Abstract
kir/Gem, Rad, Rem and Rem2 comprise the RGK (Rad/Gem/kir) family of Ras-related small G-proteins. Two important functions of RGK proteins are the regulation of the VDCC (voltage-dependent Ca2+ channel) activity and cell-shape remodelling. RGK proteins interact with 14-3-3 and CaM (calmodulin), but their role on RGK protein function is poorly understood. In contrast with the other RGK family members, Rem2 has been reported to bind neither 14-3-3 nor induce membrane extensions. Furthermore, although Rem2 inhibits VDCC activity, it does not prevent cell-surface transport of Ca2+ channels as has been shown for kir/Gem. In the present study, we re-examined the functions of Rem2 and its interaction with 14-3-3 and CaM. We show that Rem2 in fact does interact with 14-3-3 and CaM and induces dendrite-like extensions in COS cells. 14-3-3, together with CaM, regulates the subcellular distribution of Rem2 between the cytoplasm and the nucleus. Rem2 also interacts with the β-subunits of VDCCs in a GTP-dependent fashion and inhibits Ca2+ channel activity by blocking the α-subunit expression at the cell surface. Thus Rem2 shares many previously unrecognized features with the other RGK family members.
Keywords: 14-3-3, calcium channel, calmodulin (CaM), kir/Gem, Rad, Rem2
Abbreviations: CaM, calmodulin; EST, expressed sequence tag; GST, glutathione S-transferase; HA, haemagglutinin; HEK-293T, human embryonic kidney 293T cells; IRES, internal ribosomal entry site; NLS, nuclear import signal; RGK, Rad/Gem/kir; VDCC, voltage-dependent Ca2+ channel; wt, wild-type
INTRODUCTION
RGK (Rad/Gem/kir) proteins belong to a subfamily of Ras-related GTPases that consist of kir/Gem [1,2], Rad [3], Rem [4] and Rem2 [5]. Although the Ras-related core domain is conserved, RGK proteins exhibit unique structural and functional features that differ from other GTPases. These include the lack of lipid modification for membrane anchorage, the presence of N- and C-terminal extensions and an unconventional G3 motif. Another distinctive characteristic is the regulation of RGK proteins at the transcriptional level [1–4].
The physiological roles of RGK proteins have only recently been explored and they include the regulation of VDCC (voltage-dependent Ca2+ channel) activity and cell-shape remodelling. Ca2+ channels allow Ca2+ entry into cells upon membrane depolarization to regulate processes such as gene expression, muscle contraction, hormone secretion and synaptic transmission (reviewed in [6]). Association of the pore-forming α1 subunit with the auxiliary cytosolic β subunit is thought to facilitate plasma-membrane expression and modulate gating properties of VDCCs (reviewed in [7,8]). Due to its central role in calcium signalling, VDCC activity is closely regulated at different levels (reviewed in [7]). Recently, the β-subunit has been shown to interact with all RGK family members [9–11]. In the case of kir/Gem, this association is nucleotide dependent and interferes with the cell-surface expression of VDCCs [9]. In contrast, Rem2 has been reported not to prevent cell-surface expression of VDCC but instead block channel activity [11]. In addition to Ca2+ channel regulation, kir/Gem, Rad and Rem, but not Rem2 have been implicated in the reorganization of the actin cytoskeleton [12,13], which, in the case of kir/Gem and Rad, has been linked to their role as negative regulators of the Rho pathway [14].
CaM (calmodulin) and 14-3-3 regulate the function of RGK proteins. CaM, a major transducer of Ca2+ signalling, associates with kir/Gem and Rad via their C-terminal extensions in a Ca2+-dependent manner [15,16]. CaM inhibits binding of GTP by kir/Gem [15] and shows a better affinity for the GDP-bound form of Rad [16]. The role of CaM binding on RGK family function is unclear, but in the case of kir/Gem may involve the control of its subcellular localization and its association with the β-subunit [9].
14-3-3 proteins, a family of seven highly conserved isoforms (β, η, σ, ϵ, τ, γ and ζ), regulate a wide range of signalling pathways. 14-3-3 proteins bind to phosphoserine or threonine and thereby can either prevent the interaction with other proteins, regulate the subcellular distribution of proteins or protect proteins from proteolysis (reviewed in [17,18]). 14-3-3 dimers may bind to target proteins that contain two 14-3-3-binding sites (reviewed in [19]). Although kir/Gem, Rad and Rem bind 14-3-3 in a phosphorylation-dependent manner [20,21], Rem2 has been reported not to associate with 14-3-3 [22].
In the present study, we re-evaluate the functions of Rem2 and its interaction with 14-3-3 and CaM. We show that Rem2 does associate with 14-3-3 and CaM and induces changes in cell morphology. 14-3-3, in co-operation with CaM, also regulates the subcellular distribution of Rem2. Rem2 interacts with the β-subunits in a GTP-dependent fashion and thereby blocks cell-surface expression of VDCCs. Thus Rem2 shares many previously unrecognized similarities with the other members of the RGK family.
MATERIALS AND METHODS
Molecular biology
Sequence analysis of EST (expressed sequence tag) clones and genome BLAST of the rat, mouse and human sequences were performed using the NCBI online resources. An in-frame stop codon upstream of Met1 (see Figure 1A) was present in mouse EST clones, but not in EST clones from human and rat. BLAST analysis of the mouse EST sequence against the human and rat genome subsequently confirmed the presence of an in-frame stop codon upstream of Met1 in the human and rat cDNAs.
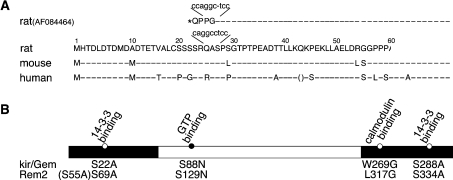
Figure 1. Identification of 14-3-3-binding sites in Rem2.
(A) N-terminal amino acid sequence of full-length rat Rem2 and a truncated form (AF084464) [21] lacking the N-terminal 69 amino acids. The nucleotide sequence of the cDNA encoding the Rem2 and the truncated form indicate that the omission of a C in the truncated form resulted in an in-frame stop codon upstream of Met70, erroneously assigned as the initiation methionine. The N-terminal amino acid sequence of Rem2 for rat, mouse and human is aligned, showing that Met1 is conserved. An in-frame stop codon is located upstream of Met1 (results not shown). Amino acids are shown in one-letter code and residues conserved between species are indicated with a dashed line. (B) Domain structure of Rem2. The Ras-like core domain (white bar), N- and C-terminal extensions (black bars) and the location of the 14-3-3, CaM (white circles) and GTP (black circle)-binding sites are shown. Mutations that affect the different binding sites are indicated, with non-functional putative 14-3-3-binding sites in brackets.
