein levels in atria of PLB−/− and SERCA2b/b. In parallel, blots were stained with the anti-PLB antibody mA1. (D) Summary of the SLN mRNA, SLN protein and PLB protein expression levels. Atria were pooled from six to eight different animals to prepare total RNA or homogenates. Analysis was performed in triplicate. Results are mean percentages±S.E.M. of expression compared with WT. *Significantly different compared with WT (P<0.05).
Preparation of the SERCA, SLN and PLB standards
SERCA, SLN and PLB were respectively enriched from rabbit EDL, pig EDL and rabbit ventricle. The purity of the standards was verified by gel electrophoresis stained with Sypro Orange (Figures 4A–4C). The concentrations of the standards were determined by comparison with a BSA and GST standard. The identity of the bands was confirmed by Western blotting. To identify SERCA on a Western blot, we used a pan-specific antibody (TRY2, epitope PDPRAVNQDKKN [11]) which does not discriminate between SERCA1 and SERCA2.
Figure 4. Purified SLN (A), PLB (B) and SERCA (C) proteins were evaluated by SDS/PAGE after staining with Sypro Orange.
In parallel, SLN, PLB and SERCA were identified via Western blotting (WB) using the SLNAP78, mA1 and TRY2 antibodies respectively. To determine the absolute amount of SLN, PLB and SERCA in the standard, the intensity of the purified protein bands on the Sypro-Orange-stained gel were compared with two protein standards of known concentrations: BSA and GST. On the gels, the amounts of BSA and GST are indicated in μg. Serial dilutions (:3) of SLN, PLB and SERCA were analysed.
The levels of SLN protein normalized to those of SERCA
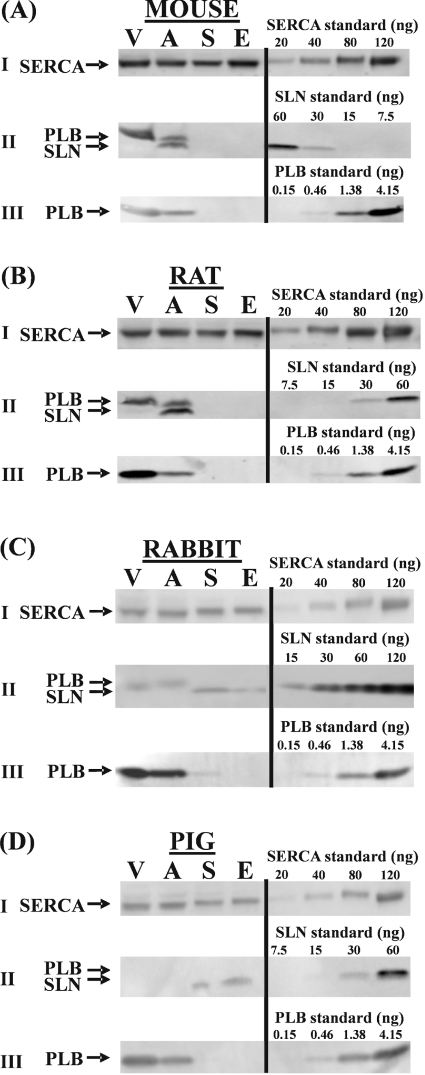
In order to assess the relative expression levels of SLN protein in the different muscle types by means of Western blotting, we adjusted the loads of homogenates applied to the electrophoresis gel to yield identical signals of SERCA. This was verified by immunostaining. In parallel, Western blots with the same ratios of protein loading were immunostained for SLN and PLB (Figure 5). The results of these analyses are summarized in Table 3. Table 3(a) expresses the amounts of SERCA per mg of protein content of the different muscle homogenates. As expected, the fast-twitch skeletal muscles express the highest relative amounts of SERCA. Except for mouse, homogenates from atrium contain 2–3-fold more SERCA than ventricle homogenates, which is again in line with earlier reports [13].
Figure 5. Western-blot analysis of total homogenates of different muscle types (V, ventricles; A, atria; S, soleus; E, EDL) of (A) mouse, (B) rat, (C) rabbit and (D) pig.
