Abstract
Based on the human cDNA sequence predicted to represent the NEU4 sialidase gene in public databases, a cDNA covering the entire coding sequence was isolated from human brain and expressed in mammalian cells. The cDNA encodes two isoforms: one possessing an N-terminal 12-amino-acid sequence that is predicted to be a mitochondrial targeting sequence, and the other lacking these amino acids. Expression of the isoforms is tissuespecific, as assessed by reverse transcription–PCR. Brain, muscle and kidney contained both isoforms; liver showed the highest expression, and the short form was predominant in this organ. In transiently transfected COS-1 cells, enzyme activity was markedly increased with gangliosides as well as with glycoproteins and oligosaccharides as substrates compared with the control levels. This differs from findings with other human sialidases. Although the isoforms were not distinguishable with regard to substrate specificity, they exhibited differential subcellular localizations. Immunofluorescence microscopy and biochemical fractionation demonstrated that an exogenously expressed haemagglutinin-tagged long form of NEU4 was concentrated in mitochondria in several human culture cell types, whereas the short form was present in intracellular membranes, indicating that the sequence comprising the N-terminal 12 amino acid residues acts as a targeting signal for mitochondria. Co-localization of the long form to mitochondria was further supported by efficient targeting of the N-terminal region fused to enhanced green fluorescent protein, and by the targeting failure of a mutant with an amino acid substitution in this region. NEU4 is possibly involved in regulation of apoptosis by modulation of ganglioside GD3, which accumulates in mitochondria during apoptosis and is the best substrate for the sialidase.
Keywords: ganglioside, glycoprotein, immunofluorescence, mitochondria, sialidase, subcellular localization
Abbreviations: EGFP, enhanced green fluorescent protein; HA, haemagglutinin; lamp-1, lysosome-associated membrane protein-1; 4MU-NeuAc, 4-methylumbelliferyl neuraminic acid; PBGD, porphobilinogen deaminase; RT, reverse transcriptase
INTRODUCTION
It has been shown that the removal of sialic acid residues from glycoproteins and glycolipids, which is catalysed by sialidase and is the initial step in the catabolism of these glycoconjugates, has a marked influence on the biological functions of these compounds. In addition to being a key enzyme in the degradation of glycoconjugates, mammalian sialidase has been considered to play important roles in various cellular processes [1–3]. Our previous studies using rat tissues revealed evidence for the existence of four types of sialidase that differ in their subcellular localization and enzymatic properties, including substrate specificity [4–6]. They were classified according to their major intracellular localization as intralysosomal sialidase, cytosolic sialidase, and membrane-associated sialidases I and II. Several rat tissues, including the liver and brain, and even isolated hepatocytes, were found to contain all four types of sialidase. Intralysosomal sialidase possesses narrow substrate specificity, such that only oligosaccharides and glycopeptides serve as substrates [4]. Sialidase found in the cytosol, in contrast, is able to hydrolyse glycoproteins and gangliosides at near neutral pH [5]. Both of these sialidases are distinct from the membrane-associated sialidases, in that the latter require detergents for solubilization and preferentially hydrolyse gangliosides [6]. Membrane sialidase I scarcely hydrolyses other substrates, including oligosaccharides or glycoproteins, whereas membrane sialidase II acts on oligosaccharides, glycoproteins and even GM2 gangliosides having an internal sialic acid residue. Sialidase I is localized mainly in plasma membranes, whereas sialidase II is localized predominantly in the mitochondrial/lysosomal membrane fractions.
The biochemical characterization of the multiple forms of sialidase suggested that each might play a unique role depending on its particular subcellular localization and catalytic properties. Recent advances in the molecular cloning of sialidases have supported this hypothesis, and facilitated the elucidation of their functional roles and expression mechanisms. Three types of sialidases, localized predominantly in the lysosomes [7–13], cytosol [14–18] and plasma membranes [17,19–22] (designated as Neu1, Neu2 and Neu3 respectively), have been cloned and characterized. However, the gene for membrane sialidase II remains unidentified. Recently, a murine Neu4 cDNA and then the human orthologue [23–25] were predicted to encode a sialidase based on the enzyme activity in the transfected cells, and a recent report [25] described the localization of the human gene product to the lysosomes.
In order to identify and characterize further the human sialidase gene, the candidate cDNA was isolated and expressed in mammalian cells. We found that this gene indeed encoded a sialidase that possessed unique properties in terms of substrate specificity and subcellular localization, similar to those expected from our previous biochemical studies of the rat enzymes. In particular, we identified the existence of two isoforms of human NEU4 sialidase that are differently expressed in various human tissues, one of which is targeted to the mitochondria.
EXPERIMENTAL
Isolation and expression of human NEU4 cDNA
A search of the GenBank™ nucleotide sequence database was conducted for the NEU4 human sialidase gene, and two sequences (NM_080741 and AJ277883) predicted to be that of the NEU4 gene were used for isolation of the cDNA. These two sequences were identical, except for the presence of sequence encoding an additional 12 amino acid residues at the N-terminus in the former. To isolate the cDNA, first-strand cDNAs were synthesized from the poly(A)+ RNA obtained from human brain (BD Biosciences Clontech, Palo Alto, CA, U.S.A.) using random primers and the murine leukaemia virus RT (reverse transcriptase) (Superscript™II RT; Invitrogen), and then applied as templates for PCR. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 63 °C for 1 min, and elongation at 72 °C for 2 min, using LA Taq DNA polymerase (Takara, Otsu, Japan). To cover the entire coding sequence, the cDNA of human brain was amplified with two primer pairs with EcoRI sites [5′-AGGCAGCCACCCATGATGAGC-3′ (forward-L), 5′-AGCATGGGGGTCCCTCGTACC-3′ (forward-S) and 5′-AGGCCTGTCAGGAGGGCCAGC-3′ (reverse)], and the products were subcloned into pBluescript, sequenced, and cloned into the pCAGGS expression vector with the chicken β-actin promoter and into the pME18 expression vector with the SRα promoter, which were generously provided by Dr Jun-ichi Miyazaki (Osaka University School of Medicine, Osaka, Japan) and Dr Kazuo Maruyama (Tokyo Medical and Dental University, Tokyo, Japan) respectively. The sequence was also amplified with a HA (haemagglutinin) epitope at the C-terminus by PCR using the cDNA in pBluescript described above as the template, and subcloned into the expression vectors.
To construct an expression vector for EGFP (enhanced green fluorescent protein) fused to the N-terminal sequence of the long form of NEU4, sequence encoding amino acids 1–33 was obtained by PCR with primers 5′-CGGAATTCGATATCGCCACCATGATGAGCTCTGCAGCC-3′ (initiation codon underlined) and 5′-GCGGATCCGCGGTCAGGCCCGTCCTCT-3′ using the long-form cDNA in pBluescript as a template, followed by digestion with EcoRI and BamHI. This fragment was ligated with pEGFP-N1 (BD Biosciences Clontech) cut with EcoRI and BamHI, and the sequence was confirmed by sequencing. A point mutation (R17A) was introduced into the long-form NEU4 cDNA by the PCR-based method of Imai et al. [26] using primers 5′-TGGGGGTCCCTGCTACCCCTTCACGGACA-3′ (mutated codon underlined) and 5′-TGCTCAGCCACCTTGGGAAG-3′, and the HA-tagged plasmid as a template. After confirmation of the DNA sequence, the mutated cDNA fragment was excised by EcoRI digestion and inserted in the EcoRI site of the pCAGGS expression vector.
