Abstract
The cDNA encoding a xyloglucan endotransglycosylase, PttXET16A, from hybrid aspen (Populus tremula×tremuloides) has been isolated from an expressed sequence tag library and expressed in the methylotrophic yeast Pichia pastoris. Sequence analysis indicated a high degree of similarity with other proteins in the XTH (xyloglucan transglycosylase/hydrolase) gene subfamily of GH16 (glycoside hydrolase family 16). In addition to the conserved GH16 catalytic sequence motif, PttXET16A contains a conserved N-glycosylation site situated proximal to the predicted catalytic residues. MS analysis indicated that the recombinant PttXET16A expressed in P. pastoris is heterogeneous due to the presence of variable N-glycosylation and incomplete cleavage of the α-factor secretion signal peptide. Removal of the N-glycan by endoglycosidase H treatment did not influence the catalytic activity significantly. Similarly, site-directed mutagenesis of Asn93 to serine to remove the N-glycosylation site resulted in an enzyme which was comparable with the wild-type enzyme in specific activity and thermal stability but had clearly reduced solubility. Hydrolytic activity was detected neither in wild-type PttXET16A before or after enzymatic deglycosylation nor in PttXET16A N93S (Asn93→Ser) mutant.
Keywords: glycoside hydrolase family 16 (GH16), hybrid aspen (Populus tremula×tremuloides), Pichia pastoris, protein glycosylation, protein MS, xyloglucan endotransglycosylase (XET)
Abbreviations: BCA, disodium 2,2′-bicinchoninate; endo H, endoglycosidase H; ESI, electrospray ionization; EST, expressed sequence tag; GH16, glycoside hydrolase family 16; MaxEnt1, Maximum Entropy™ 1 algorithm; Ptt, Populus tremula×tremuloides; XET, xyloglucan endotransglycosylase; XGO, xyloglucan oligosaccharide; XLLGol, reduced xyloglucan-derived nonasaccharide
INTRODUCTION
Xyloglucan is the principal hemicellulose in the primary cell walls of many land plants. It is composed of a β(1→4)-linked glucan backbone extensively substituted with side chains composed of xylose, galactose and, in some cases, fucose residues. The detailed structures of xyloglucans vary depending on the species and tissue of origin [1,2]. XETs (xyloglucan endotransglycosylases; EC 2.4.1.207) are unique enzymes that perform an endolytic cleavage of a xyloglucan chain with subsequent transfer of the newly created chain end to the non-reducing end of a different xyloglucan molecule [3,4]. This activity seems to play a major role in the transient cell-wall loosening required for cell-wall expansion [5,6] and may also contribute to reinforcing the connections between primary and secondary cell walls in wood-forming tissues [7].
XETs typically belong to large gene families with over 30 genes identified in different plant species [8]. These enzymes belong to the GH16 family (glycoside hydrolase family 16), which contains a variety of endolytic glucanases and galactanases (Carbohydrate-Active enZYmes server, http://afmb.cnrs-mrs.fr/CAZY/). Most of the GH16 enzymes are hydrolytic, cleaving glycosidic bonds with net retention of the configuration of the anomeric carbon, through a covalent glycosyl-enzyme intermediate [9,10]. However, enzymes with XET activity generally do not accept water in the breakdown of the glycosyl-enzyme intermediate, but instead require xyloglucan or an XGO (xyloglucan oligosaccharide) as a transglycosylation acceptor [11]. Since hydrolytic activity towards xyloglucan (EC 3.2.1.151) has been detected with some isoenzymes [12–16], kinetic partitioning of the glycosyl-enzyme to water must nevertheless occur to some extent. To reflect the observed catalytic promiscuity, a unifying name XTH (xyloglucan transglycosylase/hydrolase) has been proposed for this gene family [16].