The amount of protein on the gel was adjusted to yield equal amounts of SERCA on a Western blot stained with TRY2 anti-SERCA antibody (I). Anti-SLN (II) and anti-PLB antibody (III) were used to stain parallel Western blots loaded with the same relative protein ratios. Cross-reaction of the anti-SLN antibody with PLB is indicated in (II). To estimate the amount of SLN, PLB and SERCA, samples were compared with an absolute standard for SLN, PLB and SERCA. Results are presented in Table 3. Mouse tissue was pooled from six different animals to prepare homogenates (n=2). For other species, n=3.
Table 3. Summary of SERCA, SLN and PLB protein expression levels.
(a) Amount of SERCA in μg per mg of protein of tissue homogenate. Levels were obtained by comparison of the amount of SERCA on a Western blot with an absolute SERCA standard as shown in Figure 4. The SERCA levels in μg were corrected per mg of protein loaded on to the blot. (b) Absolute quantification of SLN and SERCA via Western blotting (Figure 4) permitted the calculations of SLN/SERCA molar ratios (molecular mass of mouse SLN=3807.27 g/mol and molecular mass of SERCA1 and SERCA2=112575.2 g/mol). (c) Absolute quantification of PLB via Western blotting (Figure 4) permitted the calculations of PLN/SERCA molar ratios (molecular mass of rabbit PLB=6094.28 g/mol). (d) SLN/PLB molar ratios are presented as means±S.E.M. For mouse, tissue was pooled from six different animals to prepare total RNA (n=2). For other species, n=3. N.D., none detected; N.A., not applicable.
| (a) μg of SERCA/mg of protein | ||||
|---|---|---|---|---|
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | 8.8±0.7 | 3.9±0.1 | 2.15±0.0 | 3.5±0.1 |
| Atria | 9.9±0.9 | 9.9±0.3 | 6.1±0.0 | 7.8±0.3 |
| Soleus | 16.3±0.1 | 18.8±1.0* | 13.9±1.3 | 15.2±0.2* |
| EDL | 141.8±3.4* | 64.9±2.8* | 96.8±7.1* | 43.3±0.8* |
| (b) mol of SLN/mol of SERCA | ||||
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | N.D. | N.D. | N.D. | N.D. |
| Atria | 1.24±0.25 | 1.61±0.24 | N.D. | N.D. |
| Soleus | N.D. | N.D. | 0.87±0.21 | 0.25±0.06 |
| EDL | N.D. | N.D. | 0.37±0.07 | 0.41±0.05 |
| (c) mol PLB/mol SERCA | ||||
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | 5.1±0.5 | 10.9±2.3 | 22.5±1.8 | 6.7±0.2 |
| Atria | 1.5±0.5* | 3.3±0.9* | 13.1±3.8* | 4.3±0.3* |
| Soleus | N.D. | N.D. | 1.35±0.9* | 0.2±0.1* |
| EDL | N.D. | N.D. | 0.14±0.05* | 0.08±0.01* |
| (d) molar ratio SLN/PLB | ||||
| Tissue | Mouse | Rat | Rabbit | Pig |
| Ventricles | N.A. | N.A. | N.A. | N.A. |
| Atria | 0.84±0.33 | 0.49±0.15 | N.A. | N.A. |
| Soleus | N.A. | N.A. | 0.64±0.35 | 1.2±0.58 |
| EDL | N.A. | N.A. | 2.74±1.04 | 5.1±1.05 |
* Significantly different compared with ventricles (P<0.05).
Table 3(b) gives the calculated molar ratios of SLN/SERCA for the four different types of muscle in each of the four animal species. SLN protein was not detected in any of the cardiac ventricles. It fell also below the detection limit in the skeletal muscles of mouse and rat, but was clearly expressed in both the fast- and slow-twitch muscle of rabbit and pig. In EDL of pig and rabbit, there was a nearly 3-fold excess of SERCA over SLN. Remarkably, rabbit soleus showed a higher SLN/SERCA molar ratio than the fast-twitch EDL, but because EDL contains relatively more SERCA than soleus, the absolute SLN content in EDL is still approx. 3-fold higher compared with soleus. The highest SLN/SERCA ratios (1.2–1.6-fold molar excess of SLN over SERCA) are found in atria of rat and mouse. In contrast with the mRNA data, SLN protein was not detected in the atria of rabbit and pig.