Point mutations were introduced into the putative 14-3-3, CaM and GTP-binding sites of mouse Rem2 by a PCR-based method. RGK and Cavβ3 were Myc and FLAG epitope-tagged respectively at their N-terminus as described previously [9]. For Ca2+ channel cell-surface expression experiments, Cav1.2 was HA (haemagglutinin)-tagged by inserting the epitope into the extracellular S5-H5 loop of domain II as reported previously [23] and introduced into a pIRES (where IRES stands for internal ribosomal entry site) vector containing the Cavβ3 subunit. Cells (90%) transfected by the pIRES vector expressed both subunits (results not shown). GST (glutathione S-transferase)–Cavβ3 and the 14-3-3 ζ dimerization mutant cDNAs were generated as described in [24]. The cDNAs for mouse Rem2, and human 14-3-3 β, γ, ϵ, σ, τ, η and ζ isoforms were obtained from IMAGE Consortium (No. 5328555, 5482228, 5532354, 5502318, 5476961, 5478108, 5531102 and 5492512 respectively). All constructs and purchased cDNAs were verified by automatic DNA sequence analysis (PerkinElmer, Indianapolis, IN, U.S.A.).
Cell culture and DNA transfection
COS-1, PC-12, HEK-293T (human embryonic kidney 293T), C2C12, GH3, HeLa, Neuro2a and NIH3T3 cells were grown and transiently transfected with wt (wild-type) or mutant cDNAs as described in [24]. MIN6 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 15% (v/v) fetal calf serum, 25 mM glucose and 71 μM 2-mercaptoethanol [25]. Biochemical and immunofluorescence experiments were performed 24–48 h after transfection.
Biochemistry
Preparation of cell homogenates, co-precipiation, pull-down and CaM-binding experiments, and SDS/PAGE (8% polyacrylamide gel) and Western-blot analyses using monoclonal anti-c-Myc (Roche, Indianapolis, IN, U.S.A.), monoclonal anti-FLAG (M2; Sigma), rabbit anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) and rabbit anti-14-3-3 (Zymed, South San Fransisco, CA, U.S.A.) antibodies were carried out essentially as described in [24].
Immunocytochemistry
Cells were stained with rabbit anti-Myc (Upstate Biotechnology, Lake Placid, NY, U.S.A.) and monoclonal anti-GST (Cell Signaling Technology, Beverly, MA, U.S.A.) followed by Cy3-labelled donkey anti-rabbit IgG (Jackson Immunoresearch Laboratories, West Grove, PA, U.S.A.) and Alexafluor 488 goat anti-mouse IgG (Molecular Probes) secondary antibodies as described in [24]. For triple-labelling experiments N-Myc RGK proteins, N-FLAG–Cavβ3 and HA–Cav1.2 proteins were detected using rabbit anti-Myc (Upstate Biotechnology), monoclonal anti-FLAG (Met2; Sigma) and rat anti-HA (Roche) antibodies, followed by Cy3-labelled donkey anti-rabbit IgG (Jackson Immunoresearch Laboratories), Alexafluor 350 goat anti-mouse IgG and Alexafluor 488 goat anti-rat IgG (Molecular Probes) secondary antibodies respectively. Cell-surface expression studies in PC-12 and HEK-293T cells were carried out 48 h after transfection. Cells were first incubated with 2 μg/ml of rat anti-HA (Roche) for 1 h at 37 °C and then washed twice in ice-cold PBS before fixation. Specimens were visualized with an Axiocam microscope (Carl Zeiss, Thornwood, NY, U.S.A.) at ×100 magnification.
Electrophysiology
Recordings were made 48 h after transfection using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA, U.S.A.) as described previously [9,24].
RESULTS
Identification of 14-3-3-binding sites in Rem2 and characterization of 14-3-3 binding
A previous study reported that 14-3-3 does not bind to Rem2 [21]. Sequence analysis of Rem2 EST clones and orthologues (Genbank® accession number: rat, CB802662; mouse, BY794767 and human, BM546743) revealed an in-frame methionine (Met1) present upstream of the initiation methionine (Met70) assigned in the rat cDNA (Genbank® accession no. AF084464) used previously [21] (Figure 1A). More detailed analysis indicated that a likely sequencing error in AF084464 (a C missing in a GC rich region), probably led to the erroneous assignment of an in-frame termination codon upstream of Met70. In contrast, Met1 is conserved across species and is preceded by an in-frame stop codon in all three species (see the Material and methods section). Thus AF084464 used in previous studies [21] encodes an N-terminally truncated form of Rem2 that lacks the first 69 amino acids.
Amino acids Ser23 and Ser289 in human kir/Gem (Ser22 and Ser288 in mouse) are known 14-3-3-binding sites [20,24]. Although the C-terminal serine is conserved in Rem2, two alternative N-terminal serine residues could act as 14-3-3-binding sites (Figure 1B). To test if these serine residues are functional 14-3-3-binding sites, Myc-tagged Rem2 or mutants where the N- or C-terminal serine residues were mutated to alanine [Rem2 S69A (where S69A stands for Ser69→Ala) and Rem2 S334A; Figure 1B] were co-expressed with GST–14-3-3 ζ in COS-1 cells and tested for their ability to co-precipitate. GST–14-3-3 ζ K49E, a 14-3-3 mutant defective in target protein binding, [26] was used as a control.
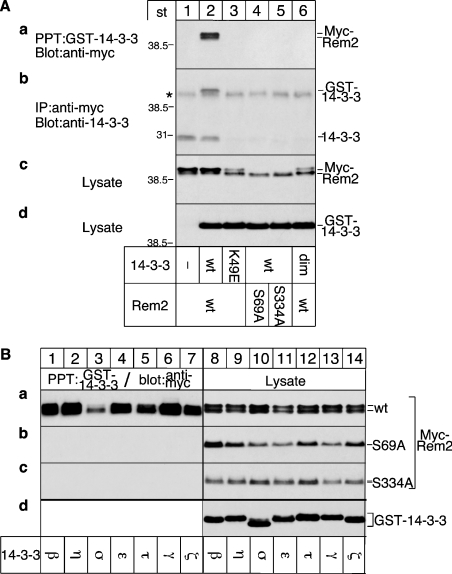
As shown in Figure 2(A), Rem2 co-precipitated with GST–14-3-3 (panel a, lane 2), but not with GST–14-3-3 K49E (panel a, lane 3). GST–14-3-3 failed to associate with Rem2 S69A and Rem2 S334A (panel a, lanes 4 and 5), demonstrating that these serine residues are functional 14-3-3-binding sites. Mutation of the other putative N-terminal 14-3-3-binding site (Ser55) did not affect 14-3-3 binding (results not shown), indicating that this serine residue is not involved in the interaction with 14-3-3. Western-blot analysis of the cell lysates confirmed that the transfected cells expressed similar levels of the wt or mutated Rem2 (Figure 2A, panel c) and GST–14-3-3 or GST–14-3-3 K49E (Figure 2A, panel d) respectively. Similar results were obtained for the inverse experiment where Rem2 was first immunoprecipitated and the associated overexpressed GST–14-3-3 and endogenous 14-3-3 were detected using an anti-14-3-3 antibody (Figure 2A, panel b, lanes 1 and 2).