The expression plasmids were transiently transfected using the Effectene (Qiagen) reagent into COS-1 cells (RIKEN Cell Bank, Tsukuba, Japan), HEK-293T cells (a gift from M. Sugai, Kyoto University School of Medicine, Kyoto, Japan) and various human cell lines grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum in 5% CO2. The human cells tested were HCT 116 (A.T.C.C., Manassas, VA, U.S.A.), DLD-1 colon carcinoma cells (HSRRB, Osaka, Japan), A431 epidermoid carcinoma cells (The Cell Bank of Tohoku University, Sendai, Japan), A172 glioblastoma cells (HSRRB) and WI-38 fibroblast cells (HSRRB). After 24–48 h, the cells were harvested for characterization. For the semi-quantitative determination of the transcripts for the two forms, first-strand cDNAs were prepared from human tissue total RNAs (BD Biosciences Clontech) with oligo(dT) primers and used as templates for PCR as described above.
Sialidase activity assay
Enzymatic studies were carried out using COS-1 cells and HEK-293 cells transiently transfected with the expression plasmid. The cells were sonicated in 9 vol. of PBS, pH 7.4, containing 1 mM EDTA, 0.5 mM PMSF, 10 μg/ml leupeptin and 0.5 μg/ml pepstatin, and centrifuged at 1000 g for 10 min. The supernatant (crude extract) was used for sialidase assays as described elsewhere [6,22]. Briefly, the routine reaction mixture contained 10–20 nmol of substrate as bound sialic acid, 0.2 mg of BSA, 10 μmol of sodium acetate (pH 4.6) and 0.2 mg of Triton X-100. After incubation at 37 °C for 10–30 min, the amount of sialic acid released was determined by a modified thiobarbituric acid method [22] or by fluorimetric HPLC with 1,2-diamino-4,5-methylene dioxybenzene [27]. Sialidase activity with 4MU-NeuAc (4-methylumbelliferyl neuraminic acid) as substrate was assayed by spectrofluorimetric measurement of the 4-methylumbelliferone released [22]. To observe the pH profile for the activity, Hepes, Mes and citrate phosphate buffers were used. Protein was determined by the dye-binding assay (Bio-Rad). One unit of sialidase was defined as the amount of enzyme catalysing the release of 1 nmol of sialic acid/h.
To confirm that the sialidase activity was derived from the expressed protein, the expression plasmid encoding HA-tagged NEU4 was transiently transfected into cells and the cell homogenates were incubated with anti-HA–agarose (Sigma) or BSA–agarose (Sigma) beads. The sialidase activity and the expressed protein were then determined in fractions adsorbed or not on to the beads by spectrofluorimetric measurement with 4MU-NeuAc as substrate and by immunoblotting with anti-HA antibody (Roche) respectively.
Subcellular localization of exogenously expressed NEU4
For immunofluorescence staining, transfected cells expressing the HA-epitope-tagged sialidase were fixed with 4% (w/v) paraformaldehyde for 10 min, permeabilized with 0.2% (w/v) Triton X-100 in PBS for 2 min, and immunostained with anti-HA monoclonal antibody, followed by anti-(rat IgG) fluorescent conjugates (Alexa 488; Molecular Probes). Cells were also stained with MitoTracker Red (Molecular Probes) or pDsRed2-Mito (BD Biosciences Clontech) for identification of the mitochondria; Lyso Tracker (Molecular Probes) and antibody against human lamp-1 (lysosome-associated membrane protein-1) (BD Biosciences Clontech) were used to identify lysosomes. Anti-(human golgin-97) antibody (Molecular Probes) and anti-calnexin antibody (BD Biosciences Clontech) were used as markers for the Golgi and endoplasmic reticulum respectively. Fluorescent images were analysed with a Zeiss LSM 5 PASCAL confocal microscope.
To support the immunofluorescence data, biochemical fractionation of crude extracts from transfected DLD-1 cells was performed at 4 °C. The cells (0.5×106) were homogenized in 9 vol. of 5 mM Hepes (pH 7.2), 1 mM EGTA, 210 mM mannitol, 70 mM sucrose and Protease Inhibitor Cocktail (Roche) using a Dounce homogenizer, and then centrifuged at 600 g for 5 min. The supernatant (crude extract) was centrifuged at 12000 g for 10 min and the pellet was suspended in the above buffer (mitochondrial/lysosomal fraction). The resulting supernatant was centrifuged further at 100000 g for 1 h and the pellet was suspended in buffer (microsomal fraction), while the supernatant was used as the cytosolic fraction. To obtain purified mitochondria, the mitochondrial/lysosomal fraction was subjected to Percoll centrifugation essentially as described in [28]. Briefly, the fraction was resuspended in 2 ml of the homogenization buffer supplemented with 8 ml of 30% Percoll in 225 mM mannitol, 1 mM EGTA and 25 mM Hepes, and centrifuged at 100000 g in a Beckman 70.1Ti rotor for 30 min. The fraction with a density of 1.052–1.075 g/ml was collected and washed twice with homogenization buffer at 6300 g for 30 min to remove the Percoll.
Submitochondrial localization was determined further using subfractions obtained from mitochondria purified as above, which were separated into the inner membrane plus matrix and the outer membrane by digitonin treatment (2 mg/10 mg of protein), and then into separate inner membrane and matrix subfractions by osmotic shock by the method of Schnaitman and Greenawalt [29]. The following enzyme markers were assayed: acid phosphatase [30] and β-galactosidase [30] (lysosomes), glucose-6-phosphatase [31] (microsomes), succinate dehydrogenase [32] (mitochondrial inner membranes), malate dehydrogenase [29] (mitochondrial matrices) and monoamine oxidase [29] (mitochondrial outer membranes).
Cell preparation and immunolabelling for electron microscopy
To confirm the biochemical data, we observed DLD-1 cells transfected with the long form of NEU4 by electron microscopy. Cultured monolayer cells were fixed with phosphate (0.1 M)-buffered fixative (mixture of 4% paraformaldehyde and 0.04% glutaraldehyde) in the dish for 15 min at room temperature. Then the cells were scraped off the dish, put into a test tube with the fixative, and centrifuged at 1000 g at 4 °C. The pellet continued to be fixed with the same fixative for up to 1 h at approx. 10 °C. It was then washed thoroughly with Dulbecco's PBS to remove the fixative, dehydrated with a graded series of ethanol, and embedded in LR White resin, hard grade (London Resin Co. Ltd). For immunolabelling, ultrathin resin sections of the cell pellet were mounted on nickel grids with the thin support film of Butvar B-98 (TAAB Laboratory Equipment Ltd), and incubated with 10% (v/v) non-immune goat serum for 20 min, followed by 1 μg/ml anti-HA antibody overnight at 4 °C in a moisture chamber. After thorough washing with Tris/HCl buffer (pH 8.4) containing 2.5% NaCl, 0.1% BSA and 0.2% Tween 20, the sections were incubated with colloidal gold (10 nm)-conjugated goat anti-(rat IgG) (BioCell International) as the second antibody for 2 h at room temperature. The antibodies and goat serum were diluted with Tris/HCl buffer (pH 8.4) containing 2.5% NaCl and 0.1% BSA. After repeated washing with distilled water and air-drying, the cells were exposed to osmium tetroxide for 30 min in a small chamber to improve the contrast of the micrograph images. They were then counterstained with uranyl acetate and Reynold's lead citrate solution, and examined in an H-9000 transmission electron microscope (Hitachi). As a control, non-immune rat IgG1 (1 μg/ml) was used instead of the first antibody.