Similar to other plants, nearly 30 different genes in the XTH family have been identified in the hybrid aspen (Populus tremula×tremuloides) EST (expressed sequence tag) library (PopulusDB server, http://www.populus.db.umu.se/). Annotation of 3000 ESTs from the wood-forming tissues of hybrid aspen revealed the presence of three distinct XTH genes [17], all of which were highly expressed during cell expansion [18]. Our preliminary results indicate that a cDNA copy of one of these genes encodes an enzyme which only exhibits XET activity (EC 2.4.1.207) [7,19]. Following the general principles of enzyme nomenclature (http://www.chem.qmul.ac.uk/iubmb/enzyme/rules.html), and the recommended nomenclature for carbohydrate-active enzymes [20], the gene was designated PttXET16A (where Ptt stands for Populus tremula×tremuloides) [7]. The translated nucleotide sequence of the cDNA revealed a family GH16 enzyme with the conserved catalytic motif typical of enzymes in the XTH family in other plants. A single N-glycosylation site (underlined) is located immediately after the conserved catalytic glutamates (in boldface) in the sequence EIDFEFLGNRT of PttXET16A. The corresponding motif in native and recombinant cauliflower (Brassica oleracea var. botrytis) XET16A has been shown to carry high-mannose-type N-glycans [21]. Enzymatic removal of the glycan from three different XTH gene products from Arabidopsis thaliana and cauliflower has been shown to result in no, partial or total loss of activity [21–23]. However, it is at present not clear whether the inactivation of some XETs upon deglycosylation is caused by protein unfolding or whether the glycan has a more direct role in the catalytic mechanism of XTH gene products [23].
Owing to the large number of XTH gene products present in plant tissues, isolation of absolutely pure enzyme in sufficient amounts for biochemical characterization is a considerable challenge. Heterologous expression of XTH genes in Escherichia coli has only yielded inactive protein [14,24], whereas active enzyme has been obtained by heterologous expression of a limited number of XTH genes from Arabidopsis, cauliflower, soya bean and tomato in insect cells [22,23,25] and in Pichia pastoris [21,26]. Thus, in spite of their apparent importance for plant development, only a few XTH gene products have been thoroughly characterized regarding their enzymatic properties. In the present study, we describe efficient heterologous expression of the PttXET16A cDNA in the methylotrophic yeast P. pastoris, as well as the in vitro characterization of the purified enzyme. We also describe the expression and enzymatic properties of a PttXET16A glycosylation site mutant, PttXET16A N93S (Asn93→Ser), and compare its properties with the deglycosylated wild-type enzyme. Finally, we present a hypothesis on the role of protein glycosylation, based on the three-dimensional structure of PttXET16A [19] and the biochemical data obtained with different XET isoenzymes.
MATERIALS AND METHODS
General
Ultrapure water was used in all experiments and refers to water purified on a Milli-Q® system (Millipore, Eschborn, Germany) with a resistivity ρ≥18.2 MΩ·cm. Proteins were analysed by SDS/PAGE using an Xcell II™ Mini Cell (Novex, San Diego, CA, U.S.A.). Precast NuPAGE gradient gels (4–12%) were purchased from Invitrogen. Molecular mass standards (LMW Calibration kit) were obtained from Amersham Biosciences. Protein gels were stained by Coomassie Brilliant Blue or silver staining [27]. Protein concentrations were determined spectrophotometrically by the Bradford method [28] using BSA as a standard. DNA sequencing was performed by the Sanger method using an ABI prism 377 DNA sequencer (PerkinElmer, Wellesley, MA, U.S.A.).
Enzyme substrates and activity assays
A mixture of XGOs was prepared by digestion of tamarind seed xyloglucan (Megazyme, Bray, Ireland) with Trichoderma reesei cellulase (Fluka, Gillingham, Dorset, U.K.) as described previously [21].