Figure 2. Identification and characterization of 14-3-3 binding sites in Rem2.
(A) Identification of 14-3-3-binding sites. (a) Cells were co-transfected with cDNAs for wt or mutated Myc-Rem2 and GST–14-3-3, GST–14-3-3 K49E ζ isoforms or a dimerization defective mutant (dim). GST–14-3-3 proteins were precipitated and associated Rem2 detected by Western blotting using Myc antibodies. (b) Cells were co-transfected with cDNAs for wt or mutated Myc-Rem2 and GST–14-3-3, GST–14-3-3 K49E or a dimerization defective mutant (dim). Rem2 was immunoprecipitated and associated GST–14-3-3 and endogenous 14-3-3 were detected by Western blotting using 14-3-3 antibodies. The IgG heavy chain, migrating just below the GST–14-3-3 in (b), is marked by an *. (c, d) Expression levels. Cell lysates were blotted with Myc (c) or GST (d) antibodies to monitor the expression level of Myc–Rem or GST–14-3-3 respectively; st, protein markers of known molecular mass. (B) Association of Rem2 with 14-3-3 isoforms. (a–c, lanes 1–7) Cells were co-transfected with cDNAs for the different GST–14-3-3 isoforms and wt or mutated Myc–Rem proteins. GST–14-3-3 proteins were precipitated and associated RGK proteins were detected by Western blotting using a Myc antibody. (a–c, lanes 8–14) Cell lysates were blotted with Myc antibodies to monitor Rem2-expression levels. (d) One representative example of cell lysates blotted with GST antibody to monitor expression levels of the GST–14-3-3 isoforms.
The presence of two 14-3-3-binding sites in Rem2 and the requirement of both sites for binding of 14-3-3 suggested that Rem2 may associate with 14-3-3 dimers. To test this hypothesis, we took advantage of a mutation in 14-3-3 ζ that has been shown to abolish dimerization without affecting binding to target proteins [17,18]. Based on co-precipitation experiments as described above, the 14-3-3 dimerization defective mutant did not associate with Rem2 (Figure 2A, panels a and b, compare lane 2 with 6), consistent with the notion that Rem2 requires two functional 14-3-3-binding sites and only associates with 14-3-3 dimers. To test whether association between Rem2 and 14-3-3 is isoform-specific, the seven known 14-3-3 isoforms were co-preciptated with Rem2 (Figure 2B). Rem2 bound all seven 14-3-3 family members via the identified 14-3-3-binding sites, although apparently with different efficiencies (Figure 2B, 1–7).
As previously observed for kir/Gem [13], Rem2 was detected as a doublet on SDS/PAGE (Figure 2A, panel c and Figure 2B, panel a), indicative of a post-translational modification. The ratios between the slower and the faster migrating bands were altered if either the 14-3-3-binding sites were mutated or 14-3-3 mutants were overexpressed (Figure 2A, panel c, compare lanes 1 and 2 with 3–6), suggesting a role of 14-3-3 proteins in this post-translational modification.
In conclusion, Rem2 can bind dimers of all seven 14-3-3 isoforms and binding requires both the N- and C-terminal 14-3-3-binding sites in Rem2.
Rem2 binds CaM
The C-terminal extension in kir/Gem interacts with CaM [9] (see Figure 1B). Since an association of CaM with Rem2 has not been established, we carried out CaM-binding experiments to determine if Rem2 associates with CaM and to analyse the role of 14-3-3-binding sites in this association. A mutation, Rem2 L317G, which corresponds to the kir/Gem W269G mutation that abolishes CaM binding [9], was generated in Rem2.
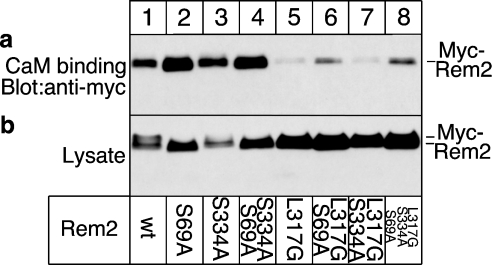
Wt Rem2 and mutants lacking one or both 14-3-3-binding sites bound CaM (Figure 3, panel a, lanes 1–4). In contrast, binding of Rem2 mutants containing the L317G substitution to CaM was significantly reduced (Figure 3, panel a, lanes 5–8), demonstrating that this mutation indeed interferes with CaM binding. Western-blot analysis of cell lysates confirmed that the transfected cells expressed similar levels of wt or mutated Rem2, although preventing CaM binding apparently stabilized the RGK protein (Figure 3, panel b, compare lanes 1–4 with 5–8).
Figure 3. Binding of CaM to Rem2.
(a) Cells were transfected with cDNA for wt or mutated Myc–Rem2. Cell homogenates were incubated with CaM beads and bound Myc–Rem2 detected by Western blotting using Myc antibody. (b) Cell lysates were blotted with Myc antibodies to monitor Rem2 expression levels.
In conclusion, Rem2 binds CaM and the L317G substitution in Rem2 severely affects this association.
14-3-3 and CaM regulate the subcellular distribution of Rem2
Since one function of 14-3-3 is to regulate the subcellular distribution of proteins it interacts with [17,18], we analysed by immunofluorescence microscopy the subcellular distribution of Rem2 in COS-1 cells expressing different combinations of wt Rem2 or mutants defective in 14-3-3 binding and GST–14-3-3 ζ (Figure 4A).
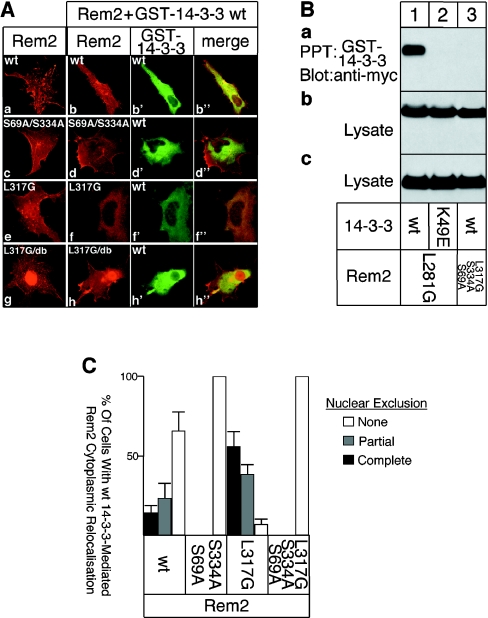
Figure 4. 14-3-3 and CaM regulate the subcellular distribution of Rem2, and Rem2 mediates changes to cell morphology.
(A) COS-1 cells were transfected with cDNAs for wt or mutated Rem2, either alone or together with GST–14-3-3. Cells were processed for immunofluorescence microscopy using Myc and GST antibodies to label Rem2 (a–h; red) and GST–14-3-3 (b′, d′ f′ and h′; green) respectively. Areas of co-localization are in yellow in the merged images (b″, d″, f″ and h″); db, double mutant. (B) Rem2 mutants defective in CaM binding associate with 14-3-3. Cells were co-transfected with cDNAs for mutated Myc–Rem2 and GST–14-3-3 or GST–14-3-3 K49E. (a) GST–14-3-3 proteins were precipitated and the associated Myc–Rem2 detected by Western blotting using Myc antibody. Cell lysates were blotted with Myc (b) or GST (c) antibodies to monitor Rem2 or GST–14-3-3 expression levels respectively. (C) Quantification of the 14-3-3 mediated cytosolic relocalization of Rem2. Transfected cells (150–200) were randomly selected and analysed in 3–5 independent experiments. The fraction of cells showing efficient (black bars), partial (grey bar) or no (white bars) nuclear clearance is plotted.
Expression of wt Rem2 induced dendrite-like extensions and the RGK protein was localized at submembranous regions in the cytosol and in the nucleus (Figure 4A, panel a). The distribution of Rem2 mutants defective in 14-3-3 binding was not markedly changed (Figure 4A, panel c). Overexpression of GST–14-3-3 (or FLAG–14-3-3; results not shown) led to a reduction of the dendrite-like extensions induced by Rem2 (Figure 4A, panels b–b″). This effect was not observed if the 14-3-3-binding sites were mutated (Figure 4A, panels d–d″). Nuclear localization of Rem2 was not visibly affected by GST–14-3-3 (Figure 4A, panels b–b″ and d′–d″). As a control, GST–14-3-3 K49E showed no effect (results not shown). Thus the reduction in Rem2-induced dendrite-like extensions by 14-3-3 correlated with the ability of 14-3-3 to bind Rem2.
The C-terminal 14-3-3-binding site in RGK proteins is in close proximity to the domain that interacts with CaM (see Figure 1A). In addition, CaM has been implicated in affecting the subcellular distribution of kir/Gem since a mutant (kir/Gem W269G) that prevents CaM binding is found in the nucleus of HEK-293T cells [9]. To explore the possibility that CaM could play a role in Rem2 localization, we co-expressed Rem2 L317G or Rem2 L317G/S69A/S334A and GST–14-3-3 and analysed their subcellular distribution.
Similar to what had previously been found for kir/Gem W269G [9], Rem2 L317G showed a more predominant nuclear localization (Figure 4A, panel e), which was even more apparent if the 14-3-3 sites were also mutated in the CaM binding deficient mutant (Figure 4A, panel g). Interestingly, overexpression of GST–14-3-3 led to the efficient translocation of Rem2 L317G from the nucleus to the cytosol, whereas no nuclear clearance of Rem2 was observed if the 14-3-3-binding sites were mutated (Figure 4A, panels h–h″ and f–f″). Quantification of these data is shown in Figure 4(C). As expected, GST–14-3-3 K49E did not affect the distribution of Rem2 carrying a mutated CaM-binding site (results not shown).
To determine whether the ability of the CaM binding deficient Rem2 mutant to bind 14-3-3 correlates with its subcellular localization, we carried out co-precipitation experiments. As expected, Rem2 L317G efficiently associated with GST–14-3-3 (Figure 4B, panel a, lane 1) but not with GST–14-3-3 K49E (Figure 4B, panel a, lane 2). Mutation of the two 14-3-3-binding sites abolished the interaction of Rem2 L317G with GST–14-3-3 (Figure 4B, panel a, lane 3). Western-blot analysis of cell lysates confirmed that the transfected cells expressed similar levels of the different Myc-Rem2 and GST–14-3-3 (Figure 4B, panels b and c). Thus the ability of the CaM binding deficient Rem2 mutants to associate with 14-3-3 correlated well with the capacity of 14-3-3 to induce their nuclear exclusion.
To ascertain that the modulation of Rem2 distribution by CaM and 14-3-3 is a general feature, we expressed wt Rem2, Rem2 L317G and Rem2 L317G/S69A/S334A in different cell lines and analysed their subcellular localization. Whereas the extent of nuclear localization of wt Rem2 differed among different cell lines, a consistent change in the localization was observed for the mutants defective in CaM and/or 14-3-3 binding, indicating that CaM and 14-3-3 regulate Rem2 localization in all cell lines in a similar fashion (Table 1). Wt Rem2 induced dendrite-like structures in all cell lines analysed except the neuroendocrine cell lines GH3, PC-12 and MIN6. In Neuro2a cells, Rem2 did not induce dendrite-like extensions but small short protrusions. Consistent with the observation for COS-1 cells, the ability to induce morphological changes was dramatically reduced when the Rem2 mutants localized to the nucleus (Table 1).
Table 1. Subcellular localization of Rem2 and its effect on cell morphology.
Wt Rem2, Rem2 L317G and Rem2 L317G/S69A/S334A were expressed in the cell lines indicated and the subcellular localization and induction of morphological changes in cells expressing moderate to high levels of Rem2 were monitored. Cells were classified into those showing only cytoplasmic (C), a diffused cytoplasmic and nuclear (C/N) or a predominant nuclear (N) distribution of Rem2. A shift towards a more nuclear or cytosolic localization is indicated in bold (N or C). In the case of Neuro2a (**), Rem2 was found at the membrane (m) and it induced short protrusions. In Hela cells, Rem2 overexpression produced filapodia-like structures that were independent of the subcellular localization of Rem2. C, cytosol; N, nucleus; and m, membrane. Morphological changes were scored by determining the fraction of cells with extensions of twice the cell diameter or longer. >75% (++++), 50–75% (+++), 5–50% (++) and <5% (−). Three independent experiments were performed and 80–100 cells with medium to high Rem2 expression scored in each experiment.
| Morphological changes | Cellular localization | Rem2 | |
|---|---|---|---|
| C2C12 (mouse myoblast) | ++++ | C/N | wt |
| +++ | C/N | L317G | |
| ++ | C/N | L317G/S69A/S334A | |
| GH3 (rat pituitary) | − | C | wt |
| − | C | L317G | |
| − | C/N | L317G/S69A/S334A | |
| HeLa (human cervix) | ++++ | C/N | wt |
| +++ | C/N | L317G | |
| − | N | L317G/S69A/S334A | |
| Neuro2a (mouse neuroblastoma) | ++(**) | m | wt |
| ++ | m/C | L317G | |
| ++ | C/N | L317G/S69A/S334A | |
| MIN6 (mouse pancreatic β-cell) | − | C | wt |
| − | C/N | L317G | |
| − | C/N | L317G/S69A/S334A | |
| NIH3T3 (mouse embryonic) | ++++ | C | wt |
| ++++ | C/N | L317G | |
| ++ | C/N | L317G/S69A/S334A | |
| PC-12 (rat pheochromocytoma) | − | C | wt |
| − | C | L317G | |
| − | C/N | L317G/S69A/S334A | |
| 293T (human embryonic kidney) | ++ | C | wt |
| − | C/N | L317G | |
| − | C/N | L317G/S69A/S334A | |
| COS1 (monkey kidney) | ++++ | C/N | wt |
| ++ | C/N | L317G | |
| − | N | L317G/S69A/S334A |
In conclusion, the above biochemical data together with the subcellular localization experiments indicate that 14-3-3 and CaM co-ordinately regulate the subcellular localization of Rem2 and its induction of morphological changes.
The Ca2+ channel β-subunit is an effector of Rem2
RGK proteins down-regulate Ca2+ channel activity through their interaction with the β-subunit [9–11]. In the case of kir/Gem, it has been shown that the β-subunit is a bona fide effector that selectively interacts with the GTP-bound form [9]. To determine if the β-subunit can interact with Rem2 in a nucleotide-dependent fashion, we carried out in vitro binding experiments (Figure 5A). Lysates of cells expressing similar amounts of wt Rem2 or Rem2 S129N and Rem2 L317G (Figure 5A, lanes 5–7) were incubated with immobilized β-subunits (GST–Cavβ3) and bound Rem2 was detected by Western-blot analysis.
Figure 5. Nucleotide-dependent binding of Rem2 to the β-subunit of VDCCs and down-regulation of Ca2+ channel activity.
(A) Pull down. Cells were transfected with cDNAs Myc–Rem2, Rem2 S129N or Rem2 L317G. Cell homogenates were incubated with immobilized recombinant GST–Cavβ3 and associated Rem2 was detected by Western blotting using Myc antibody. Recombinant GST served as a control (lane 1). Cell lysates were blotted with Myc antibodies to monitor Rem2 expression levels (lanes 5–7). (B) Electrophysiology. PC-12 cells were co-transfected with a GFP (green fluorescent protein) plasmid and cDNAs for wt or mutated Rem2 either with or without 14-3-3. GFP-positive cells were selected for electrophysiology and the average of the maximal current detected at +20 mV for endogenous Ca2+ channels was measured. For each condition, 9–16 independent experiments were carried out. An example of the I–V relationship of Ca2+ channels in PC-12 cells is shown below. A, Ampere; F, Farad. Cells transfected only with the GFP cDNA served as a control.
Rem2 bound to the β-subunits and the interaction was abolished if the GTP-binding site was mutated (compare lanes 2 and 3) but the interaction was not affected for the CaM binding mutant (lane 4). Binding of Rem2 to the β-subunit did not require the addition of the non-hydrolysable GTP analogue GTP[S], consistent with the low rate of GTP hydrolysis of this family of small G-proteins [2,16,22].
Thus the β-subunit binds preferentially to Rem2 in the GTP-bound form and hence is an effector of the small G-protein.
Rem2-mediated down-regulation of Ca2+ channel activity is not modulated by 14-3-3 or CaM
Kir/Gem [9], Rad and Rem [10] and Rem2 [11] have been shown to down-regulate the VDCC activity. To determine whether 14-3-3 or CaM binding plays a regulatory role in this function of Rem2, we expressed Rem2 and the corresponding CaM or 14-3-3 binding deficient mutants in PC-12 cells, either alone or together with GST–14-3-3, and measured endogenous VDCC Ca2+ currents.
As shown in Figure 5(B), expression of Rem2 repressed Ca2+ channel activity (lane 2). Rem2 did not require CaM binding for its function (lanes 4 and 7). Furthermore, mutation of the 14-3-3-binding sites, or overexpression of GST–14-3-3, did not interfere with RGK function (lanes 3 and 5, and 6 and 7 respectively).
These data confirm the inhibitory effect of Rem2 on Ca2+ channel activity and show that CaM and 14-3-3 binding are dispensable for this function of Rem2.
Rem2 interferes with cell-surface transport of Ca2+ channels
To determine the mechanism by which Rem2 regulates Ca2+ channel activity, we analysed the subcellular distribution of the Ca2+ channel α- and β-subunits in cells expressing either wt or mutated Rem2 proteins. PC-12 cells were co-transfected with cDNAs encoding a Cav1.2 α-subunit in which the HA-tag was inserted into an external loop [23], a FLAG-tagged Cavβ3 and Myc-tagged wt or mutated Rem2. To ensure homogenous expression of the Ca2+ channel subunits, the α- and β-subunits were expressed from an IRES containing vector (see the Materials and methods section). The expression of the RGK proteins and the α- and β-subunits was verified in permeabilized cells. Alternatively, live cells were incubated with HA antibodies before permeabilization to selectively detect the Ca2+ channels present at the cell surface (surface labelling).
As shown in Figure 6(A), the α-subunit expressed alone showed a cytoplasmic localization (panel a) and could not be detected at the cell surface (panel c). Only in the presence of the β-subunit the α-subunit was detected at the surface of unpermeabilized cells as a diffused staining (Figure 6A, panel d′), confirming the importance of the β-subunit in facilitating surface transport of the α-subunit [27]. The β-subunit was diffused in the cytoplasm and, to a lesser extent, in the nucleus (Figure 6A, panels b and d).
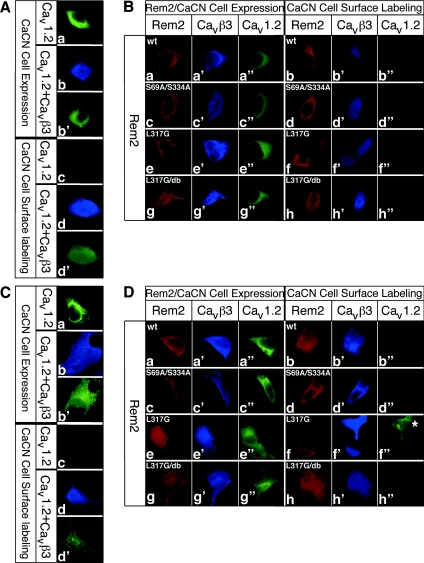
Figure 6. Rem2 blocks cell-surface expression of VDCC α-subunits in PC-12 and HEK-293T cells.
(A, C) The β-subunit facilitates surface expression of the α-subunit. PC-12 (A) or HEK-293T (C) cells were transfected with a cDNA for Cav1.2 carrying an extracellular HA tag or with an IRES-based vector carrying the cDNAs for HA–Cav1.2 and FLAG–Cavβ3 subunits. Cells were fixed, permeabililzed and processed for immunofluorescence microscopy using HA and FLAG antibodies to detect Cav1.2 (green) and Cavβ3 (blue) respectively (CaCN cell expression). Alternatively, live cells were first incubated with HA antibodies to selectively label surface exposed Cav1.2 before the fixation, permeabilization and labelling with FLAG antibodies (CaCN cell-surface labelling). (B, D) Effect of Rem2 on cell-surface expression of α-subunits. PC-12 (B) or HEK-293T (D) cells were transfected with cDNAs for wt or mutated Myc–Rem2 together with an IRES-based vector carrying the cDNAs for HA–Cav1.2 and FLAG–Cavβ3. Cells were fixed, permeabililzed and processed for immunofluorescence microscopy using Myc, HA and FLAG antibodies to detect Rem2 (red), Cav1.2 (green) and Cavβ3 (blue) respectively (Rem2/CaCN cell expression). Alternatively, live cells were first incubated with HA antibodies to selectively label surface exposed Cav1.2 before the fixation, permeabiliziation and labelling with Myc and FLAG antibodies (CaCN cell-surface labelling). db: double mutant with both 14-3-3-binding sites mutated; * in (D f″) indicates a cell that does not express Rem2 and consequently Cav1.2 is expressed at the cell surface. Independent experiments (4 or 5) were performed and 20–30 cells expressing the α- and β-CaCN subunits and the Rem2 proteins were analysed.
Rem2 (Figure 6B, panels a and b), as well as the mutants in which the 14-3-3 binding sites were mutated (Figure 6B, panels c and d) showed a similar subcellular distribution as in COS-1 cells. However, the nuclear localization of Rem2 in PC-12 cells was less pronounced than in COS-1 cells and Rem2 L317G was not detected in the nucleus (Figure 6B, panels e and f). Mutation of the 14-3-3 binding sites in the CaM binding deficient Rem2 mutant, however, resulted in a more pronounced nuclear localization (Figure 6B, panels g and h), consistent with a role of endogenous 14-3-3 in the regulation of the subcellular distribution of Rem2 in PC-12 cells. Similar distributions were obtained for HEK-293T cells (Figures 6C and 6D). Compared with HEK-293T cells, Rem2 expressed in COS-1 or PC-12 cells showed a bias for a more nuclear or cytoplasmic distribution respectively. The β-subunit predominantly co-localized with Rem2. Where Rem2 was in the cytoplasm or in the nucleus, the β-subunit was also cytoplasmic or nuclear (Figures 6B and 6D, panels a′–h′).
Cell-surface transport of the α-subunit when co-expressed with the β-subunit (Figures 6A and 6C, compare panel c with d′) was blocked upon the expression of either wt Rem2 or the different mutants (Figures 6B and 6D, panels b″–h″), showing that surface transport of the α-subunit in cells expressing Rem2 and its mutants correlated with their functional effect on Ca2+ channel activity in PC-12 cells (see above; Figure 5).
In summary, Rem2 blocks cell-surface expression of Ca2+ channels and 14-3-3 or CaM binding is apparently not involved in regulating this function of Rem2.
DISCUSSION
The functions of members of the RGK small G-protein family, kir/Gem, Rad, Rem and Rem2, have only recently started to be unravelled. Current evidence indicates that Rem2 significantly differs from the other members of the family, both in terms of its function and its regulation by 14-3-3. In contrast with kir/Gem, Rad and Rem, Rem2 has been reported to neither bind 14-3-3 nor regulate cell shape [22]. Furthermore, Rem2 has been postulated to inhibit VDCC activity by inactivating channels present at the cell surface as opposed to preventing their cell-surface expression [11]. In the present study, we show that the reported lack of binding between 14-3-3 and Rem2 in a previous study [21] is due to the use of a truncated form of Rem2 (see Figure 1A) that lacked the N-terminal 14-3-3 binding site. Using a cDNA encoding a full-length Rem2 protein, we show that Rem2 in fact does bind 14-3-3 and that binding of 14-3-3 requires the presence of both the N- and C-terminal 14-3-3-binding sites. Rem2 also regulates cell shape as also observed for kir/Gem [28] and 14-3-3 reduced the Rem2 induced dendrite-like extensions. In addition, 14-3-3 and CaM co-operatively regulate the subcellular distribution of Rem2 between the cytoplasm and the nucleus, but are not apparently involved in the inhibition of cell-surface transport of the channel.
Rem2, like kir/Gem [24], carries one N- and one C-terminal 14-3-3-binding site and interacts with all 14-3-3 isoforms. Both 14-3-3 binding sites in Rem2 are required for 14-3-3 association and Rem2 only binds 14-3-3 dimers, explaining why in a previous study [21] a truncated form of Rem2 lacking the N-terminal 14-3-3 binding site failed to detect 14-3-3 binding. 14-3-3 dimerization is thought to be required for high affinity stable binding to target proteins [19]. The association of 14-3-3 with RGK proteins may be closely regulated by kinases or phosphatases that act on the 14-3-3-binding sites. Although the C-terminal 14-3-3-binding site is common to all RGK members and does not show an obvious motif for a particular kinase, the N-terminal-binding site appears to be specific for each of the four RGK proteins. For example S69A in Rem2, but S22A in kir/Gem, is within a consensus sequence for protein kinase A phosphorylation. Thus, depending on the extracellular stimulus, 14-3-3 may associate with a specific RGK protein.
Studies on kir/Gem indicate that CaM binding might regulate the subcellular localization of RGK proteins [9]. Our results also demonstrate that Rem2 binds CaM and that in the absence of CaM binding, Rem2 partially translocates into the nucleus. Furthermore, overexpression of 14-3-3 led to the exclusion of Rem2 from the nucleus, an effect that was particularly prominent for the CaM-binding mutant. For unknown reasons, however, analysing Rem2 mutants where single 14-3-3-binding sites were inactivated did not show a strict correlation between 14-3-3 binding and nuclear clearance (results not shown). Whereas in COS-1 cells 14-3-3 overexpression was required for nuclear clearance of Rem2, in the other cell lines analysed a mutation of the 14-3-3 and/or CaM-binding sites was sufficient to alter the subcellular distribution of Rem2, showing that endogenous CaM and 14-3-3 are relevant for this process. Presumably, the balance of active CaM, availability of 14-3-3 and the phosphorylation state of the 14-3-3-binding sites in Rem2 in the different cell lines determine the steady-state distribution of the small G-protein. The mechanism by which 14-3-3 and CaM binding interfere with nuclear translocation of Rem2 is unclear but does not seem to involve Ca2+/CaM kinases (P. Béguin, R. N. Mahalakshmi, K. Nagashima, D. H. K. Cher, N. Kuwamura, Y. Yamada, Y. Seino and W. Hunziker, unpublished work). However, kir/Gem has several NLSs (nuclear import signals) that are conserved in Rem2 (P. Béguin, R. N. Mahalakshmi, K. Nagashima, D. H. K. Cher, N. Kuwamura, Y. Yamada, Y. Seino and W. Hunziker, unpublished work). Interestingly, one of these NLS is in proximity to the CaM and C-terminal 14-3-3-binding sites. It is thus conceivable that 14-3-3 and/or CaM-binding mask this NLS and thereby reduce the rate of nuclear import, as described for other proteins [17].
kir/Gem [13,14,28], Rad [14] and Rem [12] have been shown to regulate cell morphology. Similarly, Rem2 expression results in the formation of dendrite-like extensions in various cell types, except for the neuroendocrine cells GH3, PC-12 and MIN6. In the case of the Neuro2a, short protrusions were observed instead of dendrite-like extensions. When wt Rem2 induced a morphological change, this was reduced for the mutants that localized to the nucleus. Furthermore, overexpression of exogenous 14-3-3, but not a 14-3-3 mutant defective in target protein binding, reduced Rem2 mediated induction of extensions in COS-1 cells, similar to the result obtained with kir/Gem [24]. Whether, like kir/Gem and Rad [14], Rem2 also functions as a negative regulator of the Rho–Rho kinase pathway, remains to be determined.
It has been suggested that RGK proteins might be oncogenic proteins [13,28,29] by positively regulating cell migration. Rem2, which is expressed in neurons [5], could also be involved in neurite outgrowth and neuronal development, requiring a tight temporal and spatial regulation of its expression. In neuroendocrine cells, endogenous factors may repress Rem2, preventing uncontrolled neurite extensions, without affecting the role of Rem2 in calcium-regulated, VDCC-dependent secretion. Thus, whereas regulation of VDCC activity in neuronal and neuroendocrine cells might be a more general function of Rem2, its role in cell migration might be restricted to particular cell types.
Association with the β-subunit requires the GTP-bound form of Rem2 since binding to Rem2 S129N (a mutation corresponding to Rad S104N, which is locked into the GDP-bound form [16]) was severely affected. However, compared with other RGK proteins, the binding efficiency between Rem2 and Cavβ3 was low, indicating that additional factors may be required for efficient association (results not shown). By binding the β-subunit, kir/Gem, Rad, Rem and Rem2 down-regulate Ca2+ channel activity [9,11]. Cell-surface transport of VDCCs is thought to require the association of the α- with the β-subunits (reviewed in [8,30]) and kir/Gem has been shown to interfere with cell-surface transport of VDCCs by interacting with the β-subunit [9,24]. Interestingly, Rem2 has recently been shown to inactivate endogenous VDCCs in Min6 cells without blocking surface transport of the α-subunit [11]. In contrast, using an α-subunit carrying an extracellular epitope tag, we show in PC-12 and 293T cells (mouse embryonic kidney cells) that down-regulation of VDCC activity in the presence of Rem2 correlates with the absence of Ca2+ channels at the cell surface. In Min6 cells, surface expression of endogenous VDCC, which includes Ca2+ channels already present on the cell surface before the transfection of Rem2 cDNA, was analysed [11]. It is thus conceivable that these VDCCs were inactivated by Rem2. In contrast, in our study the cDNAs for the Ca2+-channel subunits and Rem2 were co-transfected into PC-12 cells and only the effect of Rem2 on newly synthesized, exogenously expressed Ca2+ channels was monitored. These two apparently conflicting findings could be reconciled by suggesting that Rem2 and possibly other RGK proteins may prevent cell-surface transport of Ca2+ channels as well as inhibit channel function by uncoupling the β-subunit from VDCCs already present at the cell surface. Indeed, it was recently shown that the α-subunit of VDCCs and kir/Gem bind to the same region in the β-subunit [31]. Mutating the CaM or 14-3-3-binding sites did not interfere with Rem2-mediated inhibition of VDCC surface expression, indicating that CaM and 14-3-3 do not play a role in regulating this function of Rem2. Whether CaM or 14-3-3 regulates Rem2-mediated inactivation of Ca2+ channels present at the cell surface remains to be analysed. Possibly, the extent to which one or the other mechanism is used to inactivate VDCCs may be determined by signalling events that control the association of CaM and 14-3-3 with the RGK proteins.
Acknowledgments
This work was supported by A*STAR (Agency for Science, Technology and Research), Biopolis Way, Centros, Singapore.
References
- 1.Maguire J., Santoro T., Jensen P., Siebenlist U., Yewdell J., Kelly K. Gem: an induced, immediate early protein belonging to the Ras family. Science. 1994;265:241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- 2.Cohen L., Mohr R., Chen Y. Y., Huang M., Kato R., Dorin D., Tamanoi F., Goga A., Afar D., Rosenberg N., et al. Transcriptional activation of a ras-like gene (kir) by oncogenic tyrosine kinases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12448–12452. doi: 10.1073/pnas.91.26.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynet C., Kahn C. R. Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 4.Finlin B. S., Andres D. A. Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J. Biol. Chem. 1997;272:21982–21988. doi: 10.1074/jbc.272.35.21982. [DOI] [PubMed] [Google Scholar]
- 5.Finlin B. S., Shao H., Kadono-Okuda K., Guo N., Andres D. A. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem. J. 2000;347:223–231. [PMC free article] [PubMed] [Google Scholar]
- 6.Dolphin A. C. Mechanisms of modulation of voltage-dependent calcium channels by G proteins. J. Physiol. 1998;506:3–11. doi: 10.1111/j.1469-7793.1998.003bx.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catterall W. A. Structure and regulation of voltage-gated Ca2+ channels. Annu. Rev. Cell Dev. Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 8.Dolphin A. C. Beta subunits of voltage-gated calcium channels. J. Bioenerg. Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- 9.Beguin P., Nagashima K., Gonoi T., Shibasaki T., Takahashi K., Kashima Y., Ozaki N., Geering K., Iwanaga T., Seino S. Regulation of Ca2+ channel expression at the cell surface by the small G-protein kir/Gem. Nature (London) 2001;411:701–706. doi: 10.1038/35079621. [DOI] [PubMed] [Google Scholar]
- 10.Finlin B. S., Crump S. M., Satin J., Andres D. A. Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14469–14474. doi: 10.1073/pnas.2437756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlin B. S., Mosley A. L., Crump S. M., Correll R. N., Ozcan S., Satin J., Andres D. A. Regulation of L-type Ca2+ channel activity and insulin secretion by the Rem2 GTPase. J. Biol. Chem. 2005 doi: 10.1074/jbc.M414261200. doi:10.1074/jbc.M414261200. [DOI] [PubMed]
- 12.Pan J. Y., Fieles W. E., White A. M., Egerton M. M., Silberstein D. S. Ges, a human GTPase of the Rad/Gem/Kir family, promotes endothelial cell sprouting and cytoskeleton reorganization. J. Cell Biol. 2000;149:1107–1116. doi: 10.1083/jcb.149.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leone A., Mitsiades N., Ward Y., Spinelli B., Poulaki V., Tsokos M., Kelly K. The Gem GTP-binding protein promotes morphological differentiation in neuroblastoma. Oncogene. 2001;20:3217–3225. doi: 10.1038/sj.onc.1204420. [DOI] [PubMed] [Google Scholar]
- 14.Ward Y., Yap S. F., Ravichandran V., Matsumura F., Ito M., Spinelli B., Kelly K. The GTP binding proteins Gem and Rad are negative regulators of the Rho-Rho kinase pathway. J. Cell Biol. 2002;157:291–302. doi: 10.1083/jcb.200111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer R., Wei Y., Anagli J., Berchtold M. W. Calmodulin binds to and inhibits GTP binding of the ras-like GTPase Kir/Gem. J. Biol. Chem. 1996;271:25067–25070. doi: 10.1074/jbc.271.41.25067. [DOI] [PubMed] [Google Scholar]
- 16.Moyers J. S., Bilan P. J., Zhu J., Kahn C. R. Rad and Rad-related GTPases interact with calmodulin and calmodulin-dependent protein kinase II. J. Biol. Chem. 1997;272:11832–11839. doi: 10.1074/jbc.272.18.11832. [DOI] [PubMed] [Google Scholar]
- 17.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio M. P., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaffe M. B. How do 14-3-3 proteins work? – Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 20.Ward Y., Spinelli B., Quon M. J., Chen H., Ikeda S. R., Kelly K. Phosphorylation of critical serine residues in Gem separates cytoskeletal reorganization from down-regulation of calcium channel activity. Mol. Cell. Biol. 2004;24:651–661. doi: 10.1128/MCB.24.2.651-661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlin B. S., Andres D. A. Phosphorylation-dependent association of the Ras-related GTP-binding protein Rem with 14-3-3 proteins. Arch. Biochem. Biophys. 1999;368:401–412. doi: 10.1006/abbi.1999.1316. [DOI] [PubMed] [Google Scholar]
- 22.Finlin B. S., Shao H., Kadono-Okuda K., Guo N., Andres D. A. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem. J. 2000;347:223–231. [PMC free article] [PubMed] [Google Scholar]
- 23.Altier C., Dubel S. J., Barrere C., Jarvis S. E., Stotz S. C., Spaetgens R. L., Scott J. D., Cornet V., De Waard M., Zamponi G. W., et al. Trafficking of L-type calcium channels mediated by the postsynaptic scaffolding protein AKAP79. J. Biol. Chem. 2002;277:33598–33603. doi: 10.1074/jbc.M202476200. [DOI] [PubMed] [Google Scholar]
- 24.Beguin P., Mahalakshmi R. N., Nagashima K., Cher D. H. K., Takahashi A., Yamada Y., Seino Y., Hunziker W. 14-3-3 and calmodulin control subcellular distribution of kir/Gem and its regulation of cell shape and calcium channel activity. J. Cell Sci. 2005;118:1923–1934. doi: 10.1242/jcs.02321. [DOI] [PubMed] [Google Scholar]
- 25.Lilla V., Webb G., Rickenbach K., Maturana A., Steiner D. F., Halban P. A., Irminger J. C. Differential gene expression in well-regulated and dysregulated pancreatic β-cell (MIN6) sublines. Endocrinology. 2003;144:1368–1379. doi: 10.1210/en.2002-220916. [DOI] [PubMed] [Google Scholar]
- 26.Fu H., Subramanian R. R., Masters S. C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 27.Lacerda A. E., Kim H. S., Ruth P., Perez-Reyes E., Flockerzi V., Hofmann F., Birnbaumer L., Brown A. M. Normalization of current kinetics by interaction between the α1 and β subunits of the skeletal muscle dihydropyridine-sensitive Ca2+ channel. Nature (London) 1991;352:527–530. doi: 10.1038/352527a0. [DOI] [PubMed] [Google Scholar]
- 28.Piddini E., Schmid J. A., de Martin R., Dotti C. G. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001;20:4076–4087. doi: 10.1093/emboj/20.15.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng Y. H., Vicent D., Zhu J., Niu Y., Adeyinka A., Moyers J. S., Watson P. H., Kahn C. R. Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and nm23. Cancer Res. 2001;61:2071–2079. [PubMed] [Google Scholar]
- 30.Catterall W. A. Structure and function of neuronal Ca2+ channels and their role in neurotransmitter release. Cell Calcium. 1998;24:307–323. doi: 10.1016/s0143-4160(98)90055-0. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki T., Shibasaki T., Beguin P., Nagashima K., Miyazaki M., Seino S. Direct inhibition of the interaction between α-interaction domain and β-interaction domain of voltage-dependent Ca2+ channels by gem. J. Biol. Chem. 2005;280:9308–9312. doi: 10.1074/jbc.M413773200. [DOI] [PubMed] [Google Scholar]