The levels of PLB protein normalized to those of SERCA
Of all the muscles studied, ventricles expressed the highest PLB protein levels (Figure 5). Table 3(c) displays the PLB/SERCA molar ratios. The highest ratio was found in the ventricles in all species. The expression levels in atria amount to only approx. two-thirds of those in the ventricle of pig and rabbit and to only approx. one-third in the rat and mouse. In the larger animals (pig and rabbit), the amount of PLB protein in the slow-twitch muscle amounts to only a few percent of the corresponding value in ventricles. In EDL, PLB levels are even lower. PLB protein levels fell below detection levels in skeletal muscles (whether slow- or fast-twitch) of rat and mouse. The SLN/PLB molar ratios are presented in Table 3(d). In the atria of rat and mouse, but also in the soleus of rabbit, absolute levels of PLB are approx. 2-fold higher than of SLN. In the EDL of rabbit and pig, there is a 3–5-fold molar excess of SLN over PLB.
SLN expression in two different mouse models
PLB−/− mice are among the most intensely studied mouse models and are characterized by a hyperdynamic state of the heart [14]. To date, it had not been investigated whether the loss in the PLB protein caused any compensatory up-regulation in SLN expression in the ventricles or atria. Therefore we checked both mRNA and protein SLN levels in the ventricles and the atria of PLB−/−. As shown in Figure 6, no difference in SLN expression levels could be found either at the mRNA [Figure 6A, WT (wild-type) 100±8.9% compared with PLB−/− 98.4±13.6%; P>0.05] or at the protein level (Figure 6C, WT 100±21.7% compared with PLB−/− 79.3±11.2%; P>0.05).
It has been shown previously that SLN mRNA levels decrease in the atria of activated H-Ras transgenic mice, which is a model for left ventricular hypertrophy [5]. This prompted us to investigate the SLN levels in SERCA2b/b mice, which also develop concentric ventricular hypertrophy [9]. SERCA2b/b mice, in which the Atp2a2 (SERCA2) gene is modified such that SERCA2a-specific splicing is excluded, express only SERCA2b in atria and ventricles, instead of the usual SERCA2a. These mice are marked with an increased cardiac expression of PLB at the protein level (Figure 6C). Figure 6(B) shows that the SERCA2b/b mice express less SLN in the atria both at the mRNA (Figure 6B, 64.3±17.9% compared with WT, P<0.05) and protein level (Figure 6C, 59.3±8.9 compared with WT, P<0.05). Again, in the ventricles, no SLN expression could be found.
DISCUSSION
Anti-SLN antibody recognizes SLN
In vitro studies have shown that PLB and SLN can form a superinhibitory ternary complex with SERCA, but it is still in debate whether this could be physiologically relevant in vivo [2]. In that respect, it was of particular interest to look for potential co-expression of SLN and PLB in vivo. Therefore we examined four different muscle types from four animal species and determined SLN, PLB and SERCA at both the mRNA and protein levels. Previous attempts to make an anti-SLN antibody were unsuccessful, and the lack of anti-SLN antibodies limited expression studies at the protein level [4,7]. To circumvent the need for anti-SLN antibodies, earlier studies made use of a N-terminally FLAG-labelled SLN (NF-SLN) instead of SLN to track its expression in HEK-293 (human embryonic kidney) cells or isolated cardiomyocytes [2,4,7]. But a disadvantage of the presence of the N-terminal FLAG is that it might change the behaviour of SLN. Now, for the first time, anti-SLN antibodies were generated which enabled us to quantify endogenous SLN protein levels in tissue homogenates. These antibodies were proved to recognize purified SLN. Because technical details of previous attempts were unpublished, it is not clear why this attempt to produce anti-SLN antibodies was more successful.
Besides SLN of rodents, the anti-SLN antibody also recognizes SLN of rabbit and pig, which may be explained by significant sequence similarity: MERSTXELXXNF. The differences in the SLN sequence might, however, result in a weaker antibody reaction with SLN of rabbit and pig than with SLN of rodents. This might limit the detection of SLN in rabbit and pig tissues (e.g. in the atria). A caveat in the estimation of absolute SLN levels is that purified SLN from pig was used as a standard for measuring SLN levels in other species. The probably lower sensitivity of the anti-SLN antibodies for rabbit SLN than for rodent SLN might lead to overestimation of SLN levels in mouse and rat.
Anti-SLN antibodies also cross-react with PLB, which can only be due to the common LXXNFC motif. Although the similarity between PLB and the SLN epitope is small, even the weakest cross-reaction may appear significant in tissues where PLB is abundantly expressed, such as in ventricle and atria. Taken together, the specificity of these antibodies for SLN is not absolute, which hampers their use in ELISA and in immunocytochemical assays, but they were proved to be useful for semi-quantitative Western-blot analysis (allowing discrimination of SLN and PLB by size).
PLB is the main regulator of SERCA2a
For all species tested, it was clear that the PLB/SERCA mRNA and protein ratios decrease from ventricle to atria, soleus and EDL. This correlates roughly with the relative expression levels of SERCA2a in these tissues. Remarkably, whereas low levels of PLB mRNA were detected in soleus and EDL of mouse, rabbit and pig (but not of rat), PLB protein could only be found in soleus and EDL of rabbit and pig, and not in skeletal muscle of mouse and rat. These observations are in line with earlier reports on absence of PLB protein from the soleus in rat [15]. The documented faster relaxation of the soleus in PLB−/− mice compared with control animals therefore seems related to an indirect effect [16].
In vivo, SLN can be co-expressed with SERCA1 and SERCA2
Results of the present study confirm earlier work which showed SLN mRNA expression in the atria of mouse [5] and rat [7], and in the soleus and EDL of rabbit. These findings are now extended to the protein level. The signal generated with our antibody on Western blots roughly correlates with SLN mRNA expression, which supports their use for the detection of SLN in tissue homogenates by Western blotting. It is clear that the SLN expression pattern differs between larger and smaller animals. SLN protein is expressed in skeletal muscle of pig and rabbit (1–3-fold excess of SERCA compared with SLN), but this is not the case for mouse or rat.
On the other hand, atria of mouse and rat present the highest SLN to SERCA protein ratios (1.5-fold molar excess) compared with all other tissues, whereas SLN protein appeared to be lacking in pig and rabbit atria. It is important to note that, when found in the atria, SLN is expressed in the absence of detectable SERCA1. Although Minamisawa et al. [5] demonstrated very weak SERCA1 mRNA levels in the atria of mouse, we show that this is likely to be irrelevant compared with the abundant SERCA2 expression. Furthermore, we failed to detect SERCA1 protein via Western blotting in microsomes of murine atria (results not shown).
Earlier studies reported SLN expression in heart of rat [17] and human [6], without making the distinction between atria and ventricles. However, more recent publications [5,7] and the present paper make it clear that SLN expression in the heart of all studied species is confined to the atria, and that in the ventricles no SLN is expressed. Also, atrial expression of SLN appears to be significant in small mammals only. In line with this observation, SLN mRNA expression in human atria was reported to be much lower compared with skeletal muscle [5]. In our present study, we did not include human muscle types, because the amino acid sequence of human SLN is significantly different from SLN in other animals, which precluded the use of the anti-SLN antibody for human.
Taken together, our data show that the traditional view of SLN as the physiological regulator of SERCA1, in contrast with PLB as the corresponding SERCA2 inhibitor, is oversimplified. In vivo, both PLB and SLN can be found in co-expression with either SERCA isoform. Since in vitro, both regulators inhibit SERCA1 and SERCA2a to the same extent, a similar situation can be expected in vivo. Indeed, artificial overexpression of SLN in the ventricle (SERCA2a) [18] or soleus (SERCA1) of rat [15] leads to impaired Ca2+ uptake and reduced contractility. And recently, adenoviral overexpression of NF-SLN in rat ventricular cardiomyocytes decreased cell shortening and the half-time of Ca2+ removal [7]. In addition, this study showed that overexpressed NF-SLN is co-localized with PLB in the sarcoplasmic reticulum of ventricular cardiomyocytes, indicating that both regulators can be present in the same subcompartments of cardiac sarcoplasmic reticulum [7].