A colorimetric assay, based on that developed by Sulová et al. [29], was used for routine measurements of XET activity; 50 μl of enzyme was added to a solution composed of 50 μl of xyloglucan (2 mg/ml in water), 50 μl of XGOs (3.5 mg/ml in water) and 50 μl of 400 mM sodium citrate buffer [citric acid (final concentration 400 mM) titrated with NaOH to pH 5.5], followed by incubation at 30 °C (for 30 min unless otherwise stated). The reactions were terminated by adding 100 μl of 1 M HCl. Immediately afterwards, 20% (w/v) Na2SO4 (800 μl) and potassium tri-iodide reagent (1% KI and 0.5% I2 in water; 200 μl) were added and the samples were kept in the dark for 30 min to allow colour development. Each sample was compared with one in which water replaced the XGO solution and enzyme activity is expressed as the difference in absorbance A620/min between samples with and without XGOs [ΔA620/min=(A620+XGO/min)−(A620−XGO/min)]; 1 unit of enzyme activity is defined as ΔA620/min. Controls without enzyme were used to confirm that the XGO mixture did not affect the absorbance. Significant endo-xyloglucanase (EC 3.2.1.151) activity was never observed, even in crude P. pastoris culture filtrates, so assay reactions to account for such activity were omitted from previous methods [21,29].
Fry's radioactive XET assay [3] was performed by the method previously adapted [21]. As before, a stock solution in 100 mM sodium citrate (pH 5.5) of 2.2 mM [1-3H]XLLGol (reduced xyloglucan-derived nonasaccharide) with a specific radioactivity of 3.7 TBq/mol was used. Values obtained by scintillation counting in counts/min were converted into molar activity values using a counting efficiency [30,31] for tritiated xyloglucan of 44% [3]; 1 unit of enzyme activity is defined as 1 nmol of XLLGol incorporated into xyloglucan/min [21]. The apparent Km value for the donor substrate was determined by varying the xyloglucan concentration between 0.3 and 5.0 g/l in 100 mM sodium citrate buffer (pH 5.5), whereas the [1-3H]XLLGol concentration was kept at 0.72 mM. To obtain the Km value for the acceptor substrate XLLGol, XET assays were performed in a reaction mixture containing 5.0 g/l xyloglucan, 83.5 kBq [1-3H]XLLGol (165 TBq/mol) and various concentrations (between 7 and 700 μM) of non-radioactive XLLGol in 100 mM sodium citrate buffer (pH 5.5). All experiments were performed at 25 °C, in duplicate, and Km values were determined by means of nonlinear regression analysis.
The BCA (disodium 2,2′-bicinchoninate) reducing sugar assay [32] was used to measure endo-xyloglucanase (EC 3.2.1.151) activity as described previously [21] versus glucose as a standard. Recombinant PttXET16A (0.09 g/l) was incubated with or without xyloglucan (1 g/l) in 0.1 M sodium acetate (pH 5.5) at 30 °C (1.2 ml). At time intervals 0, 30, 60, 120 and 240 min, 50 μl samples were withdrawn and diluted with ultrapure water (450 μl). After the addition of BCA reagent (500 μl), the sample was heated at 80 °C for 30 min, immediately cooled on ice and A560 was measured. Glucose was used to generate a seven-point standard curve over the range 0–50 μM (0–25 nmol). All measurements were performed in duplicate. The glycosylation mutant, PttXET16A N93S, was assayed similarly after an overnight incubation at 25 °C.
Recombinant expression of PttXET16A
The cDNA corresponding to the hybrid aspen XET16A gene was isolated from the hybrid aspen EST library [17] and cloned into the pPIC9 vector (Invitrogen) in-frame with the α-factor secretion signal. The insert was amplified by PCR using a proofreading DNA polymerase (Pfu; Stratagene, Cambridge, U.K.). The gene-specific primers (PttXET16A forward: 5′-TGACTACGTAGCTGCCCTGAGGAAGCCAGT-3′; PttXET16A reverse: 5′-TTAGTACGTATTATATGTCTCTGTCTCTCTTGCATTCTGG-3′) introduced SnaBI cleavage sites at both ends of the gene (underlined sequence). The pPIC9 vector and the PCR-amplified insert were each cleaved with SnaBI (MBI Fermentas, Munich, Germany) and fused by overnight ligation using T4 DNA ligase (MBI Fermentas). The ligation mixture was transformed into electrocompetent Epicurian Coli™ XL1-Blue cells (Stratagene) by electroporation (Bio-Rad Gene Pulser II; