Abstract
WbpP is the only genuine UDP-GlcNAc (UDP-N-acetylglucosamine) C4 epimerase for which both biochemical and structural data are available. This represents a golden opportunity to elucidate the molecular basis for its specificity for N-acetylated substrates. Based on the comparison of the substrate binding site of WbpP with that of other C4 epimerases that convert preferentially non-acetylated substrates, or that are able to convert both acetylated and non-acetylated substrates equally well, specific residues of WbpP were mutated, and the substrate specificity of the mutants was determined by direct biochemical assays and kinetic analyses. Most of the mutations tested were anticipated to trigger a significant switch in substrate specificity, mostly towards a preference for non-acetylated substrates. However, only one of the mutations (A209H) had the expected effect, and most others resulted in enhanced specificity of WbpP for N-acetylated substrates (Q201E, G102K, Q201E/G102K, A209N and S143A). One mutation (S144K) totally abolished enzyme activity. These data indicate that, although all residues targeted in the present study turned out to be important for catalysis, determinants of substrate specificity are not confined to the substrate-binding pocket and that longer range interactions are essential in allowing proper positioning of various ligands in the binding pocket. Hence prediction or engineering of substrate specificity solely based on sequence analysis, or even on modelling of the binding pocket, might lead to incorrect functional assignments.
Keywords: epimerase, mutagenesis, specificity, structure, substrate, UDP-N-acetylglucosamine
Abbreviations: cGne, Campylobacter jejuni Gne; eGalE, Escherichia coli GalE; hGalE, human GalE; tGalE, Trypanosoma brucei GalE; yGne, Yersinia enterocolitica Gne
INTRODUCTION
WbpP is a UDP-GlcNAc (UDP-N-acetylglucosamine) C4 epimerase [1] that is essential for the biosynthesis of the lipopolysaccharide O-antigen in Pseudomonas aeruginosa serotype O:6 [2]. This enzyme is a member of the SDR (short-chain dehydrogenase reductase) family [3] and is homologous with numerous other sugar-nucleotide C4 epimerases, including GalE, a UDP-Glc (UDP-glucose) epimerase that is found in numerous bacteria, as well as in humans. These homologues can be classified into three main groups based on their substrate specificity [4]. Group 1 represents C4 epimerases that are strongly specific for non-acetylated substrates, such as the Escherichia coli GalE (eGalE; Protein Data Bank code 1XEL), group 2 represents epimerases that can epimerize both acetylated and non-acetylated substrates equally well, such as human GalE (hGalE; Protein Data Bank code 1HZJ), and group 3 represents epimerases that exhibit a strong preference for acetylated substrates, such as WbpP. To date, WbpP remains the only genuine UDP-GlcNAc C4 epimerase that has been characterized both at the biochemical [1] and structural [4] level, and only one other group 3 C4 epimerase, WbgU from Plesiomonas shigelloides O17, that shares 64% identity with WbpP has been characterized at the biochemical level [5]. In contrast, numerous group 2 epimerases have been characterized recently [6–8].
Most studies concerning C4 epimerases have focused on refining the catalytic mechanism involved [9–16], but relatively few have examined the molecular basis for substrate specificity [6,7,17–19]. This is an important question to address, since several bacterial species possess multiple copies of closely related epimerases that most probably have slightly different substrate specificities. Other bacteria have a single copy, but are known to produce several sugar epimers [7], hence implying that the enzyme might have a rather broad substrate specificity.
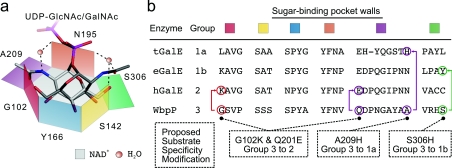
The structural characterization of WbpP in the presence of UDP-Glc or UDP-GalNAc was a first step in trying to understand the structural basis for such substrate specificity [4], using the known structures of group 1 or group 2 enzymes for comparison [16,20,21]. This work has led to a better understanding of the interactions occurring between the enzyme and its substrate in the binding pocket, and led to a structural model implicating a limited number of residues to explain the molecular basis for substrate specificity. In this model, the substrate-binding pocket can be viewed as an hexagonal-shaped box (Figure 1), in which the bottom is formed by the nicotinamide ring of the co-factor and the top is open to accommodate the ring-flipping movement that occurs during catalysis. Three of the six walls of the hexagon are represented by highly conserved residues: Asn195, Ser142 and Tyr166 in WbpP (Figure 1, orange, yellow and blue walls respectively). The latter two are part of the SYK catalytic triad. The three other walls of the hexagon comprise Ser306, Gly102 and Ala209 (Figure 1, green, red and purple walls respectively), which have been proposed to be key determinants for substrate specificity [4].
Figure 1. Model of the substrate-binding site of WbpP and other C4 epimerases.
(a) The substrate-binding pocket is represented by an hexagon. Three of the six sides are made of the three conserved Ser142 (yellow), Tyr166 (blue) and Asn195 (orange) residues in WbpP. The three other sides are made of Gly102 (red), Ala209 (purple) and Ser306 (green). (b) The corresponding residues found in other homologues are indicated, and a selection of mutations tested in the present paper is highlighted. The UDP-GlcNAc and UDP-GalNAc bound in catalytically productive conformations are represented by a solid line. The role of water molecules is signified by spheres. Adapted from [4] with permission. © 2004 American Society for Biochemistry and Molecular Biology.