SLN and PLB are co-expressed in the atria of rat and mouse and in rabbit soleus
In all species, the ventricles express the highest levels of PLB. In smaller animals, the PLB/SERCA protein ratio is three times lower in the atria compared with the ventricle, whereas, in larger animals, this trend is less pronounced (2-fold reduction). The 3-fold reduction in the PLB/SERCA protein ratio in murine atria compared with ventricles is in agreement with previous results at the mRNA level [19]. This lower ratio is the result of a higher level of SERCA and a lower level of PLB per μg of protein in atria compared with ventricles, which is an apparently general characteristic common to all mammals [13]. It explains the faster rate of Ca2+ removal in atria compared with ventricles [20]. It therefore came as a surprise to find the highest SLN/SERCA mRNA and protein ratio in the atria of rat and mouse (1.5-fold molar excess), which would allow a PLB–SLN interaction and thereby superinhibition of SERCA. Moreover, the SLN/PLB molar ratio was 0.8 and 0.5 in atria of mouse and rat respectively, indicating that PLB–SLN interaction may occur.
It should be noted that the overall impact of the high atrial SLN levels in the context of PLB is difficult to evaluate and depends on the mutual relative affinities in the ternary PLB–SLN–SERCA complex. The fact that SLN acts to dissociate PLB pentamers should thereby not be overlooked. It is also not yet known how PLB phosphorylation influences PLB–SLN interaction, nor to what extent the PLB–SLN complex resists phosphorylation. In the ventricles of SLN transgenic mice, the reduced apparent Ca2+-affinity of SERCA might at least in part be ascribed to a decrease in PLB phosphorylation [18]. Together, this could imply that the actual SERCA inhibition by the PLB–SLN complex is less pronounced than anticipated from in vitro studies.
Although SLN is probably not regulated by phosphorylation [4], it is possible that atrial SLN helps to fine-tune the β-adrenergic response. Remarkably, the β-adrenergic response is known to be different in atria compared with ventricles [19]. Koss et al. [19] and Freestone et al. [21] proposed that the lower PLB content of atria correlates with a smaller β-adrenergic response of the relaxation time. In contrast, Kaasik et al. [22] observed in isolated rat atria, a larger β effect on Ca2+-uptake and the relaxation rates than in ventricle. This discrepancy may be explained by other differences, like the higher sensitivity of atria to β stimulation [19], or by the opposite frequency dependence of β-adrenergic responsiveness in atria and ventricles [22]. Taken together, conditions in which there is a higher or a more sensitive adrenergic effect in the atria may result from the relief of combined SLN–PLB inhibition on SERCA instead of from PLB alone. But future studies in SLN targeted and transgenic mice are required to explore this hypothesis.
In addition to the atria of rodents, PLB and SLN proteins are also co-expressed in soleus and EDL of rabbit and pig. The SLN/PLB molar ratios are the highest in EDL of rabbit and pig, which contain very low levels of PLB. In soleus of rabbit and pig, a SLN/PLB ratio closer to 1 was found, suggesting a possible PLB–SLN interaction. However, it remains to be determined whether PLB and SLN are present in the same fibre type.
No SLN up-regulation in the heart of PLB−/− mice
The anti-SLN antibody was used to compare the expression of SLN in different mouse models. It was interesting to find that as a consequence of the loss of PLB, there is no compensatory up-regulation in SLN expression in the atria or ventricles of PLB−/− mice. PLB−/− can therefore serve as a good model for SLN study in the absence of PLB.
SLN may contribute to altered Ca2+ handling in left ventricular hypertrophy
Interest is growing in the role of SLN in cardiac disease. In patients with chronic atrial fibrillation, SLN mRNA is down-regulated in the right atrial myocardium [8]. Reduced SLN mRNA levels are also observed in the atria of activated H-Ras transgenes developing left ventricular hypertrophy [5]. In the heart of hyperthyroid- and thyroid-hormone-resistant mice, SLN mRNA levels are respectively decreased and increased [23]. However, a discrepancy might exist between changes at the mRNA and protein level, thus it remains to be determined whether these differences at the mRNA level are also observed at the protein level. In SERCA2b/b mice, SLN down-regulation at the mRNA level was accompanied by a similar decrease in protein level. Adult SERCA2b/b mice develop left ventricular hypertrophy as a consequence of the expression of SERCA2b in atria and ventricles instead of the usual SERCA2a. Taken together, it seems that altered SLN expression may contribute to altered Ca2+ handling in several cardiomyopathies.
In conclusion, the generation of anti-SLN antibodies enabled us for the first time to study the endogenous SLN protein in muscle types of different animals and will be an important tool for future SLN research. In larger animals, SLN is expressed in the soleus and EDL, whereas, in rodents, SLN expression is abundant in the atria, where it could interact with PLB and SERCA2a, but it is less abundant in skeletal muscle. The present study also suggests that altered SLN expression could contribute to impaired Ca2+ handling under pathological conditions.
Online data
Acknowledgments
The PLB-knockout mice were kindly provided by Dr E. Kranias (Cincinnati, OH, U.S.A.). This work was supported by the Interuniversity Attraction Poles Programme Belgian Science Policy P5/05, by grants from the Bilateral Flemish/Hungarian exchange BIL02/18 of the Flemish Community, Belgium/B-9/2 TéT, Hungary and by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen G.0166.04.
References
- 1.MacLennan D. H., Asahi M., Tupling A. R. The regulation of SERCA-type pumps by phospholamban and sarcolipin. Ann. N.Y. Acad. Sci. 2003;986:472–480. doi: 10.1111/j.1749-6632.2003.tb07231.x. [DOI] [PubMed] [Google Scholar]
- 2.Asahi M., Kurzydlowski K., Tada M., MacLennan D. H. Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs) J. Biol. Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 3.Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., MacLennan D. H. Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5040–5045. doi: 10.1073/pnas.0330962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odermatt A., Becker S., Khanna V. K., Kurzydlowski K., Leisner E., Pette D., MacLennan D. H. Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 5.Minamisawa S., Wang Y., Chen J., Ishikawa Y., Chien K. R., Matsuoka R. Atrial chamber-specific expression of sarcolipin is regulated during development and hypertrophic remodeling. J. Biol. Chem. 2003;278:9570–9575. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- 6.Odermatt A., Taschner P. E., Scherer S. W., Beatty B., Khanna V. K., Cornblath D. R., Chaudhry V., Yee W. C., Schrank B., Karpati G., et al. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics. 1997;45:541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 7.Babu G. J., Zheng Z., Natarajan P., Wheeler D., Janssen P. M., Periasamy M. Overexpression of sarcolipin decreases myocyte contractility and calcium transient. Cardiovasc. Res. 2005;65:177–186. doi: 10.1016/j.cardiores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Uemura N., Ohkusa T., Hamano K., Nakagome M., Hori H., Shimizu M., Matsuzaki M., Mochizuki S., Minamisawa S., Ishikawa Y. Down-regulation of sarcolipin mRNA expression in chronic atrial fibrillation. Eur. J. Clin. Invest. 2004;34:723–730. doi: 10.1111/j.1365-2362.2004.01422.x. [DOI] [PubMed] [Google Scholar]
- 9.Ver Heyen M., Heymans S., Antoons G., Reed T., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca2+-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction–relaxation of the heart. Circ. Res. 2001;89:838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- 10.Braz J. C., Gregory K., Pathak A., Zhao W., Sahin B., Klevitsky R., Kimball T. F., Lorenz J. N., Nairn A. C., Liggett S. B., et al. PKC-α regulates cardiac contractility and propensity toward heart failure. Nat. Med. 2004;10:248–254. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 11.Mountian I. I., Baba-Aissa F., Jonas J. C., De Smedt H., Wuytack F., Parys J. B. Expression of Ca2+ transport genes in platelets and endothelial cells in hypertension. Hypertension. 2001;37:135–141. doi: 10.1161/01.hyp.37.1.135. [DOI] [PubMed] [Google Scholar]
- 12.Ohnoki S., Martonosi A. Purification and characterization of the proteolipid of rabbit sarcoplasmic reticulum. Biochim. Biophys. Acta. 1980;626:170–178. doi: 10.1016/0005-2795(80)90208-1. [DOI] [PubMed] [Google Scholar]
- 13.Luss I., Boknik P., Jones L. R., Kirchhefer U., Knapp J., Linck B., Luss H., Meissner A., Muller F. U., Schmitz W., et al. Expression of cardiac calcium regulatory proteins in atrium v ventricle in different species. J. Mol. Cell. Cardiol. 1999;31:1299–1314. doi: 10.1006/jmcc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 14.Luo W., Grupp I. L., Harrer J., Ponniah S., Grupp G., Duffy J. J., Doetschman T., Kranias E. G. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of β-agonist stimulation. Circ. Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 15.Tupling A. R., Asahi M., MacLennan D. H. Sarcolipin overexpression in rat slow twitch muscle inhibits sarcoplasmic reticulum Ca2+ uptake and impairs contractile function. J. Biol. Chem. 2002;277:44740–44746. doi: 10.1074/jbc.M206171200. [DOI] [PubMed] [Google Scholar]
- 16.Slack J. P., Grupp I. L., Luo W., Kranias E. G. Phospholamban ablation enhances relaxation in the murine soleus. Am. J. Physiol. 1997;273:C1–C6. doi: 10.1152/ajpcell.1997.273.1.C1. [DOI] [PubMed] [Google Scholar]
- 17.Gayan-Ramirez G., Vanzeir L., Wuytack F., Decramer M. Corticosteroids decrease mRNA levels of SERCA pumps, whereas they increase sarcolipin mRNA in the rat diaphragm. J. Physiol. 2000;524:387–397. doi: 10.1111/j.1469-7793.2000.t01-2-00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asahi M., Otsu K., Nakayama H., Hikoso S., Takeda T., Gramolini A. O., Trivieri M. G., Oudit G. Y., Morita T., Kusakari Y., et al. Cardiac-specific overexpression of sarcolipin inhibits sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koss K. L., Ponniah S., Jones W. K., Grupp I. L., Kranias E. G. Differential phospholamban gene expression in murine cardiac compartments: molecular and physiological analyses. Circ. Res. 1995;77:342–353. doi: 10.1161/01.res.77.2.342. [DOI] [PubMed] [Google Scholar]
- 20.Minajeva A., Kaasik A., Paju K., Seppet E., Lompre A. M., Veksler V., Ventura-Clapier R. Sarcoplasmic reticulum function in determining atrioventricular contractile differences in rat heart. Am. J. Physiol. 1997;273:H2498–H2507. doi: 10.1152/ajpheart.1997.273.5.H2498. [DOI] [PubMed] [Google Scholar]
- 21.Freestone N. S., Ribaric S., Scheuermann M., Mauser U., Paul M., Vetter R. Differential lusitropic responsiveness to β-adrenergic stimulation in rat atrial and ventricular cardiac myocytes. Pflugers Arch. 2000;441:78–87. doi: 10.1007/s004240000397. [DOI] [PubMed] [Google Scholar]
- 22.Kaasik A., Paju K., Minajeva A., Ohisalo J. Decreased expression of phospholamban is not associated with lower β-adrenergic activation in rat atria. Mol. Cell. Biochem. 2001;223:109–115. doi: 10.1023/a:1017945810355. [DOI] [PubMed] [Google Scholar]
- 23.Miller L. D., McPhie P., Suzuki H., Kato Y., Liu E. T., Cheng S. Y. Multi-tissue gene-expression analysis in a mouse model of thyroid hormone resistance. Genome Biol. 2004;5:R31. doi: 10.1186/gb-2004-5-5-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.