Abstract
RCP (Rab coupling protein) belongs to the recently identified Rab11-FIPs (Rab11 family of interacting proteins). All the Rab-FIP members have the ability to bind Rab11 tightly via a Rab-binding domain located near their C-termini. RCP belongs to the class I Rab11-FIP subfamily, characterized by the presence of a conserved C2 domain near its N-terminus. The function of this protein in Rab11-dependent membrane trafficking remains to be fully understood. In the present study, we have identified three putative PEST (Pro, Glu, Ser/Thr-rich) sequences in RCP. PEST motifs play a role in targeting a protein for proteolytic degradation. We have demonstrated that RCP undergoes calcium-dependent degradation which can be prevented by specific calpain inhibitors. Using a mutant, lacking the three PEST sequences, RCPΔPEST, we demonstrated that they are necessary for the cleavage of RCP by calpains. When expressed in A431 cells, RCPΔPEST displays significantly greater localization to the plasma membrane, compared with the wild-type protein. Similarly, treatment with the calpain inhibitor, calpeptin, results in the redistribution of endogenous RCP to the periphery of the cell. We propose that once the Rab11/RCP-regulated cargo has been delivered from the endocytic recycling compartment to the plasma membrane, RCP is inactivated by calpain-mediated proteolysis.
Keywords: calpain, endocytic recycling compartment, Pro, Glu and Ser/Thr-rich motif (PEST motif), Rab coupling protein, Rab11
Abbreviations: Arf, ADP-ribosylation factor; DMEM, Dulbecco's modified Eagle's medium; ERC, endocytic recycling compartment; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; PEST, Pro, Gly and Ser/Thr-rich; Rab11-FIP, Rab11-family interacting protein; RBD, Rab-binding domain; RCP, Rab coupling protein; TBS, Tris-buffered saline; Tfn, transferrin; TfnR, Tfn receptor; TGN, trans-Golgi network; ZO-1, zonula occludens-1
INTRODUCTION
Membrane transport in eukaryotic cells is a complex process that is regulated by a large and diverse array of proteins. The Rab family of small GTPases, comprising approx. 60 members, are key regulators of intracellular transport. Each Rab GTPase appears to control a specific membrane transport pathway. Individual GTPases have been implicated in regulating the budding, movement and the delivery of transport vesicles along distinct transport pathways. Their subcellular localization defines the pathway that they regulate [1]. For example, Rab4, localized predominantly at the sorting endosome, regulates the transport of cargo from this compartment to either the degradative or the recycling pathway [2–4]. Rab11 is localized to the ERC (endocytic recycling compartment) [5] and the TGN (trans-Golgi network) [6] and regulates transport from the ERC to the plasma membrane, or traffic between the TGN and ERC [7,8].
The regulation of these transport pathways is far from fully understood, and a major challenge is the identification and characterization of effector proteins through which the Rab GTPases mediate their functions. We, and others, have identified several proteins that interact with Rab11. These include Rab11BP/Rabphilin-11 [9,10], myosin Vb [11] and the Rab11-FIP (Rab11-family interacting protein) family [12–16]. To date, the Rab11-FIP family comprises six members, all characterized by the presence of a conserved RBD (Rab-binding domain) at their C-terminus. The family can be subdivided further into three classes, based on their amino acid sequence [17,18]. Class III comprises just one member, Rab11-FIP1. Rab11-FIP3 and Rab11-FIP4 make up the class II Rab11-FIPs. They both possess an ERM (ezrin/radixin/moesin) domain, calcium-binding EF hand motifs, and a proline-rich region. They are localized to the ERC, TGN and centrosomes [15,16,19,20]. Rab11-FIP3 and Rab11-FIP4 were also identified as Arf (ADP-ribosylation factor) GTPase-binding proteins and were termed arfophilin-1 and arfophilin-2 respectively [20–22]. They are believed to play a role in membrane delivery to the cleavage furrow during cell division [19,20,23]. The class I Rab11-FIPs, including Rab11-FIP2, Rip11 and RCP (Rab coupling protein), are characterized by the presence of a conserved C2 domain near the N-terminus [12,14,15], which mediates their binding to phospholipids at the plasma membrane [24]. The three class I Rab11-FIPs are mainly localized to the ERC, but their specific roles in intracellular trafficking remain to be clearly understood. Rab11-FIP2 was found to interact with myosin Vb and appears to function in plasma membrane recycling [25]. Indeed, the expression of a dominant-negative mutant of Rab11-FIP2 reduced Tfn (transferrin) [26] and chemokine receptor CXCR2 recycling [27]. These data suggest that the Rab11-FIP2–Rab11 complex could mediate the movement of recycling vesicles along actin filaments via binding to myosin Vb. RCP was identified as a Rab4-binding protein in a yeast two-hybrid screen, and subsequent analysis revealed that it could also bind Rab11 with high affinity, suggesting that it may serve as a Rab4/Rab11 coupling protein [14]. RCP is also involved in regulating the traffic of cargo from the ERC to the plasma membrane. Indeed, overexpression of a C-terminal truncation mutant of RCP inhibited the recycling of radiolabelled Tfn to the plasma membrane [14]. In addition, RCP was found to regulate recycling from phagosomes in macrophages [28] and has been implicated in endosomal sorting [29].
In the present study, we identified three PEST (Pro, Gly and Ser/Thr-rich) motifs in RCP, located between the C2 domain and the RBD. PEST sequences have been shown to serve as targets for calcium-activated proteases, calpains [30–34] and also the proteasome complex [32,35,36]. To determine if the putative PEST motifs in RCP target it for regulated degradation, and to identify the mechanism of this degradation, we first set out to determine whether RCP is selectively degraded in the presence of calcium. We observed that treating cells with increasing concentrations of calcium triggers the degradation of RCP, and this degradation can be abolished by addition of specific calpain inhibitors. The involvement of PEST sequences in this calcium-regulated degradation was validated using an RCP mutant in which the PEST sequences were removed (RCPΔPEST). Interestingly, in immunofluorescence studies, we found that this mutant was localized predominantly to the cell periphery where it co-localized with Rab11. Finally, we propose a model for a role of the PEST sequences in RCP function.
MATERIALS AND METHODS
cDNA construct and cloning
pcDNA3.1/HisB-RCPWT was generated by subcloning the BamHI fragment from pEGFP-C3/RCP [24] into the BamHI site of pcDNA3.1/HisB (Invitrogen). pcDNA3.1/HisB-RCPΔPEST was made in two steps. First, pcDNA3.1/HisB-RCP(2-181) was created: cDNA encoding RCP(2-181) was generated by PCR using RCPFWD (5′-AAAAGGATCCATCCCTAATGGTCTCGGCT-3′) and RCPC2REV (5′-AAAAGAATTCTTCTTCCCTCTGATCTTGTC-3′) and ligated into the BamHI/EcoRI site of pcDNA3.1/HisB. Secondly, cDNA encoding RCP(379-649) was obtained by digesting pcDNA3.1/HisB-H13 [14] with EcoRI, and the resulting fragment was subcloned into the EcoRI site of pcDNA3.1/HisB-RCP(2-181). All the constructs generated by PCR were confirmed by double-strand sequencing.
Cell culture and immunofluorescence
A431 cells were maintained in culture in DMEM (Dulbecco's modified Eagle's medium) (BioWhittaker) supplemented with 10% (v/v) foetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin, 2 mM glutamine and 25 mM Hepes, pH 7.4, at 37 °C in a water-saturated atmosphere containing 5% CO2. Transfections were carried out using Effectene transfection reagent (Qiagen), according to the manufacturer's instructions. At 24 h post-transfection, the cells were fixed with 3% (w/v) paraformaldehyde and mounted with Mowiol (Calbiochem). For antibody labelling, the cells were permeabilized with 0.05% (w/v) saponin supplemented with 0.2% (w/v) BSA. Primary antibodies were used at the following dilutions: rabbit affinity-purified anti-RCP [14] (1:200), mouse anti-ZO-1 (zonula occludens-1) (1:150) (BD Transduction Laboratories), mouse anti-Xpress (1:1500) (Invitrogen), rabbit anti-Rab11A (1:1500) (Zymed Laboratories), rabbit affinity-purified anti-Rab11-FIP3 [19] (1:150), mouse anti-TfnR (Tfn receptor) (1:1000) (Roche Molecular Biochemical). With the exception of the Cy3-labelled donkey anti-rabbit secondary antibody, purchased from Jackson Immunoresearch and used at 1:2000 dilution, secondary antibodies were from Molecular Probes and were used at 1:200 dilution: Alexa Fluor® 488 goat anti-mouse, Alexa Fluor® 594 goat anti-mouse and Alexa Fluor® 488 goat anti-rabbit. Images were acquired on a Zeiss LSM 510 confocal microscope using a PlanApo 63×, 1.4 NA (numerical aperture) oil-immersion objective. Images were processed using Image Examiner software (Carl Zeiss) and imported into Adobe Illustrator for printing.
Alexa Fluor® 594–Tfn uptake and chase
A431 cells, grown on glass coverslips as a monolayer, were washed twice with DMEM/25 mM Hepes then pre-treated with 30 μg/ml calpeptin (Calbiochem) or solvent alone (DMSO) for 3 h in DMEM/25 mM Hepes. After this pre-treatment, cells were incubated in DMEM/25 mM Hepes/3.3 μg/ml Alexa Fluor® 594–Tfn (Molecular Probes) plus calpeptin (30 μg/ml) or DMSO for 45 min at 37 °C (uptake). The uptake samples were briefly washed with ice-cold PBS and immediately fixed with 3% (w/v) paraformaldehyde. For the chase samples, Alexa Fluor® 594–Tfn was allowed to recycle for 30 min at 37 °C in DMEM/25 mM Hepes/333 μg/ml holotransferrin/0.1 mM desferroxamine (Sigma) with 30 μg/ml calpeptin or DMSO. Following the chase, the cells were washed with ice-cold PBS and fixed with 3% (w/v) paraformaldehyde.
Analysis of RCPWT and RCPΔPEST phenotypes
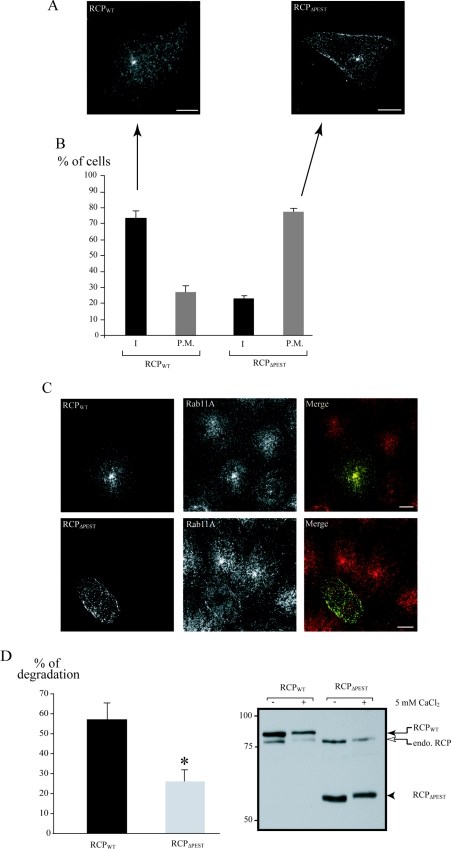
A431 cells were transfected with either pcDNA3.1/HisB-RCPWT or pcDNA3.1/HisB-RCPΔPEST. At 24 h after transfection, cells were fixed and processed for immunofluorescence. The recombinant proteins were detected with the anti-Xpress antibody. The intracellular localization phenotype (I) was defined as a vesicular pattern with no fusion protein localization at the periphery of the cell (see Figure 5A, RCPWT). The plasma membrane localization phenotype (P.M.) was defined as a presence of the fusion protein at the periphery of the cell (see Figure 5A, RCPΔPEST). For both constructs, we analysed 50 transfected cells per experiment in simple blind conditions. The results are expressed as the mean (from three independent experiments) of the percentage±S.D. of cells representing each phenotype.
Figure 5. RCPΔPEST localizes to the plasma membrane.
A431 cells were transfected with either pcDNA3.1/HisB-RCPWT or pcDNA3.1/HisB-RCPΔPEST. At 24 h post-transfection, cells were fixed and immunostained with anti-Xpress antibody alone (A) or in conjunction with anti-Rab11A antibody (C). Scale bar, 10 μm. (B) Proportion of cells displaying the two localization phenotypes. A total of 50 cells per transfection experiment were analysed and scored into two groups according to their localization pattern (I, intracellular; P.M., plasma membrane) (see the Materials and methods section). Results are the mean percentages±S.D. for three independent experiments. (D) A431 cells transfected with the indicated construct were lysed, divided into two equal volumes and incubated in the presence or absence of 5 mM CaCl2 for 25 min at room temperature. The samples were analysed by SDS/PAGE and immunoblotted with anti-RCP. The intensity of the bands corresponding to RCPWT and RCPΔPEST were quantified and the percentage degradation in the presence of calcium was calculated as described in the Materials and methods section. Results are the mean percentage degradation±S.E.M. for three independent experiments. *P<0.01 (Student's t test). On the right, a representative blot is shown (RCPWT, black arrow; RCPΔPEST, black arrowhead; endogenous RCP, open arrow). The positions of the molecular-mass markers (in kDa) are also indicated.
In vitro RCP degradation
Confluent A431 cells in six-well plates were washed twice with PBS then lysed in 150 μl/well of lysis buffer (10 mM Tris/HCl, pH 7.4, 100 mM NaCl, 1 mM dithiothreitol and 0.5% Triton X-100) containing CaCl2 with or without various inhibitors [5 μM calpastatin (Calbiochem), 30 μg/ml calpeptin, 5 mM EGTA and 10 μM lactacystin (Calbiochem)] for 25 min on a rocking platform at room temperature (20 °C). Reactions were stopped by adding 75 μl of SDS sample buffer, and samples were denatured for 5 min at 95 °C. Each sample (20 μl) was used for SDS/PAGE and Western blotting analysis.
In vivo RCP degradation
Confluent A431 cells in six-well plates were washed twice with DMEM/25 mM Hepes and treated with DMEM/25 mM Hepes with or without various drugs at the following concentrations: 1 μM ionomycin (Calbiochem), 30 μg/ml calpeptin and 5 mM EGTA. After the indicated incubation times, the cells were washed with PBS/5 mM EGTA and lysed in 150 μl of lysis buffer/5 mM EGTA for 15 min at 4 °C. SDS sample buffer (75 μl) was added, proteins were denatured for 5 min at 95 °C, and 20 μl of each sample was used for SDS/PAGE and Western blotting analysis.
SDS/PAGE and Western blotting
Proteins were resolved on SDS/10% PAGE, transferred on to nitrocellulose, and the membrane was blocked for 1 h at room temperature in blocking buffer [TBS (Tris-buffered saline: 10 mM Tris/HCl, pH 8, and 150 mM NaCl)/0.1% (v/v) Tween 20/5% (w/v) non-fat dried milk] then probed with rabbit polyclonal anti-RCP [14] antibody at 1:1000 dilution in blocking buffer for 1 h. The membrane was washed with TBS/0.1% (v/v) Tween 20 and incubated with horseradish-peroxidase-conjugated antirabbit antibody (Sigma) at 1:2000 in blocking buffer for 1 h. Secondary antibody was revealed using chemiluminescence reagent (Pierce). After RCP revelation, the membrane was washed extensively and probed with mouse anti-β-actin (Sigma) antibody at 1:20000 dilution in blocking buffer for 1 h. The membrane was washed with TBS/0.1% (v/v) Tween 20 and incubated with horseradish-peroxidase-conjugated anti-mouse antibody at 1:2000 in blocking buffer for 1 h. Secondary antibody was revealed using chemiluminescence reagent.
RCPWT and RCPΔPEST in vitro calcium-induced degradation
A431 cells in a six-well plate were transfected with pcDNA3.1/HisB-RCP or pcDNA3.1/HisB-RCPΔPEST. At 24 h post-transfection, cells were scraped in 400 μl/well of lysis buffer and divided in two equal volumes, transferred into 1.5 ml tubes and incubated in the presence or absence of 5 mM CaCl2 for 25 min at room temperature. The reaction was stopped by adding SDS sample buffer, with denaturation for 5 min at 95 °C. Western-blot analysis was carried out as described above. The immunoreactivity intensity of RCPWT and RCPΔPEST (both revealed by anti-RCP antibody) was quantified by densitometry using GeneTools (Syngene) analysis software. The percentage of degradation of RCPWT or RCPΔPEST in the presence of calcium was calculated as follows:
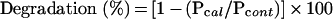 |
where Pcal=immunoreactivity intensity of RCPWT or RCPΔPEST in cells incubated in the presence of 5 mM CaCl2, Pcont=immunoreactivity intensity of RCPWT or RCPΔPEST in cells incubated in the absence of calcium.
RESULTS
PEST sequences are found in RCP and class II Rab-FIPs
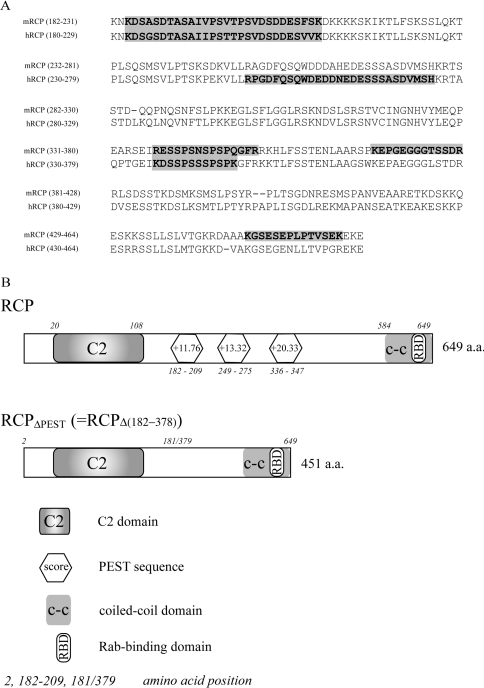
Vac17p, a vacuole-specific receptor for the yeast class V myosin Myo2p, undergoes regulated degradation. This degradation is perturbed by the removal of a PEST domain located in the centre of the Vac17p sequence [37]. The degradation of Vac17p has been proposed to be a means of regulating the function of Myo2p. Rab11-FIP2 has recently been shown to interact directly with the mammalian class V myosin, myosin Vb, and is likely to act as an organelle-specific receptor for this motor protein [25]. To determine whether a similar regulatory mechanism exists in mammals, we set out to determine whether Rab11-FIP2 possesses any PEST motifs. Using the PESTfind software (http://emb1.bcc.univie.ac.at/embnet/tools/bio/PESTfind/) [32], we obtained no significant PEST motif ‘hits’ in the Rab11-FIP2 sequence; however, RCP, and both class II Rab11-FIPs, Rab11-FIP3 and Rab11-FIP4, possess multiple significant PEST motifs (Table 1; a score greater than +5 is considered significant). As the highest scoring motifs were found in RCP (Table 1), we have initially focused our analysis on this protein. PEST sequences are found in numerous bacterial, plant, and mammalian proteins [32], so we set out to determine whether there was a conservation of these sequences among Rab11-FIP proteins throughout evolution. Indeed, we have found PEST sequences in Rab11-FIP orthologues from mouse, rat, Fugu and Drosophila melanogaster. Figure 1(A) represents a partial ClustalW alignment of mouse RCP (mRCP) and human RCP (hRCP). We used PESTfind to determine the presence of PEST sequences and found that mRCP has four putative PEST motifs (score: +10.95, +6.67, +10.27 and +10.24) which are partially aligned with those of hRCP (Figure 1A). In Fugu, we found four putative class II Rab11-FIP orthologues, two of which have putative PEST sequences. The Drosophila orthologue of class I Rab11-FIPs, CG6606-PA, contains two potential PEST sequences (results not shown). We also found that the Drosophila orthologue of the class II Rab11-FIPs, Nuf (Nuclear Fallout), has one potential PEST sequence (score: +6.81). These data suggest that the presence of PEST sequences in Rab11-FIP proteins is conserved throughout evolution.
Table 1. PEST sequences in Rab11-FIPs family.
The amino acid sequences of each human Rab11-FIP family member were analysed using the PESTfind software. Identified PEST motifs, corresponding amino acid numbers (in brackets) and scores are indicated.
| Protein | PEST sequence (amino acids) | Score |
|---|---|---|
| RCP | PEST1 (182–209) | +11.76 |
| PEST2 (249–275) | +13.32 | |
| PEST3 (336–347) | +20.33 | |
| Rab11-FIP2 | No PEST found | |
| Rip11 | No PEST found | |
| Rab11-FIP3 | PEST1 (1–40) | +11.76 |
| PEST2 (111–144) | +9.88 | |
| PEST3 (191–206) | +7.85 | |
| PEST4 (267–333) | +7.53 | |
| PEST5 (333–363) | +9.73 | |
| PEST6 (437–465) | +7.98 | |
| Rab11-FIP4 | PEST1 (129–175) | +12.10 |
| PEST2 (192–204) | +6.31 | |
| PEST3 (204–239) | +13.91 |
Figure 1. PEST sequence are evolutionarily conserved in RCP homologues.
(A) ClustalW alignment of the central amino acid sequences of mouse RCP (mRCP) and human RCP (hRCP). The potential PEST sequences are shown against a grey background. (B) Schematic representation of RCP and RCPΔPEST. Representation of human RCP (RCP) with its C2 domain, coiled-coil (c-c) domain, RBD and predicted PEST sequences. RCPΔPEST represents an RCP truncation lacking the three PEST sequences and comprises the N-terminal domain including the C2 domain (amino acids 2–181) fused with the C-terminus (amino acids 379–649) end containing the RBD.
Endogenous RCP is degraded by calpains in A431 cells
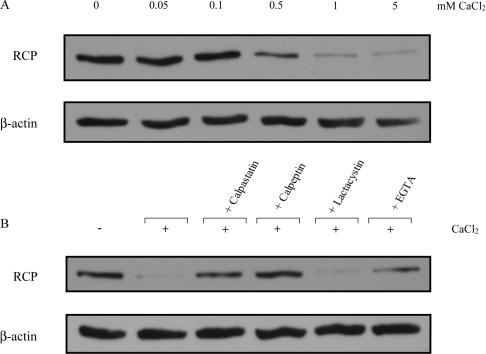
Proteins that contain PEST sequences are generally targeted by the proteasome [32,35,36] or by members of the calpain protease family [30–34]. Since calpains are activated by calcium, we investigated whether there was a calcium-dependent degradation of RCP. We prepared cell lysates from the human A431 squamous cell carcinoma cell line in the presence of increasing concentrations of CaCl2. Under these conditions, a dose-dependent degradation of RCP was observed (Figure 2A). Partial degradation was observed with 0.5 mM CaCl2, and degradation was almost complete at 5 mM CaCl2. When cell lysates were incubated with EGTA, a calcium chelator, degradation was prevented (Figure 2B, rightmost lane), demonstrating that the presence of calcium triggered the degradation event. To determine if calpains mediate RCP degradation, calpain inhibitors were added to the lysis buffer in addition to 5 mM CaCl2. Both calpeptin and calpastatin, a more specific peptide inhibitor, prevented RCP degradation. In contrast, the proteasome inhibitor lactacystin failed to inhibit degradation (Figure 2B). These results indicate that calpains and not the proteasome play a role in the proteolysis of RCP. Since the concentration of calcium required to degrade RCP was in the millimolar range, it is likely that M-calpain is preferentially involved.
Figure 2. Endogenous RCP is degraded by calpain in vitro.
(A) A431 cells were lysed in buffer containing increasing amounts (0–5 mM) of CaCl2 for 25 min. (B) A431 cells were lysed in buffer containing 5 mM CaCl2 plus the indicated inhibitor for 25 min at room temperature. The samples were analysed by SDS/PAGE and immunoblotted with anti-RCP or anti-β-actin antibody as a control for equal loading. The results shown are representative of three independent experiments.
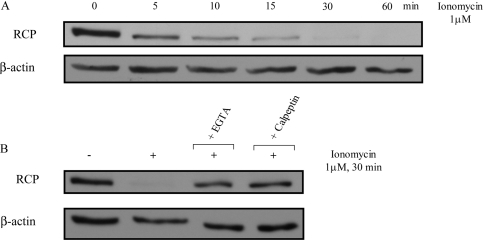
To demonstrate that the calcium-regulated degradation of RCP occurs in living cells, and is not a consequence of the lysis procedure, the Ca2+ ionophore, ionomycin, was added to the culture medium. Upon exposure to 1 μM ionomycin for increasing times, we observed a concomitant increase in the degradation of RCP. This degradation occurred rapidly, as soon as 5 min, and was complete after 30 min (Figure 3A). The degradation of RCP following ionomycin exposure involved calcium entry into the cells, since pre-treatment with the cell-impermeant calcium chelator, EGTA (5 mM for 15 min) before ionomycin addition, prevented RCP degradation (Figure 3B). Similarly, treating the cells with calpeptin (30 μg/ml for 60 min) before ionomycin exposure abolished degradation (Figure 3B). Thus RCP undergoes proteolysis by calpain in a calcium-dependent manner, both in vitro and in living cells.
Figure 3. Endogenous RCP is degraded by calpain in vivo.
(A) A431 cells were treated with 1 μM ionomycin for increasing times (0–60 min) and resuspended in lysis buffer containing 5 mM EGTA to avoid any further degradation of RCP. (B) A431 cells were pre-treated with 5 mM EGTA for 15 min, or 30 μg/ml calpeptin for 60 min, before treatment with 1 μM ionomycin for 30 min. The cells were then resuspended in lysis buffer containing 5 mM EGTA. The samples were analysed by SDS/PAGE and immunoblotted with anti-RCP or anti-β-actin antibody as a control for equal loading. The results shown are representative of three independent experiments.
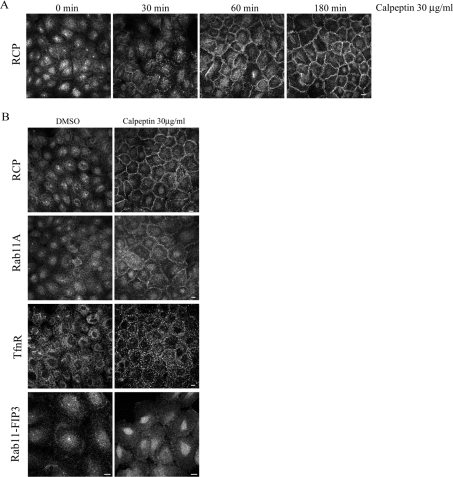
RCP vesicles accumulate at the plasma membrane upon calpeptin treatment in A431 cells
We examined the localization of endogenous RCP in A431 cells treated with calpeptin for increasing lengths of time. After 30 min of calpeptin treatment, RCP localization at the ERC was significantly reduced. At 3 h after calpeptin addition, the majority of RCP had translocated from the ERC and intracellular vesicles to the periphery of the cell, where it displayed a striking ‘honey-comb’ pattern in A431 monolayers (Figure 4A). Close examination of RCP in these cells revealed that it localized to vesicles in the vicinity of the plasma membrane (labelled with ZO-1) (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/389/bj3890223add.htm). This peripheral localization of RCP in cells treated with a calpain inhibitor is consistent with a model in which RCP is targeted for degradation by calpain upon delivering its cargo to the plasma membrane. In agreement with this model is the recent proposal that M-calpain is activated by ERK (extracellular-signal-regulated kinase) at the plasma membrane, and that this is the location where it performs its cellular functions [38].
Figure 4. RCP is redistributed to the plasma membrane following calpeptin treatment.
(A) A431 cells were treated with calpeptin (30 μg/ml) or solvent (DMSO) alone for the indicated times. The cells were then fixed and labelled with anti-RCP. (B) Cells treated with solvent alone (DMSO) or 30 μg/ml calpeptin for 3 h were fixed and labelled for RCP, Rab11A, TfnR or Rab11-FIP3. Scale bar, 10 μm.
We also investigated the effect of calpeptin on the localization of a number of endosomal markers. As expected, when A431 cells are treated with calpeptin for 3 h, we observed a shift of Rab11 staining from the ERC to the cell periphery, similar to RCP (Figure 4B). Rab11-FIP3, which, like RCP, also has putative PEST sequences, did not translocate to the plasma membrane upon calpeptin treatment (Figure 4B). Rab11-FIP3 has not been implicated in ERC to plasma membrane transport [19] and therefore may undergo regulated degradation at a different location within the cell. Since RCP has been shown to be involved in Tfn trafficking in HeLa cells [14,29] and RAW macrophages [28], we investigated the effect of calpeptin on the TfnR pattern. Treatment with calpeptin induced the formation of large TfnR-positive vesicles near the cell surface (Figure 4B). However, these vesicles were not positive for RCP (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/389/bj3890223add.htm). These data suggest that the primary role of RCP may not be in the recycling of Tfn in A431 cells. To investigate this, we compared further the internalization and recycling of Alexa Fluor® 594–Tfn in cells treated with, or without, calpeptin. A431 cells were pre-treated with calpeptin for 3 h and were then allowed to internalize Alexa Fluor® 594–Tfn continuously for 45 min. The Tfn accumulated in large peripherally located vesicles, whereas, in control cells, the Tfn labelled a larger number of smaller vesicles throughout the cytoplasm, and concentrated in the perinuclear region (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/389/bj3890223add.htm). In both control and calpeptin-treated cells, there was no significant co-localization between the Alexa Fluor® 594–Tfn and RCP (results not shown). The recycling of Tfn back out of the cell was somewhat inhibited in the calpeptin-treated cells, as can be seen when the fluorescent transferrin was chased with excess unlabelled ligand after the initial uptake. In control cells, virtually no Alexa Fluor® 594–Tfn remained after the chase, whereas in the calpeptin-treated samples, there was still some fluorescent Tfn visible in the enlarged vesicles (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/389/bj3890223add.htm). Since RCP does not display any significant co-localization with Tfn at any point along its transport pathway, we believe that RCP is not involved in regulating Tfn recycling. This is consistent with the observation that Rab11A is not involved in the basolateral recycling of Tfn in polarized cells, rather it regulates transcytosis and apical recycling [39]. The minor inhibition of Tfn recycling out of the cell upon calpeptin treatment is probably due to the inhibitor affecting other components of the Tfn-trafficking machinery.
RCPΔPEST is localized to the plasma membrane
If the PEST sequences in RCP do serve as calpain recognition/cleavage motifs, we hypothesized that the calpeptin effects on endogenous RCP localization would be mimicked by an RCP mutant lacking the PEST sequences. To test this hypothesis, we generated an RCP mutant in which the PEST motifs had been removed, RCPΔPEST (Figure 1B). Wild-type and mutant RCP constructs were transfected into A431 cells and the recombinant proteins were expressed for 24 h. We found that deletion of the PEST sequences in RCP increased its resistance to degradation in the presence of calcium (Figure 5D; 57.1±8% for RCPWT compared with 26.1±5.8% for RCPΔPEST), suggesting that these motifs are targeted by calpain for RCP degradation. For immunofluorescence studies, the cells were fixed, and the recombinant proteins were visualized with an antibody that recognizes a small epitope (Xpress) fused to their N-termini. The majority of cells expressing RCPWT displayed a punctate vesicular localization, with a concentration of the fusion protein in the pericentrosomal region, a pattern very similar to that of endogenous RCP (Figure 5A). In contrast, the RCPΔPEST fusion protein displayed significant localization to the cell periphery in vesicles adjacent to the plasma membrane (Figure 5A), a situation comparable with endogenous RCP after calpeptin treatment. For both RCPWT and RCPΔPEST, we counted the number of cells that displayed plasma membrane labelling, and expressed the results as a percentage of the total number of transfected cells counted. We defined two phenotypes, intracellular (I) and plasma membrane (P.M.). The majority of RCPWT-expressing cells displayed an intracellular phenotype, with only 26.6±4.6% showing any plasma membrane labelling. In contrast, 77.3±2.3% of cells expressing RCPΔPEST displayed a plasma membrane phenotype (Figure 5B). Thus these data correlate with our inhibitor studies, in that the inability of calpain to cleave RCP results in the accumulation of RCP-positive vesicles in the vicinity of the plasma membrane. As expected, we found that expression of RCPΔPEST also causes a redistribution of Rab11 vesicles to the plasma membrane vicinity (Figure 5C).
DISCUSSION
Rab11 is a small GTPase that is involved in regulating the transport of molecules from the ERC to the plasma membrane. It is also believed that Rab11 plays a role in transport between ERC and TGN. The precise molecular mechanisms by which Rab11 controls endosomal recycling remain to be clarified. One approach used by several groups has consisted of identifying new Rab11-interacting proteins through which it mediates its functions. This approach has led to the identification of the Rab11-FIP family which comprises Rip11, Rab11-FIP2, RCP, Rab11-FIP1, Rab11-FIP3 and Rab11-FIP4. While these proteins display a strong interaction with Rab11, their specific roles in Rab11-mediated membrane transport remain unclear. It seems that the class I Rab11-FIPs (Rip11, RCP and Rab11-FIP2) are involved in the receptor recycling pathways [14,26,27,29,40]. For the class II Rab11-FIPs (Rab-FIP3 and Rab11-FIP4), their ability to bind Arf GTPases in addition to Rab11 suggests that they act as ‘molecular links’ that couple the pathways regulated by these small GTPases. Recent data provide evidence for a role of these proteins in membrane delivery during cell division [19,20,23]. To understand further the function of the Rab11-FIPs, we set out to study how the function of these proteins could be regulated.
In the present paper, we report that RCP contains PEST sequences that act as signals for its regulated degradation. PEST motifs have been reported to act as targets for calpain proteases [30–34] or the proteasome [32,35,36]. We provide several lines of evidence in the present study indicating that calpain proteases, and not the proteasome, degrade RCP in a calcium-dependent manner. An RCP-truncation mutant that lacks the PEST motifs is resistant to calcium-induced degradation, strongly implicating these domains as serving to target the protein for calpain proteolysis [31]. Calpains are Ca2+-activated proteases that exist as two major ubiquitously expressed isoforms, M-calpain and μ-calpain. They were named with reference to the concentrations of calcium required for their activation in vitro. M-calpain is activated by millimolar Ca2+ (0.2–1 mM) and μ-calpain is activated by micromolar concentrations (5–50 μM) [41]. In our system, the A431 cell line, the high calcium concentration requirement for inducing RCP proteolysis suggests an involvement of M-calpain. Nevertheless, we cannot rule out the involvement of μ-calpain, since M- and μ-calpain can cleave the same substrates [42]. Since the high concentrations of calcium required to activate M-calpain in vitro are never found within the cell, it is likely that there are other pathways involved in activating this isoform in vivo. Recently, it has been shown that such a mechanism could involve EGF (epidermal growth factor) and ERK [38].
We demonstrated previously that the C2 domain of the class I Rab11-FIPs preferentially binds the phospholipids PtdIns-(3,4,5)P3 and phosphatidic acid. Both are present in the plasma membrane, and we proposed that they serve as a target for the Rab11-FIP-containing transport vesicles that emanate from the ERC. Indeed, when PtdIns(3,4,5)P3 or phosphatidic acid production is stimulated, these proteins can translocate to the plasma membrane [24]. Interestingly, we observed a similar phenomenon in cells treated with calpeptin in which RCP redistributed from intracellular vesicles to the cell periphery (Figure 4). This effect is also mimicked by the RCPΔPEST mutant, which displays more prominent localization at the plasma membrane and peripheral vesicles in comparison with RCPWT (Figure 5). These results strongly suggest that calpain cleavage of RCP occurs at the plasma membrane. Indeed, at the resting state, calpains (this general name refers to the two most studied: M- and μ-calpain) are localized in the cytosol and, upon an increase of intracellular calcium, they translocate to the plasma membrane, where they are activated by calcium and phospholipids. Once activated, they cleave their substrates either in the cytoplasm or at the plasma membrane [43]. We propose a model in which RCP mediates the transport or docking of recycling vesicles to the plasma membrane. Once delivery of the cargo has been accomplished, RCP function is down-regulated as a result of cleavage by calpain at the plasma membrane. Moreover, when calpain action is inhibited (either by calpeptin treatment or by deletion of the PEST motifs), RCP is predominantly localized to vesicles in the vicinity of the plasma membrane. This suggests that calpain cleavage is a necessary prerequisite for vesicles to fuse with the plasma membrane (Figure 6).
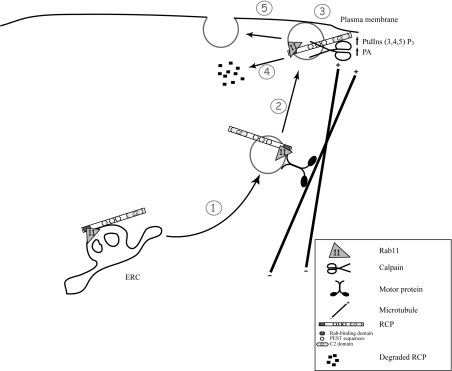
Figure 6. Proposed model of RCP functional cycle.
This model is based on findings presented in the present paper and in previous studies [14,24,49]. (1) Recycling cargo is transported to the plasma membrane in RCP- and Rab11-positive recycling vesicles, created by budding from the ERC. These vesicles move along the microtubule cytoskeleton in conjunction with an as yet unknown motor protein. (2) Localized synthesis of phosphatidic acid (PA) and PtdIns(3,4,5)P3 at the plasma membrane provides a signal for vesicle docking mediated by the C2 domain of RCP. (3) Vesicles are docked to the plasma membrane and RCP is then cleaved by calpains (4), allowing the fusion of the vesicle with the plasma membrane (5).
Calpain-dependent cleavage of their substrates results in either the inactivation or the activation of the target proteins. For instance, M-calpain cleavage of brain calmodulin-dependent phosphodiesterase results in its activation independently of calmodulin [44]. In contrast, p53 cleavage by calpain suppresses its ability to regulate gene expression [45–47]. As depicted in Figure 1, the three PEST motifs in RCP are positioned between the two functional domains (C2 domain and Rab-binding domain), in the region of amino acids 182–347. Thus calpain cleavage at these sites will cause separation of these two important functional domains. An important question that arises is whether the two resulting fragments have a functional role. For the C-terminus of the protein, containing the RBD, it is possible that it may serve as a Rab11 inhibitor and consequently as negative feedback of the Rab11-dependent recycling process. Indeed, when a truncated mutant of RCP, containing just the RBD, is expressed in HeLa cells, a condensation of the TfnR compartment is observed and Tfn recycling is inhibited [14]. It should be noted that we were unable to detect degradation products of RCP either in vitro or in vivo. A similar observation was made by Bordone and Campbell [48] with DNA ligase III. Furthermore, our data demonstrating that RCP- positive vesicles accumulate near the plasma membrane under conditions where calpain activity is inhibited, suggest that RCP cleavage acts in a positive fashion regarding the tethering and/or fusion between the recycling vesicles and plasma membrane.
The signals leading to calpain activation are still a matter of debate, mainly because of the calcium requirement. As previously mentioned, the high concentrations of calcium required to activate M-calpain in vitro are never achieved in vivo. Recently, Glading et al. [38] demonstrated that M-calpain could be activated by calcium-independent phosphorylation mechanisms. Indeed, these authors showed that ERK, activated by EGF, could phosphorylate and subsequently activate M-calpain [38]. These data are interesting in the light of our previous results demonstrating that EGF promotes RCP translocation to the plasma membrane and thus may serve as a signal for the RCP-positive recycling vesicles to deliver their content to the plasma membrane [24]. One possibility is that EGF could recruit calpain to the site of RCP docking at the plasma membrane where it can then cleave RCP.
In conclusion, we have demonstrated that RCP possesses three PEST motifs, which function to target it for degradation. We have shown further that this degradation is mediated by calpain proteases, and is likely to occur at the plasma membrane. This fits in with a model in which RCP–Rab11 complexes regulate the transport of cargo from the ERC to the plasma membrane and, upon delivery, RCP is down-regulated via proteolysis by calpains (Figure 6). Our next avenue of investigation will be to identify the specific calpain isoform(s) that target RCP and to determine the signalling pathways that are involved.
Online data
Acknowledgments
We thank the members of the laboratory and especially Sara Hanscom for her excellent technical assistance. This work was supported by European Union (Marie Curie Development Host Fellowship, Grant # HPMD-CT-2000-00024) and Science Foundation Ireland (SFI) (Investigator Grant # 02/IN.1/B070).
References
- 1.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 2.van der Sluijs P., Hull M., Webster P., Male P., Goud B., Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 3.Van Der Sluijs P., Hull M., Zahraoui A., Tavitian A., Goud B., Mellman I. The small GTP-binding protein rab4 is associated with early endosomes. Proc. Natl. Acad. Sci. U.S.A. 1991;88:6313–6317. doi: 10.1073/pnas.88.14.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCaffrey M. W., Bielli A., Cantalupo G., Mora S., Roberti V., Santillo M., Drummond F., Bucci C. Rab4 affects both recycling and degradative endosomal trafficking. FEBS Lett. 2001;495:21–30. doi: 10.1016/s0014-5793(01)02359-6. [DOI] [PubMed] [Google Scholar]
- 5.Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R. G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbe S., Huber L. A., Zerial M., Tooze S. A., Parton R. G. Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 1993;334:175–182. doi: 10.1016/0014-5793(93)81707-7. [DOI] [PubMed] [Google Scholar]
- 7.Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M., Sabatini D. D. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcke M., Johannes L., Galli T., Mayau V., Goud B., Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng J., Ren M., Gravotta D., De Lemos-Chiarandini C., Lui M., Erdjument-Bromage H., Tempst P., Xu G., Shen T. H., Morimoto T., et al. Identification of a putative effector protein for rab11 that participates in transferrin recycling. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2840–2845. doi: 10.1073/pnas.96.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammoto A., Ohtsuka T., Hotta I., Sasaki T., Takai Y. Rab11BP/Rabphilin-11, a downstream target of rab11 small G protein implicated in vesicle recycling. J. Biol. Chem. 1999;274:25517–25524. doi: 10.1074/jbc.274.36.25517. [DOI] [PubMed] [Google Scholar]
- 11.Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Jr, Mercer J. A., Bahler M., Goldenring J. R. Myosin Vb is associated with plasma membrane recycling systems. Mol. Biol. Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prekeris R., Klumperman J., Scheller R. H. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell. 2000;6:1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 13.Prekeris R., Davies J. M., Scheller R. H. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay A. J., Hendrick A. G., Cantalupo G., Senic-Matuglia F., Goud B., Bucci C., McCaffrey M. W. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- 15.Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- 16.Wallace D. M. E., Lindsay A. J., Hendrick A. G., McCaffrey M. W. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem. Biophys. Res. Commun. 2002;299:770–779. doi: 10.1016/s0006-291x(02)02720-1. [DOI] [PubMed] [Google Scholar]
- 17.Meyers J. M., Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J. Biol. Chem. 2002;277:49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- 18.Wallace D. M., Lindsay A. J., Hendrick A. G., McCaffrey M. W. The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem. Biophys. Res. Commun. 2002;292:909–915. doi: 10.1006/bbrc.2002.6736. [DOI] [PubMed] [Google Scholar]
- 19.Horgan C. P., Walsh M., Zurawski T. H., McCaffrey M. W. Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem. Biophys. Res. Commun. 2004;319:83–94. doi: 10.1016/j.bbrc.2004.04.157. [DOI] [PubMed] [Google Scholar]
- 20.Hickson G. R., Matheson J., Riggs B., Maier V. H., Fielding A. B., Prekeris R., Sullivan W., Barr F. A., Gould G. W. Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol. Biol. Cell. 2003;14:2908–2920. doi: 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin O. H., Couvillon A. D., Exton J. H. Arfophilin is a common target of both class II and class III ADP-ribosylation factors. Biochemistry. 2001;40:10846–10852. doi: 10.1021/bi0107391. [DOI] [PubMed] [Google Scholar]
- 22.Shin O. H., Ross A. H., Mihai I., Exton J. H. Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J. Biol. Chem. 1999;274:36609–36615. doi: 10.1074/jbc.274.51.36609. [DOI] [PubMed] [Google Scholar]
- 23.Wilson G. M., Fielding A. B., Simon G. C., Yu X., Andrews P. D., Hames R. S., Frey A. M., Peden A. A., Gould G. W., Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsay A. J., McCaffrey M. W. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 2004;117:4365–4375. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- 25.Hales C. M., Vaerman J.-P., Goldenring J. R. Rab11 family interacting protein 2 associates with myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- 26.Lindsay A. J., McCaffrey M. W. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J. Biol. Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 27.Fan G.-H., Lapierre L. A., Goldenring J. R., Sai J., Richmond A. Rab11-family interacting protein 2 and myosin Vb Are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol. Biol. Cell. 2004;15:2456–2469. doi: 10.1091/mbc.E03-09-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damiani M. T., Pavarotti M., Leiva N., Lindsay A. J., McCaffrey M. W., Colombo M. I. Rab coupling protein associates with phagosomes and regulates recycling from the phagosomal compartment. Traffic. 2004;5:785–797. doi: 10.1111/j.1600-0854.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- 29.Peden A. A., Schonteich E., Chun J., Junutula J. R., Scheller R. H., Prekeris R. The RCP–Rab11 complex regulates endocytic protein sorting. Mol. Biol. Cell. 2004;15:3530–3541. doi: 10.1091/mbc.E03-12-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dice J. F. Molecular determinants of protein half-lives in eukaryotic cells. FASEB J. 1987;1:349–357. doi: 10.1096/fasebj.1.5.2824267. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilagyi A., Banoczi Z., Hudecz F., Friedrich P. On the sequential determinants of calpain cleavage. J. Biol. Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- 32.Rechsteiner M., Rogers S. W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 33.Shumway S. D., Maki M., Miyamoto S. The PEST domain of IκBα is necessary and sufficient for in vitro degradation by μ-calpain. J. Biol. Chem. 1999;274:30874–30881. doi: 10.1074/jbc.274.43.30874. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda M., Itoh T. Slac2-a/Melanophilin contains multiple PEST-like sequences that are highly sensitive to proteolysis. J. Biol. Chem. 2004;279:22314–22321. doi: 10.1074/jbc.401791200. [DOI] [PubMed] [Google Scholar]
- 35.Allaman-Pillet N., Storling J., Oberson A., Roduit R., Negri S., Sauser C., Nicod P., Beckmann J. S., Schorderet D. F., Mandrup-Poulsen T., Bonny C. Calcium- and proteasome-dependent degradation of the JNK scaffold protein islet-brain 1. J. Biol. Chem. 2003;278:48720–48726. doi: 10.1074/jbc.M306745200. [DOI] [PubMed] [Google Scholar]
- 36.Spencer M. L., Theodosiou M., Noonan D. J. NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J. Biol. Chem. 2004;279:37069–37078. doi: 10.1074/jbc.M402507200. [DOI] [PubMed] [Google Scholar]
- 37.Tang F., Kauffman E. J., Novak J. L., Nau J. J., Catlett N. L., Weisman L. S. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature (London) 2003;422:87–92. doi: 10.1038/nature01453. [DOI] [PubMed] [Google Scholar]
- 38.Glading A., Bodnar R. J., Reynolds I. J., Shiraha H., Satish L., Potter D. A., Blair H. C., Wells A. Epidermal growth factor activates M-calpain (calpain II), at least in part, by extracellular signal-regulated kinase-mediated phosphorylation. Mol. Cell. Biol. 2004;24:2499–2512. doi: 10.1128/MCB.24.6.2499-2512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Kumar R., Navarre J., Casanova J. E., Goldenring J. R. Regulation of vesicle trafficking in Madin–Darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 2000;275:29138–29146. doi: 10.1074/jbc.M004410200. [DOI] [PubMed] [Google Scholar]
- 40.Cullis D. N., Philip B., Baleja J. D., Feig L. A. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 41.Dayton W. R., Schollmeyer J. V., Lepley R. A., Cortes L. R. A calcium-activated protease possibly involved in myofibrillar protein turnover: isolation of a low-calcium-requiring form of the protease. Biochim. Biophys. Acta. 1981;659:48–61. doi: 10.1016/0005-2744(81)90270-9. [DOI] [PubMed] [Google Scholar]
- 42.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki K., Sorimachi H., Yoshizawa T., Kinbara K., Ishiura S. Calpain: novel family members, activation, and physiologic function. Biol. Chem. Hoppe Seyler. 1995;376:523–529. doi: 10.1515/bchm3.1995.376.9.523. [DOI] [PubMed] [Google Scholar]
- 44.Kakkar R., Raju R. V., Sharma R. K. In vitro generation of an active calmodulin-independent phosphodiesterase from brain calmodulin-dependent phosphodiesterase (PDE1A2) by M-calpain. Arch. Biochem. Biophys. 1998;358:320–328. doi: 10.1006/abbi.1998.0858. [DOI] [PubMed] [Google Scholar]
- 45.Kubbutat M., Vousden K. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol. Cell. Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pariat M., Carillo S., Molinari M., Salvat C., Debussche L., Bracco L., Milner J., Piechaczyk M. Proteolysis by calpains: a possible contribution to degradation of p53. Mol. Cell. Biol. 1997;17:2806–2815. doi: 10.1128/mcb.17.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonen H., Shkedy D., Barnoy S., Kosower N. S., Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- 48.Bordone L., Campbell C. DNA ligase III is degraded by calpain during cell death induced by DNA-damaging agents. J. Biol. Chem. 2002;277:26673–26680. doi: 10.1074/jbc.M112037200. [DOI] [PubMed] [Google Scholar]
- 49.Lindsay A. J., McCaffrey M. W. Characterisation of the Rab binding properties of Rab coupling protein (RCP) by site-directed mutagenesis. FEBS Lett. 2004;571:86–92. doi: 10.1016/j.febslet.2004.06.068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.