One of these walls is occupied by a bulky residue, such as Tyr299 in eGalE, that cannot catalyse conversion of acetylated substrates, whereas it is replaced by a much smaller residue in enzymes that convert acetylated substrates: cysteine in hGalE [19] and Yersinia enterocolitica Gne (yGne) [6], leucine in Campylobacter jejuni Gne (cGne) [7] and Ser306 in WbpP [2] (Figure 1, green wall). Hence it was hypothesized that widening the binding pocket would allow catalysis of both types of substrates, whereas narrowing it would limit catalysis to non-acetylated substrates. Data concerning a C307Y mutant in hGalE and a C297Y mutant in yGne showed that narrowing the size of the binding pocket resulted in significant loss of activity with acetylated substrates, whereas conversion of non-acetylated substrates was not affected [6,18], hence supporting this simple hypothesis. But earlier data concerning a Y299C mutation in eGalE challenged it, since the mutation resulted in significant loss of catalysis of non-acetylated substrates [17], probably because they could assume unproductive conformations in the widened pocket. Our own data on a S306Y WbpP mutant that was expected to be unable to process acetylated substrates, but have enhanced ability to process non-acetylated substrates, also challenge this hypothesis, since the mutation totally abolished the activity of the enzyme [4]. This suggested that this residue might actually have some role in the catalytic process itself. Indeed, Ser306 could assist the tighter packing of the binding pocket by forming hydrogen bonds with active-site water molecules (Figure 2a) to other residues that are lining the walls of our hexagonal model of the binding site, such as Ser143 and Asn195. These data point to the fact that models of the substrate-binding pocket, including that of WbpP, need further experimental assessment for validation to yield a good understanding of the determinants of substrate specificity.
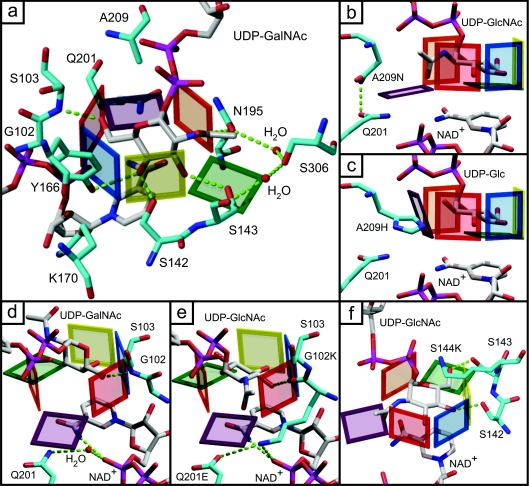
Figure 2. Highlights of interactions between the residues chosen for mutagenesis and neighbouring residues, substrate or co-factor.
(a) Overall view of the sugar-binding pocket in the presence of UDP-GalNAc. (b) and (c) Effect of the A209N (b) and A209H (c) mutations on the ability of WbpP to bind different substrates. (d) Detail of interactions between the amine group of Ser103 and the 6′-OH of UDP-GalNAc, and of the co-ordination of the nicotinamide ring of NAD by an active-site water molecule. (e) Effect of the G102K mutation on the interaction with UDP-GlcNAc and on co-ordination of the nicotinamide ring of NAD. (f) Effect of the S144K mutation that amounts to adding a lid on top of the hexagonal binding site.
The only other available experimental data addressing substrate specificity deal with the fifth wall of the hexagon model of the substrate binding site that is made of Ala209 in WbpP. All other epimerases have a bulkier residue at the equivalent position, such as asparagine in eGalE and hGalE, or histidine in Trypanosoma brucei GalE (tGalE), which partially or totally closes the hexagon and limits the ability to accommodate acetylated substrates (Figure 1, purple wall). Hence, an A209H mutant was designed to assess if re-introducing a bulkier residue in this position would limit the ability of WbpP to epimerize acetylated residues as highlighted in Figure 2(c). This is what we have observed [4], hence validating partially the predictions regarding this residue.
In summary, previous work has identified a limited number of residues that are potentially responsible for substrate specificity, but very few mutagenesis and biochemical studies are available to test and validate these structural models experimentally. In the present study, we report the biochemical characterization of a panel of WbpP mutants designed to test the model of the control of substrate specificity that was established based on structural analysis of the catalytic site of WbpP. Our data show that substrate specificity is not governed uniquely by residues located in the substrate-binding pocket and that the network of interactions with other remotely located residues might also play an essential role in refining the final structure of the substrate-binding pocket. Hence, although predictions based on structural analyses of the catalytic site remain valuable to understand the catalytic mechanism, further extensive mutagenesis involving larger domains of the protein might be necessary to understand fully what governs substrate specificity in these enzymes. This is important, since these enzymes not only control metabolic functions such as the Leloir pathway [23,24], but are also key enzymes for protein glycosylation both in eukaryotes and prokaryotes, or for production [2] or secretion of virulence factors [7,25]. Therefore understanding the molecular basis for substrate specificity might lead to the development of novel therapeutic agents useful to modulate the activity of epimerases as needed.
EXPERIMENTAL
Mutagenesis
All mutants were constructed using the QuikChange® procedure (Stratagene, La Jolla, CA, U.S.A.) using E. coli DH5α cells. The template used for mutagenesis was the His-WbpP-pET construct described previously [1]. The primers used for mutagenesis are listed in Table 1. The entire length of the gene was sequenced for each mutant to ensure the presence of the correct mutation and to allow elimination of constructs with unwanted secondary mutations. The DNA sequencing was carried out at the Robarts Institute sequencing facility (London, Ontario, Canada).
Table 1. Sequence of the primers used for site-directed mutagenesis.
Bases corresponding to mutated codons are underlined.
| Mutant | Primer sequence |
|---|---|
| A209H top | 5′-ATGGTGCCTATGCGCATGTCATACCAAAATG-3′ |
| A209H bottom | 5′-CATTTTGGTATGACATGCGCATAGGCACCAT-3′ |
| A209N top | 5′-ATGGTGCCTATGCGAATGTCATACCAAAATG-3′ |
| A209N bottom | 5′-CATTTTGGTATGACATTCGCATAGGCACCAT-3′ |
| G102K top | 5′-CATCAAGCTGCCTTGAAGTCGGTACCGCGTTC-3′ |
| G102K bottom | 5′-GAACGCGGTACCGACTTCAAGGCAGCTTGATG-3′ |
| S306Y top | 5′-GGATGTCGTCACTACCTCGCTGATATCAG-3′ |
| S306Y bottom | 5′-CTGATATCAGCGAGGTAGTGACGTACATCC-3′ |
| Q201E top | 5′-GTGTTCGGTCGTCGAGAAGATCCCAATGGTGC-3′ |
| Q201E bottom | 5′-GTTACACAAGCCAGCAGCTCTTCTAGGGTTACCACG-3′ |
| S143A top | 5′-CACTTATGCGGCAAGTGCCTCTACCTATGGAGATC-3′ |
| S143A bottom | 5′-GATCTCCATAGGTAGAGGCACTTGCCGCATAAGTG-3′ |
| S144K top | 5′-CTTATGCGGCAAGTAGCAAGACCTATGGAGATCATC-3′ |
| S144K bottom | 5′-GATGATCTCCATAGGTCTTGCTACTTGCCGCATAAG-3′ |
Protein expression and purification
Appropriate clones were transformed into the expression strain BL21(DE3)pLys grown in Luria–Bertani broth (Invitrogen), with selection with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Induction of protein expression was carried out overnight using 0.1 mM IPTG (isopropyl β-D-thiogalactoside) at 30 °C. Cultures (300 ml) were harvested by centrifugation at 6000 g for 30 min, and the pellet was resuspended in 40 ml of binding buffer (20 mM imidazole, 20 mM Tris/HCl, pH 8, and 0.1 M NaCl). The cells were lysed by passage three times through a French Press and purified by nickel chelation using a Poros® MC 20 column (4.6 mm×100 mm; Applied Biosystems) fitted on an AKTA purifier (Amersham Biosciences). After injection, the column was washed with 10 column vol. of binding buffer and elution was carried out using a linear gradient to 100% of elution buffer (1 M imidazole, 20 mM Tris/HCl, pH 8, and 0.1 M NaCl) in 10 column vol. Purified fractions were dialysed overnight in 50 mM Tris/HCl, pH 8, and the protein concentration was determined using the Bio-Rad protein concentration reagent according to the manufacturer's instructions.
Activity assays
Typically, the reactions were carried out in 35 μl volumes and contained 1.5, 6 or 12 μg of enzyme and 0.5 mM of substrate in 50 mM Tris/HCl, pH 8. NAD+ was added at a final concentration of 0.1 mM, whenever necessary. The reactions were incubated at 37 °C and quenched by boiling for 5 min at the appropriate time point. Time courses were performed over 1 h and 30 min for the acetylated substrates, and over 6 h for the non-acetylated substrates. The percentage of substrate conversion was assessed by capillary electrophoresis using a PACE-MDQ system (Beckman) as described previously [1]. For each mutant, the analyses were performed twice using an independent batch of overexpressed enzymes. The results are the average of both experiments.
CD analyses
For CD analyses, the samples were dialysed in 50 mM sodium phosphate buffer (pH 8) overnight, and their concentration was adjusted to 0.1 g/l as determined using the Bio-Rad protein concentration reagent. The spectra were recorded on a Jasco J-810 spectropolarimeter from 190 to 250 nm with a 0.2 nm step, at 10 °C and using a 0.1 cm pathlength cell.
Fluorescence analyses
The samples were dialysed in 50 mM sodium phosphate buffer (pH 8) overnight, as above, and their concentration was adjusted to 0.125 g/l as determined using the Bio-Rad protein concentration reagent. The emission spectra were recorded using 120 μl of sample in a 96-well plate (MicroFluor2) using a Cary Eclipse Varian fluorimeter. The spectra were recorded from 300 to 400 nm, with an excitation wavelength of 280 nm. The emission and excitation slits were set on 10 and 20 nm respectively.
RESULTS AND DISCUSSION
Based on structural data, it was postulated that residues Ala209, Gln201, Gly102, Ser144 and Ser143 might significantly contribute to the specificity of WbpP for N-acetylated substrates [4]. A model highlighting the potential role of these residues was proposed, and seven mutants, including six single mutants (A209H, A209A, Q201E, G102K, S143A and S144K) and one double mutant (Q201E/G102K), were constructed and assessed for substrate specificity to test the model experimentally.
For each mutant, analyses were performed on two independent batches of overexpressed enzymes, and the wild-type protein was purified and analysed alongside the mutants. Activity on the N-acetylated substrates was tested in a time course over 90 min to highlight kinetic differences that would not be apparent with endpoint equilibrium data. The experiments were performed using two different quantities of enzymes (1.5 and 6 μg of enzyme per reaction), to reveal subtle differences in enzymatic efficiency. The less efficient catalysis of non-acetylated substrates was tested using substantially higher amounts of enzyme (12 μg) over a longer period of time (6 h).
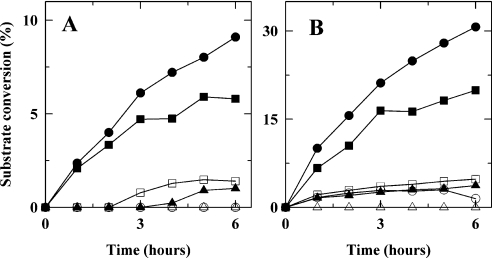
First, to investigate the role of Ala209 in letting better access for acetylated substrates to the binding pocket than the histidine or asparagine residues that are found in group 1a (histidine), group 1b (asparagine) or group 2 (asparagine) C4 epimerases, the A209N and A209H mutants were constructed and analysed. Our previously reported equilibrium data [4] indicated that the A209H mutant had reduced ability to convert the acetylated substrates, but retained a good ability to catalyse UDP-Glc and UDP-Gal (UDP-galactose). The equilibrium (Figure 3) and kinetic analyses (Figures 4 and 5) confirm these findings, indicating that the A209H mutation triggers a switch from group 3 to group 2, instead of the expected switch from group 3 to group 1a, since it has similar, although low, catalytic efficiency on acetylated and non-acetylated substrates. The role of the Ala209 residue on substrate specificity was tested further by using an A209N mutant. Although the asparagine side chain is bulkier than that of the original alanine, the A209N mutation is not predicted to restrict the size of the binding pocket, but rather to allow the ‘purple wall’ to assume an open configuration suitable for catalysis of N-acetylated substrates, as seen in hGalE. This is because the asparagine side chain is predicted to form a hydrogen bond with residue Gln201 (Figure 2b). Accordingly, a significant level of conversion of acetylated substrates could still be obtained with this A209N mutant (Figure 3). Although close to wild-type levels of conversion of UDP-GlcNAc and UDP-GalNAc were obtained after 1 h and 30 min of reaction using 6 μg of enzyme per reaction, the time-course data indicate that catalysis is much slower with the A209N mutant. Indeed, whereas equilibrium was reached within 15 min with the wild-type, equilibrium was only reached with this mutant after 1 h with UDP-GlcNAc (Figure 4B) and was not totally reached after 1 h and 30 min with UDP-GalNAc (Figure 4D). The use of a limited amount of enzyme (1.5 μg) enhanced those differences with wild-type further for both substrates (Figures 4A and 4C). It should be noted that, although limited catalysis was obtained with this A209N mutant under these conditions, it was still significant (8.5% for UDP-GlcNAc and 16.1% for UDP-GalNAc), whereas the A209H mutant showed absolutely no activity in these conditions. Moreover, in contrast with the A209H mutant, the A209N mutant was strongly impaired in its ability to use non-acetylated substrates (Figures 3 and 5). Hence, although the A209N mutation was expected to lead to a switch from a group 3 to group 2 as in hGalE (a further switch to group 1b that also have asparagine in this position would require an extra S306Y mutation), it did not do so. Rather, it enhanced the specificity of WbpP for acetylated substrates, and this was accompanied by a decrease in catalytic efficiency. The equivalent residue in eGalE and hGalE is known to swing back and forth, depending on the substrate. It is possible that within the WbpP background, the asparagine residue of the A209N mutant is not swinging, but forms a hydrogen bond with Gln201. Hence it is rather stationary in an open conformation that accommodates acetylated substrates (Figure 2b), but does not allow non-acetylated ones to assume a catalytically productive position.
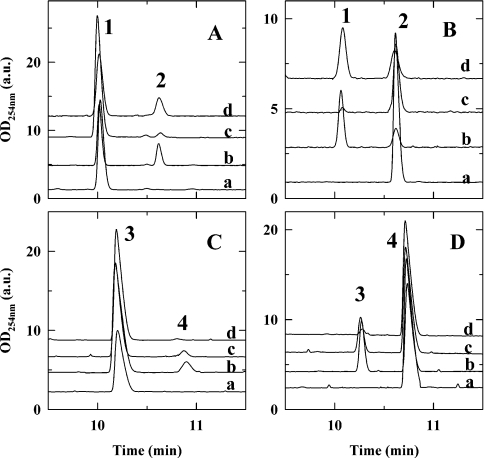
Figure 3. Capillary electrophoresis profiles showing catalysis of the N-acetylated and non-acetylated substrates by the A209N and A209H mutants compared with wild-type.
Conversion of UDP-GlcNAc (A), UDP-GalNAc (B), UDP-Glc (C) and UDP-Gal (D). For all panels: trace a, no enzyme; trace b, wild-type WbpP; trace c, A209H mutant; trace d, A209N mutant. Substrate conversion percentages reported in Figures 4 and 5 were obtained from such profiles by integration of the surface area of the UDP-GlcNAc (peak 1), UDP-GalNAc (peak 2), UDP-Glc (peak 3) or UDP-Gal (peak 4) peak. These profiles show that the A209N mutant is more specific for the acetylated substrates than the wild-type enzyme, whereas the A209H mutant is more specific for the non-acetylated substrates.
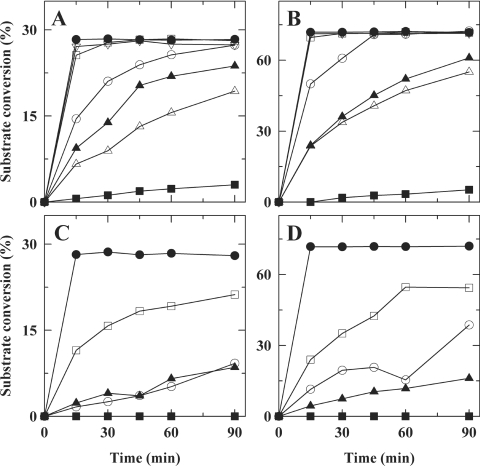
Figure 4. Time course of catalysis of UDP-GlcNAc (A and B) and UDP-GalNAc (C and D) by wild-type and mutant enzymes using either 1.5 μg (A and C) or 6 μg (B and D) of enzyme per reaction.
For each mutant tested, a large reaction mixture was prepared and split in six reactions that were incubated at 37 °C. One reaction was withdrawn at the appropriate time, quenched by boiling and analysed by capillary electrophoresis to determine conversion percentages. ●, Wild-type; ■, A209H; ▲, A209N; △, G102K; □, Q201E; ○, Q201E/G102K; ▽, S143A.
Figure 5. Time course of catalysis of UDP-Glc (A) and UDP-Gal (B) by wild-type and mutant enzymes using 12 μg of enzyme per reaction.
The procedure was the same as for Figure 4. ●, Wild-type; ■, A209H; ▲, A209N; △, G102K; □, Q201E; ○, Q201E/G102K.
Secondly, the role of Gly102 (Figure 1, red wall) in determining substrate specificity was investigated by analysis of G102K, Q201E and Q201E/G102K mutants. The rationale for making these mutations was based on three main reasons. The primary reason was that, in the WbpP•NAD+•UDP-GalNAc ternary complex structure, the amine group of Ser103 interacts with 6′-OH of GalNAc (Figure 2d). This is slightly different from the way eGalE and hGalE (subunit B) interact with UDP-Glc or UDP-GlcNAc via the carbonyl oxygen of a lysine residue, which is situated in the equivalent position of Gly102 of WbpP. However, there is an indication in the electron density of the WbpP•NAD+•UDP-GalNAc ternary complex that a peptide flip occurs between Gly102 and Ser103 to facilitate a similar interaction between the carbonyl oxygen of Gly102 and UDP-GlcNAc. This is inferred on the basis that UDP-GalNAc in the WbpP•NAD+•UDP-GalNAc ternary complex only represents approx. 70% of the total substrate, while the remaining is constituted by UDP-GlcNAc [4]. When UDP-GlcNAc is modelled into the ternary complex structure in the catalytically productive conformation, indeed the peptide flip allows the carbonyl oxygen of Gly102 to face the active site and interact with 3′-OH of GlcNAc (Figure 2e). Although a peptide flip can be an energetically costly process, a peptide flip involving a glycine residue in the active site of p38α MAPK (mitogen-activated protein kinase) has been shown to result in a higher specificity towards certain inhibitors [26]. Therefore it is possible that WbpP utilizes a peptide flip between Gly102 and Ser103 to differentiate the two N-acetylated substrates, GlcNAc and GalNAc. This could be one of the reasons why there is a higher catalytic preference for UDP-GalNAc [1], although GlcNAc and GalNAc are both N-acetylated.
The second reason is that Gly102 is systematically replaced by a bulkier and charged lysine residue in eGalE and hGalE, and by a bulkier leucine or isoleucine residue in tGalE and cGne respectively. This suggests a role in determining substrate specificity, despite the fact that the side chains of these residues all face away from the substrate (Figure 1, red wall, and Figures 2d and 2e). Hence we can hypothesize that the impact of these residues on substrate specificity is indirect and involves interactions with other residues that are not in direct contact with the substrate, but have a stabilizing effect on the walls of the binding pocket.
The third reason for making the G102K, Q201E and Q201E/G102K mutants is that in hGalE, Lys92, which is equivalent to Gly102 in WbpP, is forming a salt bridge with Glu199, which is equivalent to Gln201 in WbpP (Figure 1, red wall). Constraints resulting from establishing a salt bridge between the glutamate and lysine residues might help fine tune the positioning of the smaller substrates in the binding site of group 1b and 2 epimerases, and the absence of such constraint in WbpP might allow these substrates to assume non-productive positioning. Hence introducing salt bridging opportunities in a Q201E/G102K double mutant of WbpP might allow catalysis of non-acetylated substrates. This would turn WbpP into a group 2 epimerase. The assumption that substrate specificity would be altered via indirect interactions is reasonable, since naturally occurring mutations in hGalE observed in patients with epimerase-deficiency galactosaemia do not map within the clusters of residues that are directly involved in substrate binding based on crystallography data [27–30]. Also, these mutations have different effects on enzyme activity, affecting catalysis of acetylated and non-acetylated substrates equally (G90E) or not (V94M) [28,29]. Hence this indicates that the shape of the substrate-binding pocket is dependent on interactions stemming from residues located remotely from the binding pocket and that are very important in fine-tuning substrate specificity. For example, the hydrophobic side chain of Val94 is located near the catalytic Tyr157 of hGalE and serves as a molecular fence to limit rotation of the sugar within the active site [27].
As we anticipated the carbonyl of Gly102 to be in interaction with GlcNAc and not GalNAc, the G102K substitution was expected to have most effect on GlcNAc catalysis, and this is what we observed. Indeed, whereas wild-type levels of UDP-GalNAc conversion could be obtained with the G102K mutant (Figure 4D), conversion of UDP-GlcNAc only reached 50% of wild-type levels (Figure 4B). These data showing a reduced activity of the G102K mutant towards UDP-GlcNAc while maintaining the same level of activity towards UDP-GalNAc agree with the proposal of a peptide-flipping mentioned above that would be responsible for discrimination between UDP-GlcNAc and UDP-GalNAc. A significantly larger side chain of G102K may have reduced the ability of this mutant to perform a peptide flip upon binding UDP-GlcNAc and UDP-GalNAc.
Also, catalysis proceeded much slower with the G102K mutant than with the wild-type with both acetylated substrates (Figure 5). Overall, this could suggest that the lysine might interfere with the N-acetyl group of UDP-GlcNAc in the absence of any glutamate counterpart to establish a salt bridge, and that the introduction of the lysine residue slightly distorts the substrate-binding site in the absence of glutamate. This would result in improper alignment of the carbonyl group of the substituting residue with the 3′-OH of GlcNAc and, to a lesser extent, in misalignment of its neighbouring Ser103 residue with the 6′-OH of GalNAc, both resulting in lower catalytic efficiency, especially with UDP-GlcNAc. This latter explanation based on distortion of the binding site upon introduction of the G102K mutation would be consistent with the fact that this mutation also resulted in a total loss of activity on UDP-Glc and UDP-Gal. Hence, overall, the G102K mutation rendered WbpP more specific for the acetylated substrates.
Analysis of the Q201E mutant did not reveal any defect in conversion of N-acetylated substrates (Figures 4B and 4D), unless fairly low amounts of enzyme were used in the assays (Figures 4A and 4C). Even under these conditions, 75% of wild-type levels of conversion were achieved, although much slower than with the wild-type enzyme. Interestingly, and in agreement with our hypothesis, we observed that the Q201E mutation was able to rescue almost entirely the defects caused by the G102K mutation in a Q102E/G102K mutant, since wild-type levels of conversion of acetylated substrates could be obtained (Figure 4). This indicates that salt bridging through the double mutation G102K/Q201E must have prevented the slight distortion of the binding site that is probably occurring upon introduction of the single G102K mutation. Indeed, when coupled to the Q201E mutation, the G102K mutation is predicted to result in minimal structural disturbance of the active site as the amino group of Lys102 facilitates co-ordination of the nicotinamide ring of NAD which is otherwise carried out by an active-site water molecule in wild-type (Figures 2d and 2e). This also validates our hypothesis that substrate binding is fine-tuned by interactions between residues that are in direct contact with the substrate and structurally neighbouring residues which are not in direct contact with the substrate.
Our data also show that the single Q201E mutation severely impairs the ability of WbpP to catalyse conversion of non-acetylated substrates (Figure 5), rendering it more specific for N-acetylated substrates than the wild-type, as seen for the single G102K mutant. Once again, the single mutation turned out more deleterious for catalysis of the non-acetylated substrates. Their catalysis seems more sensitive to binding site distortion probably because there are no counterbalancing interactions available to maintain them positioned properly, such as those provided by Gly102 and Ser103 to acetylated substrates. As expected, a slight rescue effect of the G102K mutation was observed in the double mutant for conversion of UDP-Gal (Figure 5B). Against expectations, this was not observed for UDP-Glc (Figure 5A), but the reason why is not clear at this stage.
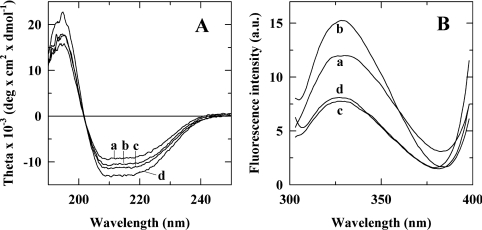
Because the G102K mutant showed a global reduction of activity on all substrates, the mutation could have induced significant perturbation of the global protein structure. Such a hypothesis can be excluded for the other mutants that exhibit significant activity on one set of substrates (acetylated or not) and only lose activity on the other set. Global perturbation of the protein structure would abolish activity on all substrates and not specifically on one. Hence, to refine our interpretation of the catalytic properties observed in the G102K mutant, CD and fluorescence spectra were recorded to assess whether this mutation had altered global protein folding (Figure 6). The spectra of the wild-type, Q201E and double mutant were also recorded as controls. No significant differences could be observed between the CD spectrum of the G102K mutant with those of the wild-type, Q201E or double mutant (Figure 6A). All spectra exhibited the minima at 208 and 222 nm that are typical of a predominantly α-helical structure, consistent with the crystal-structure-based prediction of 45.7% α-helices compared with 15.8% β-sheets, 14.7% turns and 23.8% loops. The fluorescence spectrum of G102K exhibited a slight (3 nm) shift of its maximum emission towards shorter wavelengths and also had slightly reduced fluorescence intensity compared with wild-type (Figure 6B). These differences are subtle and could simply reflect a limited change in the polarity of the environment of nearby aromatic residues such as Tyr166 and Tyr207 upon introduction of the mutation. Hence, overall, the lower catalytic efficiency of the G102K mutant is not due to altered global protein folding, since the secondary and tertiary structures appear wild-type-like, based on CD and fluorescence analyses. The single Q201E mutant had increased fluorescence intensity without shift of the wavelength of the emission maximum compared with wild-type. The effects could be due to alterations of the local environment of neighbouring Phe27 and Tyr336 residues, and to a lesser extent to modification of the environment of the Trp335 and Phe197 residues which are located slightly further away (5–6 Å; 1 Å=0.1 nm). Such alterations were abolished by establishment of the lysine/glutamate salt bridge in the double mutant Q201E/G102K, since its fluorescence spectrum was almost superimposable with that of G102K. Overall, none of these mutations has introduced any significant protein secondary- or tertiary-structure changes that could be responsible for differences observed at the catalytic level.
Figure 6. CD (A) and fluorescence (B) spectra of the wild-type (traces a) and Q201E (traces b), G102K (traces c) and Q201E/G102K (traces d) mutants.
Both analyses indicate that the mutations did not induce any significant alterations of the protein's secondary and tertiary structures.
As a final analysis on this set of mutants, the impact of addition of NAD+ on catalysis was examined. This is because residue 102 is also in close proximity with NAD+, hence potentially playing a role in catalysis by allowing proper positioning of the sugar ring with regards to the co-factor. The catalytic efficiency of the mutants was not restored by the addition of excess amounts of NAD+ in the reactions (results not shown), indicating that the mutations had not interfered with the ability to bind NAD+.
This analysis of the G102K, Q201E and Q201E/G102K mutants suggests that the side chains of these residues are not involved in directly controlling access of large substrates to the binding site via a simple steric hindrance mechanism. Rather, they seem to be involved in establishing additional interactions that result in a more or less flexible binding pocket, hence allowing proper fitting of the substrates within the cavity, so that they can acquire the proper positioning and avoid assuming unproductive configurations.
Finally, we investigated whether two residues, Ser143 and Ser144, located close to the catalytic triad residue Ser142, played any role in catalysis or substrate specificity. Residue Ser143 (Figure 1, yellow wall) was considered to be a good candidate to determine substrate specificity, since its hydroxy group forms a hydrogen bond with the 6′-OH of UDP-GlcNAc (Figure 2f) or the 3′-OH of GalNAc (Figure 2a), along with the carbonyl oxygen of Tyr193. It is replaced by alanine in eGalE and hGalE. Hence the S143A mutant was constructed and assessed for substrate specificity. Loss of hydrogen bonding via the S143A mutation in WbpP should result in reduced catalytic efficiency by allowing the substrates to assume non-productive configurations. The effect is anticipated to be significant for the non-acetylated substrates, but rather minor or absent for the N-acetylated substrate, since the presence of the additional stabilizing interactions of the latter via Gly102 or Ser103 should counterbalance the effect of the S143A mutation. This is what we observed, since the S143A mutant showed normal activity on N-acetylated substrates (Figures 4B and 4D), but showed no activity on UDP-Gal and UDP-Glc. This, once again, enhanced the specificity of WbpP for N-acetylated substrates (Figure 5).
The hydroxy group of Ser144 and its backbone amine group are both hydrogen bonding to the hydroxy group of Ser142. These hydrogen bonds may help to stabilize the state of Ser142, so that this residue can form a low-barrier hydrogen bond with the 4′-OH of the sugar as a hydrogen bond donor. The hGalE and eGalE have threonine in this position, but the same mechanism applies. Hence we did not construct a S144T mutation. Instead, we constructed a S144K mutant, since this serine residue is replaced by lysine in the highly homologous UDP-GlcNAc C6 dehydratase, FlaA1 [31]. Based on preliminary structural data obtained for FlaA1 (N. Ishiyama, C. Creuzenet, J. S. Lam and A. M. Berghuis, unpublished work), the lysine residue might prevent rotation of the substrate in the binding pocket. Hence a S144K mutation amounts to closing the top lid in the hexagon model (Figure 2f). The following scenarios are possible: (i) the mutation might block the catalysis totally by not allowing the return of the abstracted proton from the co-factor, (ii) it might only allow the enzyme to accommodate the smaller substrates that could still rotate in the cavity, or (iii) it might result in dehydration rather than epimerization of UDP-GlcNAc. We observed that the S144K mutation resulted in a total loss of epimerase activity on all substrates and no dehydratase activity was observed either (results not shown). Hence Ser144 appears to be essential for activity, but at present we cannot tell from these analyses if the mutant was unable to bind the substrates or was unable to reduce the 4-keto intermediate during catalysis for lack of proper rotation. The possibility that the mutation resulted in a major structural defect cannot be excluded, but was not investigated further.
Conclusion
Overall, our data show that most mutations of residues of the substrate-binding pocket of WbpP tested in the present study have resulted in enhancing the specificity of WbpP for N-acetylated substrates (Table 2). These observations were not anticipated, since several of the substituting amino acids were chosen with the intention to turn WbpP into a mimic of group 1a or 2 epimerases, and the residues that were mutated are strictly conserved in the only other genuine UDP-GlcNAc C4 epimerase characterized to date, WbgU [5]. Hence it could be expected that they represent the best combination that evolution has led to for efficient catalysis of N-acetylated substrates. This indicates clearly that the determinants of substrate specificity are not exclusively located directly in the surface of the substrate-binding pocket and that interactions with multiple other residues are important in the process. This is in agreement with our previous structure-based finding that the binding pocket is actually made of several fragments of protein that are intricately connected by hydrogen bonds through active-site water molecules [4]. The rescue effects shown with the double mutant Q201E/G102K highlighted the fact that positioning of these substrate-binding residues with regards to the substrate is very finely tuned by interactions with other neighbouring residues, and amino acid pairs that allow optimal catalysis regardless of the substrate seem to have evolved. For example the Gln201/Gly102 pair found in WbpP is as good as the equivalent glutamate/lysine pair found in eGalE, but does not define substrate specificity, since the WbpP Q201E/G102K mutant is still highly specific for the N-acetylated substrates. This is illustrated perfectly by our previous phylogenetic analysis [4], which indicated that binding pocket residues have co-evolved with the rest of the sequence, with early evolution of groups 1a, 2 and 3. Hence understanding and predicting substrate specificity is more complex than just reasoning in terms of steric hindrance, size of the binding pocket and accessibility for large molecules, since fine positioning between the amino acids that bind the substrate directly, the sugar ring and the co-factor molecule is necessary to allow for productive catalysis.
Table 2. Summary of the effects of the mutations analysed in the present study on the substrate specificity of WbpP.
| Catalysis on | ||||||
|---|---|---|---|---|---|---|
| Mutant | UDP-GlcNAc | UDP-GalNAc | UDP-Glc | UDP-Gal | Specificity expected | Specificity observed |
| A209H | Very limited | Very limited | Slightly reduced | Slightly reduced | Group 1a | Group 2 |
| A209N | Slightly reduced | Slightly reduced | Very limited | Very limited | Group 2 | Enhanced group 3 |
| G102K | Slightly reduced | Slightly reduced | Abolished | Abolished | None | Enhanced group 3 |
| Q201E | Maintained | Maintained | Very limited | Very limited | None | Enhanced group 3 |
| Q201E/G102K | Slightly reduced | Slightly reduced | Abolished | Very limited | Group 2 | Enhanced group 3 |
| S143A | Maintained | Maintained | Abolished | Abolished | Enhanced group 3 | Enhanced group 3 |
| S144K | Abolished | Abolished | Abolished | Abolished | Group 2 or dehydration | Not applicable |
In summary, we have demonstrated that residues of the substrate-binding pocket are critical for catalysis, as anticipated, but did not directly determine substrate specificity, so that we cannot predictably change substrate specificity by simply modifying them individually. The few examples of successful alterations of substrate specificity conferred by single point mutations that were reported in the present study (A209H) and elsewhere [17,18] should not hide the complexity of the problem. Hence this report cautions against hasty functional assignments based on sequence and even on structural modelling in the absence of biochemical data.
Acknowledgments
We thank Rob Urbanic for making the S143A mutant and Deanna Yaniw for making the A209N mutant. We thank Dr M. Valvano (UWO) for the use of his spectrofluorimeter and Dr S. Dunn (UWO) for access to the CD facility. This work was funded through an NSERC (Natural Sciences and Engineering Research Council of Canada)-CHRP (Collaborative Health Research Projects) grant (# 251007-02) to J.S.L., A.M.B. and C.C. C.C. is the recipient of an NSERC University Faculty Award and a Premier's Research Excellence Award from the province of Ontario. N.I. is the recipient of a doctoral studentship from the Canadian Cystic Fibrosis Foundation. J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology, and A.M.B. holds a Canada Research Chair in Structural Biology.
References
- 1.Creuzenet C., Belanger M., Wakarchuk W. W., Lam J. S. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 2000;275:19060–19067. doi: 10.1074/jbc.M001171200. [DOI] [PubMed] [Google Scholar]
- 2.Belanger M., Burrows L. L., Lam J. S. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology. 1999;145:3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 3.Jornvall H., Persson B., Krook M., Atrian S., Gonzalez-Duarte R., Jeffery J., Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 4.Ishiyama N., Creuzenet C., Lam J. S., Berghuis A. M. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 2004;279:22635–22642. doi: 10.1074/jbc.M401642200. [DOI] [PubMed] [Google Scholar]
- 5.Kowal P., Wang P. G. New UDP-GlcNAc C4 epimerase involved in the biosynthesis of 2-acetamino-2-deoxy-L-altruronic acid in the O-antigen repeating units of Plesiomonas shigelloides O17. Biochemistry. 2002;41:15410–15414. doi: 10.1021/bi026384i. [DOI] [PubMed] [Google Scholar]
- 6.Bengoechea J. A., Pinta E., Salminen T., Oertelt C., Holst O., Radziejewska-Lebrecht J., Piotrowska-Seget Z., Venho R., Skurnik M. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 2002;184:4277–4287. doi: 10.1128/JB.184.15.4277-4287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernatchez S., Szymanski C. M., Ishiyama N., Li J., Jarrell H. C., Lau P. C., Berghuis A. M., Young N. M., Wakarchuk W. W. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J. Biol. Chem. 2005;280:4792–4802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 8.Soldo B., Scotti C., Karamata D., Lazarevic V. The Bacillus subtilis Gne (GneA, GalE) protein can catalyse UDP-glucose as well as UDP-N-acetylglucosamine 4-epimerisation. Gene. 2003;319:65–69. doi: 10.1016/s0378-1119(03)00793-5. [DOI] [PubMed] [Google Scholar]
- 9.Thoden J. B., Frey P. A., Holden H. M. High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol. Protein Sci. 1996;5:2149–2161. doi: 10.1002/pro.5560051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoden J. B., Frey P. A., Holden H. M. Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism. Biochemistry. 1996;35:5137–5144. doi: 10.1021/bi9601114. [DOI] [PubMed] [Google Scholar]
- 11.Thoden J. B., Frey P. A., Holden H. M. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 12.Thoden J. B., Hegeman A. D., Wesenberg G., Chapeau M. C., Frey P. A., Holden H. M. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:6294–6304. doi: 10.1021/bi970025j. [DOI] [PubMed] [Google Scholar]
- 13.Thoden J. B., Gulick A. M., Holden H. M. Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10685–10695. doi: 10.1021/bi9704313. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Thoden J. B., Kim J., Berger E., Gulick A. M., Ruzicka F. J., Holden H. M., Frey P. A. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 15.Thoden J. B., Holden H. M. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1998;37:11469–11477. doi: 10.1021/bi9808969. [DOI] [PubMed] [Google Scholar]
- 16.Thoden J. B., Wohlers T. M., Fridovich-Keil J. L., Holden H. M. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 17.Thoden J. B., Henderson J. M., Fridovich-Keil J. L., Holden H. M. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase: teaching an old dog new tricks. J. Biol. Chem. 2002;277:27528–27534. doi: 10.1074/jbc.M204413200. [DOI] [PubMed] [Google Scholar]
- 18.Schulz J. M., Watson A. L., Sanders R., Ross K. L., Thoden J. B., Holden H. M., Fridovich-Keil J. L. Determinants of function and substrate specificity in human UDP-galactose 4′-epimerase. J. Biol. Chem. 2004;279:32796–32803. doi: 10.1074/jbc.M405005200. [DOI] [PubMed] [Google Scholar]
- 19.Thoden J. B., Wohlers T. M., Fridovich-Keil J. L., Holden H. M. Human UDP-galactose 4-epimerase: accommodation of UDP-N-acetylglucosamine within the active site. J. Biol. Chem. 2001;276:15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 20.Bauer A. J., Rayment I., Frey P. A., Holden H. M. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 Å resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- 21.Shaw M. P., Bond C. S., Roper J. R., Gourley D. G., Ferguson M. A., Hunter W. N. High-resolution crystal structure of Trypanosoma brucei UDP-galactose 4′-epimerase: a potential target for structure-based development of novel trypanocides. Mol. Biochem. Parasitol. 2003;126:173–180. doi: 10.1016/s0166-6851(02)00243-8. [DOI] [PubMed] [Google Scholar]
- 22. Reference deleted.
- 23.Frey P. A. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 24.Leloir L. F. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. 1951;33:186–190. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- 25.Winans K. A., Bertozzi C. R. An inhibitor of the human UDP-GlcNAc 4-epimerase identified from a uridine-based library: a strategy to inhibit O-linked glycosylation. Chem. Biol. 2002;9:113–129. doi: 10.1016/s1074-5521(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald C. E., Patel S. B., Becker J. W., Cameron P. M., Zaller D., Pikounis V. B., O'Keefe S. J., Scapin G. Structural basis for p38α MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity. Nat. Struct. Biol. 2003;10:764–769. doi: 10.1038/nsb949. [DOI] [PubMed] [Google Scholar]
- 27.Thoden J. B., Wohlers T. M., Fridovich-Keil J. L., Holden H. M. Molecular basis for severe epimerase deficiency galactosemia: X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase. J. Biol. Chem. 2001;276:20617–20623. doi: 10.1074/jbc.M101304200. [DOI] [PubMed] [Google Scholar]
- 28.Wohlers T. M., Christacos N. C., Harreman M. T., Fridovich-Keil J. L. Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia. Am. J. Hum. Genet. 1999;64:462–470. doi: 10.1086/302263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlers T. M., Fridovich-Keil J. L. Studies of the V94M-substituted human UDPgalactose-4-epimerase enzyme associated with generalized epimerase-deficiency galactosaemia. J. Inherit. Metab. Dis. 2000;23:713–729. doi: 10.1023/a:1005682913784. [DOI] [PubMed] [Google Scholar]
- 30.Quimby B. B., Alano A., Almashanu S., DeSandro A. M., Cowan T. M., Fridovich-Keil J. L. Characterization of two mutations associated with epimerase-deficiency galactosemia, by use of a yeast expression system for human UDP-galactose-4-epimerase. Am. J. Hum. Genet. 1997;61:590–598. doi: 10.1086/515517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creuzenet C., Schur M. J., Li J., Wakarchuk W. W., Lam J. S. FlaA1, a new bifunctional UDP-GlcNAc C6 dehydratase/C4 reductase from Helicobacter pylori. J. Biol. Chem. 2000;275:34873–34880. doi: 10.1074/jbc.M006369200. [DOI] [PubMed] [Google Scholar]