Northern blotting
Multiple-tissue Northern blots of poly(A)+ RNAs were purchased from Clontech Laboratories, hybridized with the 32P-labelled NEU4 cDNA probe containing the entire open reading frame in a solution containing 5×SSPE, 5×Denhardt's solution, 0.5% SDS, 50% (v/v) formamide and 50 μg/ml salmon sperm DNA, and then washed in 2×SSC/0.1% SDS followed by 0.2×SSC/0.1% SDS at 42 °C.
Quantification of NEU4 transcripts and comparison with levels of other sialidases
Quantitative analysis was performed by real-time PCR using a LightCycler rapid thermal cycler system (Roche). Total RNAs from human tissues were purchased from Clontech. First-strand cDNAs were synthesized using oligo(dT) 12–18 primers and murine leukaemia virus RT (Superscript™II RT; Invitrogen). A standard curve for NEU4 cDNA was generated by serial dilution of the pME18S vector containing the gene encoding the entire open reading frame. For the NEU1 [8] and NEU3 [20] cDNAs, pME18S vectors containing the respective genes covering the entire coding sequences were employed. For real-time PCR, the sequence-specific primers for NEU4 were 5′-CCGTCTTCCTCTTCTTCATCGC-3′ (forward) and 5′-CATTGCAGTAGAGGAAGCTGCC-3′ (reverse), which yielded a 411 bp fragment. The primers for NEU1 were 5′-TGAGAACGACTTCGGTCTGGTG-3′ (forward) and 5′-CCAGGAAACACCATCATCCTTG-3′ (reverse), which yielded a 403 bp fragment; those for NEU3 were 5′-GACTGGTCATCCCTGCGTAT-3′ (forward) and 5′-GAGCCATGATTCTGACGGTGTT-3′ (reverse), yielding a 469 bp fragment; and those for NEU2 were 5′-TGAGCACGCAGAGCTGATTGTC-3′ (forward) and 5′-CACTGCAAAGGTGGACCACTC-3′ (reverse), yielding a 340 bp fragment. For a long form of NEU4, the primers used were 5′-CCACCCATGATGAGCTCTGCAG-3′ (forward) and 5′-GCGATGAAGAAGAGGAAGACGG-3′ (reverse). To correct for differences in RNA quality and quantity between samples, a housekeeping isoform of PBGD (porphobilinogen deaminase) [33] was used. The primers used to amplify the reference gene were 5′-ACTTTCCAAGCGGAGCCATGTC-3′ (forward) and 5′-TCATGAGGGTTTTCCCGCTTGC-3′ (reverse), yielding a 379 bp fragment. The PCR reaction was carried out in glass capillary reaction vessel (Roche) in 20 μl of a reaction mixture containing 0.5 μM primers, cDNA, and QuantiTect SYBR Green PCR master mix (Qiagen). Amplification of the cDNAs involved a 15 min denaturation step, followed by 45 cycles at 94 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. Fluorescence from SYBR Green bound to the PCR product was detected, and the specificity of the reactions was confirmed by melting curve analysis and, subsequently, by agarose gel electrophoresis.
RESULTS
Isolation and identification of human NEU4 cDNA
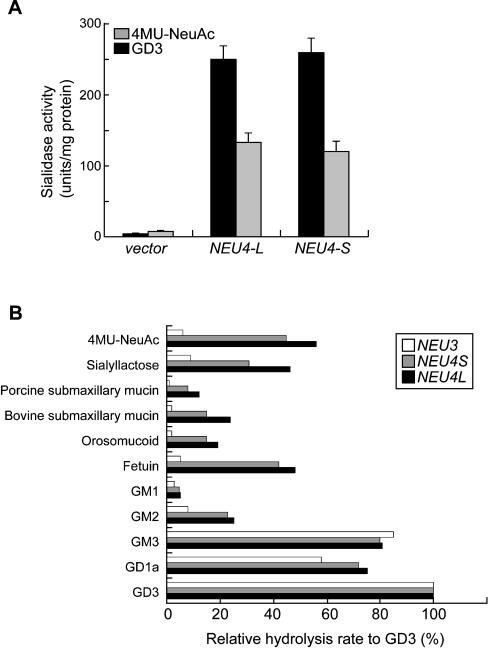
A search of the GenBank™ databases revealed the existence of six NEU4-related sequences (BC012899, AJ277883, AK091038, AK096992, NM_080741 and NT005416). They were completely identical in their overlapping regions, and the genes contained four exons and mapped to chromosome 2q37.3. Based on these sequences, we isolated two cDNAs from the human brain by PCR. As shown in Figure 1, the two differed only in their length at the N-terminus. The long form encoded an additional 12 amino acid residues at the N-terminus, which was predicted to be a mitochondrial targeting sequence by the MitoProt II, PSORT II server and TargetP v 1.0 prediction programs [34–36]; the short form did not have this sequence. It was also predicted that the long form can be cleaved at the 19th amino acid from the N-terminus [34] (Figure 1), although at present we do not have evidence that the targeting sequence of the long form is processed on import. One of the characteristic features of the NEU4 cDNA sequence is the very high content of GC (70%). The deduced protein for the long form comprises 496 amino acids, with a molecular mass of 52937 Da, and the sequence includes an Arg-Ile(Val)-Pro sequence and three typical and two atypical Asp boxes [37], the consensus sequence for sialidases, but no potential N-glycosylation site. Comparison of the nucleotide sequence with those of other mammalian sialidases revealed significant identity: 40% with NEU3 [20], 35% with NEU2 [15] and 24% with NEU1 [8]. When the expression vectors for the two forms were introduced into COS-1 or HEK-293T cells, both forms exhibited marked sialidase activity against 4MU-NeuAc, and interestingly also against gangliosides, as shown in Figure 2(A), indicating that the cDNAs encode a sialidase.
Figure 1. Deduced amino acid sequence of the human NEU4 sialidase.
The typical Asp boxes are boxed, and atypical ones are indicated by a broken line. The YRIP motif is double underlined. The putative initiating methionines for the two forms are in shaded boxes. This sequence is based on two sequences (NM_080741 and AJ277883) in the GenBank™ nucleotide sequence databases. The asterisk indicates a potential cleavage site.
Figure 2. Expression of the NEU4 sialidase in COS-1 cells.
(A) Sialidase activity in cells transfected with the expression vector was assayed using 4MU-NeuAc or gangliosides as substrates. The values are means±S.D. for four independent experiments. NEU4L and NEU4S indicate the long and short forms respectively of NEU4. (B) Substrate specificity of the sialidase expressed in COS-1 cells. Sialidase activities toward various sialic acid-containing glycoconjugates were examined in the cell homogenates and expressed as the percentage sialic acid release relative to the desialylation of ganglioside GD3.
Characterization of the enzymatic properties of NEU4 sialidase
Although the two forms of NEU4 described above did not differ significantly from each other in terms of their enzymatic properties, comparisons with other human sialidases revealed marked differences in substrate specificity. Over 70% of the activity of the expressed sialidase in the crude extract was recovered in the particulate fraction, with less than 20% of the activity remaining in the cytosolic fraction, suggesting that the enzyme is probably almost entirely membrane-bound. Testing with several buffers, including Hepes, Mes and citrate phosphate buffers, using gangliosides as substrate showed that the human enzyme had two peaks in its pH curve at pH 4.5–4.8 and at pH 6.0–7.0, the former giving maximum activity and the latter representing a small peak with considerable activity (55–60% of the maximum). As shown in Figure 2(B), the sialidase acted efficiently on glycoproteins and oligosaccharides as well as on 4MU-NeuAc and gangliosides, unlike the NEU1 and NEU3 sialidases. Both isoforms of NEU4 revealed this broad substrate specificity, in that all substrates tested except ganglioside GM1 were hydrolysed effectively. In particular, submaxillary mucins were cleaved to a significant degree, which seems to be a unique characteristic of this sialidase among the mammalian sialidases. The relative rates of hydrolysis of GD1a and GM3 compared with GD3 by the enzyme were similar to those of NEU3, which almost specifically hydrolyses gangliosides and scarcely showed any activity towards sialyl-lactose, fetuin or α1-acid glycoprotein. With gangliosides as the substrate, Triton X-100 stimulated the enzymatic activity by over 10-fold at optimal concentrations (0.05–0.1%); sodium cholate gave much less stimulation. The Km value for GM3 in the presence of Triton X-100 (17.2 μM) was comparable with that of NEU3 (23.3 μM).
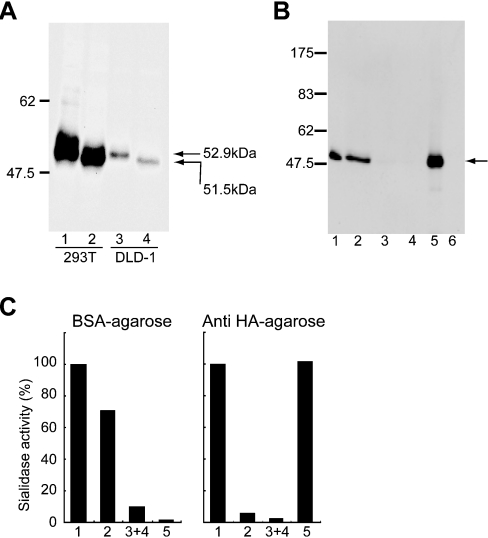
Both forms of the expressed HA-tagged sialidase were analysed by immunoblotting with anti-HA antibody (Figure 3A). Protein bands with molecular masses of 53 kDa and 51.5 kDa were revealed, in accordance with their respective sequences. To verify further that the activity was derived from the expressed proteins, homogenates of the transfected cells were subjected to anti-HA–agarose chromatography. The expressed protein of 53 kDa (the long form) was found mainly in the fraction bound to the anti-HA–agarose beads (Figure 3B), accompanied by the sialidase activity (Figure 3C), while the activity mostly did not bind to the beads on BSA–agarose chromatography. These data indicate that the expressed protein encoded by the cDNA directly catalyses the sialidase reaction. Experiments using the expression plasmid for the short form gave similar results.
Figure 3. Association of sialidase activity with the expressed NEU4 protein.
(A) Immunoblotting of the two forms of the HA-tagged NEU4 protein. The expression plasmid for the long or short form was transiently transfected into HEK-293T and DLD-1 cells, and the homogenates were subjected to SDS/8%-PAGE and immunoblotted with anti-HA antibody. Bands of 53 kDa and 51.5 kDa were obtained for the long and short forms respectively. (B) Enrichment of HA-tagged NEU4 protein after anti-HA–agarose affinity chromatography. The homogenates (fraction 1) of cells transfected with the expression plasmid for HA-tagged NEU4 were incubated with anti-HA–agarose, and the fractions that did not bind (fraction 2) or did bind (fraction 5) to the beads and the washings (fractions 3 and 4) were examined by immunoblotting. Numbers to the left in (A) and (B) denote the positions of molecular-mass markers (kDa). (C) Percentage recovery of sialidase activity in the fractions treated as above with anti-HA–agarose or BSA–agarose beads. Sialidase activity was estimated with 4MU-NeuAc as substrate.
Subcellular localization of NEU4 sialidase
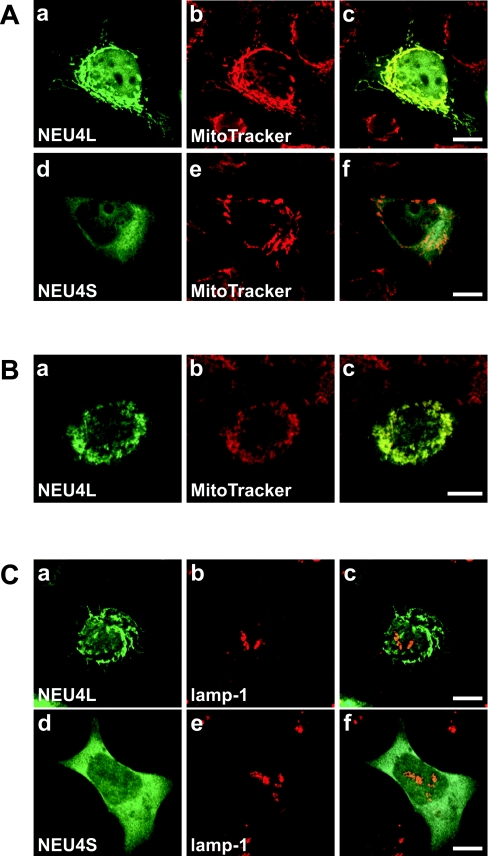
HA-tagged sialidase exogenously expressed in COS-1 and several human cells was examined by immunofluorescence staining using anti-HA antibody. COS-1 cells transfected with either NEU4 form exhibited a diffuse distribution in the intracellular membranes, although staining was coupled with lysosomal markers (Lyso Tracker and human lamp-1) only in cells with extremely high expression of exogenous NEU4. A lysosomal localization in the latter case was also observed in COS-7 cells. Unlike these cells, the human cells showed a mitochondrial localization pattern for the long form of NEU4. As shown in Figure 4(A), co-localization with MitoTracker was observed in DLD-1 cells expressing the long form, whereas the same cells transfected with the short form showed a diffuse distribution. Similar results were obtained with A431 cells (Figure 4B) and other types of human cells, including HCT 116 and A172 cells. However, immunofluorescence with anti-(human lamp-1) failed to show co-localization of any of these forms with lamp-1 (Figure 4C). Staining was not coupled with endoplasmic reticulum or Golgi markers (anticalnexin and anti-human golgin-97 antibodies respectively; results not shown). These results indicate that the long form of NEU4 is apparently localized, as predicted by the presence of the N-terminal 12 amino acids, only to the mitochondria in human cells. The short form, lacking this sequence, is not localized in mitochondria, but rather is found on intracellular membranes, as it was biochemically membrane-bound.
Figure 4. Co-localization of the exogenously expressed NEU4 long form with mitochondria.
(A) Subcellular localization of the HA-tagged long (a; NEU4L) and short (d; NEU4S) forms of NEU4 expressed in human DLD-1 cells. Localization was determined by immunofluorescence staining using anti-HA antibody. Mitochondria in the same cell as in (a) and (d) were identified by MitoTracker staining (b and e respectively). The merged images are shown in (c) and (f), demonstrating apparent localization of the long form in mitochondria. (B) Mitochondrial localization of the exogenous HA-tagged long form of NEU4 in A431 cells. The merged image (c) demonstrates co-localization with MitoTracker. (C) Lack of co-localization of exogenous NEU4 with lysosomes. Using anti-lamp-1 antibody, neither of the two forms co-localized significantly with lamp-1 as a lysosomal marker. Bars=10 μm.
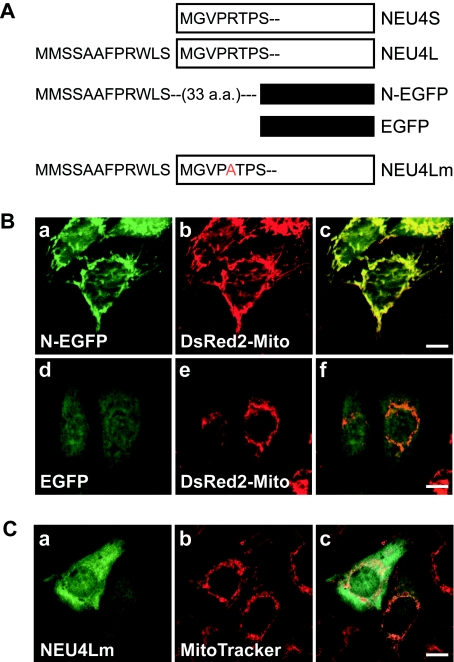
We then examined whether the N-terminal 12 amino acid residues in the long form can actually function as a potential mitochondrial targeting sequence, using the plasmid constracts shown in Figure 5(A). Fusion of EGFP to sequence encoding N-terminal amino acids 1–33 of the long form resulted in protein import into mitochondria, while EGFP lacking the sequence (Figure 5B), or fused to only amino acids 1–12 (results not shown), showed a diffuse distribution. Furthermore, substitution of arginine (at position 17) by alanine near the putative cleavage site at the long-form N-terminus prevented the mitochondrial targeting (Figure 5C), supporting evidence that the N-terminus of the long form of NEU4, covering the putative cleavage site, contains a mitochondrial targeting signal.
Figure 5. The N-terminus of the NEU4 long form is sufficient for mitochondrial targeting.
(A) Schematic representation of the long (NEU4L) and short (NEU4S) NEU4 isoforms, EGFP fused to the N-terminus of NEU4L (N-EGFP) and a NEU4L mutant (NEU4Lm), illustrating the unique N-termini containing a putative mitochondrial targeting sequence. These vectors were expressed in DLD-1 cells and processed for immunofluorescence staining. (B) EFGP fused to the N-terminus of the long form of NEU4 (N-EFGP) accumulated in mitochondria (a, c), whereas the control EGFP showed a diffuse pattern (d, f). In this experiment, DsRed2-Mito was used as a mitochondrial marker. (C) A mutant (NEU4Lm; mutation R17A) was not targeted to mitochondria (a, c).
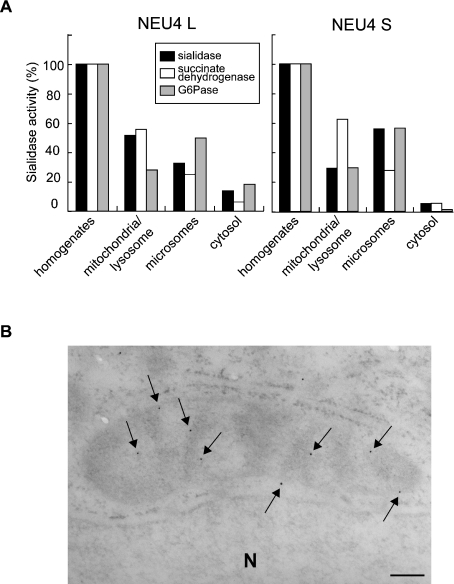
To ascertain whether the sialidase protein encoded by the long-form cDNA is localized in the mitochondria as a functionally active protein, homogenates of transfected DLD-1 cells were fractionated by differential centrifugation and assayed for sialidase activity (Figure 6). Over 50% of the activity for the long form was recovered in the mitochondrial/lysosomal fractions, where succinate dehydrogenase (a mitochondrial marker) was concentrated. In contrast, the major activity of the short form was detected in the microsomal fraction, along with a high recovery of glucose-6-phosphatase activity (a microsomal marker). To obtain further evidence for this localization, mitochondria were purified from the mitochondrial/lysosomal fraction by Percoll density centrifugation, and the sialidase activity in the purified mitochondrial fraction was determined (Table 1, upper part). This procedure allowed the efficient removal of contaminating organelles such as lysosomes. Sialidase activity was recovered in the mitochondria much more abundantly than were lysosomal marker enzymes, although recovery was not as high as that of succinate dehydrogenase. These results obtained by biochemical fractionation are consistent with the immunofluorescence data, indicating that the long form of NEU4 is localized to mitochondria. On the other hand, the short form did not co-localize with any of the marker proteins tested, including an endoplasmic reticulum protein, calnexin.
Figure 6. Subcellular distribution of NEU4 exogenously expressed in DLD-1 cells.
(A) DLD-1 cells were transfected with an expression plasmid containing the long (NEU4L) or the short (NEU4S) form of NEU4. Subcellular fractionation was carried out as described in the Experimental section. Using GM3 as substrate, sialidase activities were 29.5 and 24.8 units/mg of protein after transfection, with NEU4L and NEU4S respectively; the value after mock transfection was 6.1 units/mg of protein. (B) Electron microscopy of mitochondria in the transfected DLD-1 cells. Immunogold particles (arrows) were located along and/or in mitochondrial inner and outer membranes. Moderately electron-dense portions show the tangential or oblique section of the inner and outer membranes in the mitochondrion. Less electron-dense portions show mitochondrial matrices. Bar=200 nm. N, nucleus.
Table 1. Mitochondrial concentration of sialidase activity in colon cancer DLD-1 cells transfected with the NEU4 long form.
Mitochondria were purified from NEU4L-transfected DLD-1 cells by Percoll density gradient centrifugation. They were fractionated into outer membrane and inner membrane plus matrix with digitonin treatment, and then the matrix was separated from the inner membrane by osmotic shock, as described in the Experimental section. Each fraction was assayed for sialidase activity using ganglioside GM3 as substrate.
| Recovery (%) | ||||||
|---|---|---|---|---|---|---|
| Sialidase | Acid phosphatase | β-Galactosidase | Succinate dehydrogenase | Monoamine oxidase | Malate dehydrogenase | |
| Fractions | ||||||
| Post-nuclear supernatant | 100 | 100 | 100 | 100 | ||
| 12000 g supernatant | 47.5 | 60.9 | 82.1 | 22.5 | ||
| 12000 g pellet | 41.6 | 32.7 | 23.2 | 53.0 | ||
| Purified mitochondria | 28.5 | 12.3 | 9.1 | 49.4 | ||
| Submitochondrial fractions | ||||||
| Outer membrane | 72.0 | 39.3 | 81.8 | 34.5 | ||
| Inner membrane+matrix | 28.0 | 60.1 | 18.2 | 64.5 | ||
| Inner membrane | 23.0 | 58.6 | 19.0 | |||
| Matrix | 3.2 | 0.2 | 43.3 | |||
To determine the submitochondrial localization of the long form, purified mitochondria were separated into three fractions. Digitonin treatment separated the inner membrane plus matrix from the outer membrane, and then the matrix was separated from the inner membrane by osmotic shock. Sialidase activity was detected primarily in the inner and outer membrane fractions, but to a lesser extent in the matrix (Table 1, lower part). These biochemical data were supported by electron microscopy showing that immunogold particles (arrows) were located along and/or in the mitochondrial inner and outer membranes (Figure 6B).
Estimation of the level of expression of the NEU4 gene and comparison with that of other human sialidases
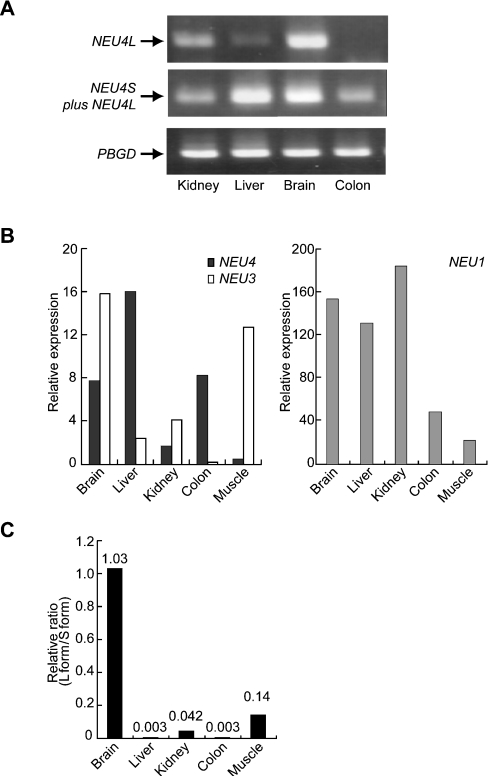
The relative abundance of mRNAs in human tissues was determined by Northern blotting. The human NEU4 sialidase gene was expressed predominantly in the liver, and at relatively low levels in the kidney, heart and brain; an approx. 2.2 kb transcript was the major form in these tissues (results not shown). To determine whether and how the two forms are actually expressed, several human tissues were examined by RT-PCR using forward primers corresponding to their N-terminal sequences and a common reverse primer. The brain and kidney yielded two cDNA fragments (496 bp and 484 bp) using primers for the long and short forms respectively, and the fragments were identified as the two forms of the NEU4 gene by sequencing. The liver expressed the short form predominantly, and the the long form was barely detectable in the colon. The results from representative PCR reactions are shown in Figure 7(A). We then quantified total NEU4 transcripts in these tissues by quantitative real-time PCR. In order to obtain relative values for gene expression, PCR reactions were carried out for PBGD as a reference gene [38]. To compare relative expression among tissues, the data for the NEU4 gene were then normalized to the values obtained for PBGD. For further understanding of the expression level of NEU4 compared with those of other human sialidase genes, quantification of the sialidases was performed based on standard curves generated by serial dilution of the plasmid DNAs containing the respective sialidase genes. The quantities of the transcripts encoding NEU4 and other human sialidases were expressed as relative amounts normalized to the amount of the reference gene PBGD (Figure 7B). Analysis of mRNA levels showed that NEU4 mRNA was expressed in a tissue-specific manner, and was expressed at highest levels in the liver, as assessed by Northern blotting, and at relatively high levels in the brain and colon. When the expression of all sialidases was analysed comparatively, NEU4 appeared to be expressed at a lower level than NEU3 (Figure 7B, left panel), and at below one-tenth the level of NEU1 (Figure 7B, right panel). Although we also attempted to estimate the expression level of the NEU2 gene in these tissues, amplification of the partial sequence by PCR failed. Furthermore, we did not succeed in obtaining the entire open reading frame even with cDNA transcribed from poly(A)+ RNA obtained from skeletal muscle, which has been reported to be the predominant tissue for expression of this enzyme [15]. The relative expression levels of the two forms of NEU4 were then quantified (Figure 7C). The level of the long form was estimated with specific primers using the NEU4 plasmid for standard curve generation, and that of the short form was obtained by subtracting the value for the long form from the total NEU4 level. Consistent with the data shown in Figure 7(A), the long form/short form ratio was nearly 1 in brain, lower in kidney and muscle, and extremely low in liver and colon.
Figure 7. Differential expression of the NEU4 gene in human tissues.
(A) Expression of the two forms of NEU4 was determined by semi-quantitative RT-PCR using respective forward primers corresponding to their N-terminal sequences and a common reverse primer, as described in the Experimental section. Expression of PBGD was also measured as a reference gene. (B) Comparison with the expression levels of NEU1 and NEU3 in several human tissues assessed by quantitative real-time PCR. Quantities of the transcripts encoding NEU4 and other human sialidases were determined as described in the Experimental section and expressed as relative amounts normalized to that of the reference gene PBGD. (C) Relative levels of expression of the two forms of NEU4 as determined by quantitative real-time PCR. Relative expression of the long form compared with the short form is shown. The values are based on the assumption that the efficiency of PCR is comparable among these sialidase genes.
DISCUSSION
On the basis of sequence information for cDNA predicted as representing that of human NEU4 obtained from public databases, we isolated from human brain a cDNA identified as encoding a sialidase that was unique with regard to its substrate specificity and subcellular localization. The sialidase was found to effectively hydrolyse gangliosides, glycoproteins and oligosaccharides, as well as the synthetic substrate 4MU-NeuAc. This broad specificity differentiated it clearly from NEU1 and NEU3, because the rat orthologue Neu1, despite acting efficiently on glycopeptides and oligosaccharides, hydrolyses gangliosides to only a low extent [4,13], whereas NEU3 almost specifically hydrolyses gangliosides [20,21]. Although NEU2 [15] and its rat orthologue [14] have broad specificity, they are active only at near-neutral pH. In addition, interestingly, NEU4 cleaved a significant amount of sialic acids from mucins, which seems to be a property unique to NEU4. Our previous studies of rat sialidases demonstrated four types differing in their subcellular localization and substrate specificity [3,5,6]; subsequently, we isolated cDNAs corresponding to three of them, namely the rat orthologues Neu1 [13], Neu2 [14] and Neu3 [22], whereas the fourth gene, corresponding to the sialidase found in the lysosomal/mitochondrial membrane fractions, remained unisolated. The sialidase partially purified and characterized from rat brain as a putative lysosomal/membrane sialidase was found to possess substrate specificity identical with that of NEU4.
Comparison of NEU4 with the other sialidases in terms of their expression levels in various human tissues by quantitative RT-PCR revealed that NEU4 is expressed at a relatively low but significant level in tissues. Interestingly, the two forms of NEU4 are actually expressed in several tissues. In contrast with the short form, which is ubiquitously expressed, the long form is barely detectable in liver or colon, but is detected to the same extent as the short form in the brain, and at lower levels in kidney and muscle. On the other hand, NEU2 expression was not detectable at all in these tissues, including in skeletal muscle, which is supposed to be the predominant site of expression of this enzyme [15], indicating the extremely low levels, if any, in human tissues. It is also conceivable that the high content of GC (64%) in the nucleotide sequences may cause some difficulty in cDNA synthesis, or that transcription of the gene encoding the enzyme may have been down-regulated for some reason, because NEU2 cDNA covering the open reading frame was not derived from the cDNA transcribed from tissue RNA, but was constructed by the ligation of two genomic fragment sequences [15].
With regard to subcellular localization, it has been reported that the major localization sites of the three sialidases cloned previously are the lysosomes, cytosol and plasma membranes for NEU1, NEU2 and NEU3 respectively. Immunofluorescence studies with HA-tagged NEU4 revealed that the long form is localized in the mitochondria in several human cell types, including DLD-1 and A431 cells. Analyses using EGFP fused to the N-terminus of the long form and point mutation of the signal sequence provided evidence that the N-terminus contains a potential mitochondrial targeting sequence. In contrast, the ubiquitously expressed short form was not concentrated in any special organelle, such as lysosomes or endoplasmic reticulum, but showed a diffuse localization probably in the intracellular membranes, as the activity was assayed as being mainly membrane-bound. In agreement with the subcellular localization, NEU4 possesses considerable activity towards gangliosides even at near-neutral pH, as well as at acidic pH. Although it is not known why introduction into COS-1 cells of the gene encoding the long form resulted in a different localization pattern, it is possible that a peptidase responsible for cleavage of the N-terminal mitochondrial targeting sequence is overexpressed in these cells, leading to abrogation of the targeting. With regard to the short form of NEU4, our data are consistent with the observations by Monti et al. [24] that transfection of a cDNA corresponding to the short form resulted in a diffuse pattern in membranous networks inside the cells. Inconsistent with these findings is a report that exogenous NEU4 was localized in the lysosomal lumen in COS-7 cells and human fibroblasts [25], on utilizing a cDNA probably corresponding to the long form. We do not know at present why such a discrepancy exists. However, as described in the Results, some of our data did show fluorescence staining with HA-tagged NEU4 coupled to Lyso Tracker, but only when extremely high expression was obtained, possibly due to degradation of the abundantly expressed exogenous proteins. Whatever reasons may exist for the discrepancy, it is possible that the localization is not fixed and can be changed under different physiological conditions. Although previous studies on mammalian sialidases have revealed a close relationship between their subcellular localization and function, these enzymes may be expected to be targeted to other organelles in response to a variety of cellular events, as reported previously [39]. It is therefore important to observe the localization of NEU4 under various cellular conditions.
Although the function of the NEU4 sialidase is still unclear, its substrate specificity and intracellular localization described above suggest that it may be involved in the mitochondrial apoptosis pathway. Previous observations [40] indicate that mitochondrion is a key destination for the apoptogenic ganglioside GD3, as shown by the reports that apoptosis induced by ceramide exposure [41] and tumour necrosis factor-α [42] caused targeting of GD3 to the mitochondria. In line with these observations, NEU4 may regulate the level of mitochondrial GD3, the best substrate for this sialidase. If regions containing the N-terminal 12 amino acid residues can be cleaved physiologically from the long form of NEU4, this would be expected to lead to trafficking out of the mitochondria, followed by accumulation of GD3 and, subsequently, induction of apoptosis. Especially in the nervous system, where the long form is enriched, it may function as a mitochondrial targeting sialidase in apoptosis. This is different from the possible function of NEU3, which is thought to participate in cell signalling at the cell surface (e.g. insulin signalling). Determination of the physiological significance of the presence of the two forms of NEU4 and the molecular mechanism for the possible cleavage of the long form may facilitate a clearer understanding of the functions of this novel sialidase.
Acknowledgments
This study was supported in part by The Intelligent Cosmos Foundation, and by a Grant-in-Aid from The Japan Medical Association.
References
- 1.Saito M., Yu R. K. Biochemistry and function of sialidases. In: Rosenberg A., editor. Biology of the Sialic Acids. New York: Plenum; 1995. pp. 261–313. [Google Scholar]
- 2.Monti E., Preti A., Venerando B., Borsani G. Recent development in mammalian sialidase molecular biology. Neurochem. Res. 2002;27:649–663. doi: 10.1023/a:1020276000901. [DOI] [PubMed] [Google Scholar]
- 3.Miyagi T., Wada T., Yamaguchi K., Hata K. Sialidase and malignancy: a minireview. Glycoconj. J. 2004;20:189–198. doi: 10.1023/B:GLYC.0000024250.48506.bf. [DOI] [PubMed] [Google Scholar]
- 4.Miyagi T., Tsuiki S. Rat-liver lysosomal sialidase. Solubilization, substrate specificity and comparison with the cytosolic sialidase. Eur. J. Biochem. 1984;141:75–81. doi: 10.1111/j.1432-1033.1984.tb08159.x. [DOI] [PubMed] [Google Scholar]
- 5.Miyagi T., Tsuiki S. Purification and characterization of cytosolic sialidase from rat liver. J. Biol. Chem. 1985;260:6710–6716. [PubMed] [Google Scholar]
- 6.Miyagi T., Sagawa J., Konno K., Handa S., Tsuiki S. Biochemical and immunological studies on two distinct ganglioside-hydrolyzing sialidases from the particulate fraction of rat brain. J. Biochem. (Tokyo) 1990;107:787–793. doi: 10.1093/oxfordjournals.jbchem.a123126. [DOI] [PubMed] [Google Scholar]
- 7.Bonten E., van der Spoel A., Fornerod M., Grosveld G., d'Azzo A. Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev. 1996;10:3156–3169. doi: 10.1101/gad.10.24.3156. [DOI] [PubMed] [Google Scholar]
- 8.Milner C. M., Smith S. V., Carrillo M. B., Taylor G. L., Hollinshead M., Campbell R. D. Identification of a sialidase encoded in the human major histocompatibility complex. J. Biol. Chem. 1997;272:4549–4558. doi: 10.1074/jbc.272.7.4549. [DOI] [PubMed] [Google Scholar]
- 9.Pshezhetsky A. V., Richard C., Michaud L., Igdoura S., Wang S., Elsliger M. A., Qu J., Leclerc D., Gravel R., Dallaire L., Potier M. Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat. Genet. 1997;15:316–320. doi: 10.1038/ng0397-316. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo M. B., Milner C. M., Ball S. T., Snoek M., Campbell R. D. Cloning and characterization of a sialidase from the murine histocompatibility-2 complex: low levels of mRNA and a single amino acid mutation are responsible for reduced sialidase activity in mice carrying the Neu1a allele. Glycobiology. 1997;7:975–986. doi: 10.1093/glycob/7.7.975. [DOI] [PubMed] [Google Scholar]
- 11.Igdoura S. A., Gafuik C., Mertineit C., Saberi F., Pshezhetsky A. V., Potier M., Trasler J. M., Gravel R. A. Cloning of the cDNA and gene encoding mouse lysosomal sialidase and correction of sialidase deficiency in human sialidosis and mouse SM/J fibroblasts. Hum. Mol. Genet. 1998;7:115–121. doi: 10.1093/hmg/7.1.115. [DOI] [PubMed] [Google Scholar]
- 12.Rottier R. J., Bonten E., d'Azzo A. A point mutation in the neu-1 locus causes the neuraminidase defect in the SM/J mouse. Hum. Mol. Genet. 1998;7:313–321. doi: 10.1093/hmg/7.2.313. [DOI] [PubMed] [Google Scholar]
- 13.Kato T., Wang Y., Yamaguchi K., Milner C. M., Shineha R., Satomi S., Miyagi T. Overexpression of lysosomal-type sialidase leads to suppression of metastasis associated with reversion of malignant phenotype in murine B16 melanoma cells. Int. J. Cancer. 2001;92:797–804. doi: 10.1002/ijc.1268. [DOI] [PubMed] [Google Scholar]
- 14.Miyagi T., Konno K., Emori Y., Kawasaki H., Suzuki K., Yasui A., Tsuiki S. Molecular cloning and expression of cDNA encoding rat skeletal muscle cytosolic sialidase. J. Biol. Chem. 1993;268:26435–26440. [PubMed] [Google Scholar]
- 15.Monti E., Preti A., Rossi E., Ballabio A., Borsani G. Cloning and characterization of NEU2, a human gene homologous to rodent soluble sialidases. Genomics. 1999;57:137–143. doi: 10.1006/geno.1999.5749. [DOI] [PubMed] [Google Scholar]
- 16.Fronda C. L., Zeng G., Gao L., Yu R. K. Molecular cloning and expression of mouse brain sialidase. Biochem. Biophys. Res. Commun. 1999;258:727–731. doi: 10.1006/bbrc.1999.0698. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa T., Yamaguchi K., Wada T., Takeda A., Itoyama Y., Miyagi T. Molecular cloning of mouse ganglioside sialidase and its increased expression in Neuro2a cell differentiation. J. Biol. Chem. 2000;275:8007–8015. doi: 10.1074/jbc.275.11.8007. [DOI] [PubMed] [Google Scholar]
- 18.Kotani K., Kuroiwa A., Saito T., Matsuda Y., Koda T., Kijimoto-Ochiai S. Cloning, chromosomal mapping, and characteristic 5′-UTR sequence of murine cytosolic sialidase. Biochem. Biophys. Res. Commun. 2001;286:250–258. doi: 10.1006/bbrc.2001.5374. [DOI] [PubMed] [Google Scholar]
- 19.Miyagi T., Wada T., Iwamatsu A., Hata K., Yoshikawa Y., Tokuyama S., Sawada M. Molecular cloning and characterization of a plasma membrane-associated sialidase specific for gangliosides. J. Biol. Chem. 1999;274:5004–5011. doi: 10.1074/jbc.274.8.5004. [DOI] [PubMed] [Google Scholar]
- 20.Wada T., Yoshikawa Y., Tokuyama S., Kuwabara M., Akita H., Miyagi T. Cloning, expression, and chromosomal mapping of a human ganglioside sialidase. Biochem. Biophys. Res. Commun. 1999;261:21–27. doi: 10.1006/bbrc.1999.0973. [DOI] [PubMed] [Google Scholar]
- 21.Monti E., Bassi M. T., Papini N., Riboni M., Manzoni M., Venerando B., Croci G., Preti A., Ballabio A., Tettamanti G., Borsani G. Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane. Biochem. J. 2000;349:343–351. doi: 10.1042/0264-6021:3490343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa T., Feijoo C. C., Wada T., Itoyama Y., Miyagi T. Differential expression of three sialidase genes in rat development. Biochem. Biophys. Res. Commun. 2001;280:726–732. doi: 10.1006/bbrc.2000.4186. [DOI] [PubMed] [Google Scholar]
- 23.Comelli E. M., Amado M., Lustig S. R., Paulson J. C. Identification and expression of Neu4, a novel murine sialidase. Gene. 2003;321:155–161. doi: 10.1016/j.gene.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Monti E., Bassi M. T., Bresciani R., Civini S., Croci G. L., Papini N., Riboni M., Zanchetti G., Ballabio A., Preti A., et al. Molecular cloning and characterization of NEU4, the fourth member of the human sialidase gene family. Genomics. 2004;83:445–453. doi: 10.1016/j.ygeno.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Seyrantepe V., Landry K., Trudel S., Hassan J. A., Morales C. R., Pshezhetsky A. V. Neu4, a novel human lysosomal lumen sialidase, confers normal phenotype to sialidosis and galactosialidosis cells. J. Biol. Chem. 2004;279:37021–37029. doi: 10.1074/jbc.M404531200. [DOI] [PubMed] [Google Scholar]
- 26.Imai Y., Matsushima Y., Sugimura T., Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara S., Yamaguchi M., Takemori Y., Nakamura M., Ohkura Y. Highly sensitive determination of N-acetyl- and N-glycolylneuraminic acids in human serum and urine and rat serum by reversed-phase liquid chromatography with fluorescence detection. J. Chromatogr. 1986;377:111–119. doi: 10.1016/s0378-4347(00)80766-5. [DOI] [PubMed] [Google Scholar]
- 28.Giulivi C., Poderoso J. J., Boveris A. Production of nitric oxide by mitochondria. J. Biol. Chem. 1998;273:11038–11043. doi: 10.1074/jbc.273.18.11038. [DOI] [PubMed] [Google Scholar]
- 29.Schnaitman C., Greenawalt J. W. Enzymatic properties of the inner and outer membranes of rat liver mitochondria. J. Cell Biol. 1968;38:158–175. doi: 10.1083/jcb.38.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyagi T., Sato K., Hata K., Taniguchi S. Metastatic potential of transformed rat 3Y1 cell lines is inversely correlated with lysosomal-type sialidase activity. FEBS Lett. 1994;349:255–259. doi: 10.1016/0014-5793(94)00682-2. [DOI] [PubMed] [Google Scholar]
- 31.Baginski E. S., Foa P. P., Zak B. Glucose-6-phosphatase. In: Bergmeyer H. U., editor. Methods of Enzymatic Analysis. New York: Academic Press, Inc.; 1974. pp. 875–880. [Google Scholar]
- 32.Pennington R. J. Biochemistry of dystrophic muscle. Mitochondrial succinate-tetrazolium reductase and adenosine triphosphatase. Biochem. J. 1961;80:649–654. doi: 10.1042/bj0800649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussey D. J., Moore S., Nicola M., Dobrovic A. Fusion of the NUP98 gene with the LEDGF/p52 gene defines a recurrent acute myeloid leukemia translocation. BMC Genet. 2001;2:20. doi: 10.1186/1471-2156-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claros M. G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakai K., Horton P. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 36.Emanuelsson O., Nielsen H., Brunak S., von Heijine G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 37.Roggentin P., Rothe B., Kaper J. B., Galen J., Lawrisuk L., Vimr E. R., Schauer R. Conserved sequences in bacterial and viral sialidases. Glycoconj. J. 1989;6:349–353. doi: 10.1007/BF01047853. [DOI] [PubMed] [Google Scholar]
- 38.Raich N., Romeo P. H., Dubart A., Beaupain D., Cohen-Solal M., Goossens M. Molecular cloning and complete primary sequence of human erythrocyte porphobilinogen deaminase. Nucleic Acids Res. 1986;14:5955–5968. doi: 10.1093/nar/14.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukong K. E., Seyrantepe V., Landry K., Trudel S., Ahmad A., Gahl W. A., Lefrancois S., Morales C. R., Pshezhetsky A. V. Intracellular distribution of lysosomal sialidase is controlled by the internalization signal in its cytoplasmic tail. J. Biol. Chem. 2001;276:46172–46181. doi: 10.1074/jbc.M104547200. [DOI] [PubMed] [Google Scholar]
- 40.Malisan F., Testi R. GD3 ganglioside and apoptosis. Biochim. Biophys. Acta. 2002;1585:179–187. doi: 10.1016/s1388-1981(02)00339-6. [DOI] [PubMed] [Google Scholar]
- 41.Rippo M. R., Malisan F., Ravagnan L., Tomassini B., Condo I., Costantini P., Susin S. A., Rufini A., Todaro M., Kroemer G., Testi R. GD3 ganglioside directly targets mitochondria in a bcl-2-controlled fashion. FASEB J. 2000;14:2047–2054. doi: 10.1096/fj.99-1028com. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Ruiz C., Colell A., Morales A., Calvo M., Enrich C., Fernandez-Checa J. C. Trafficking of ganglioside GD3 to mitochondria by tumor necrosis factor-α. J. Biol. Chem. 2002;277:36443–36448. doi: 10.1074/jbc.M206021200. [DOI] [PubMed] [Google Scholar]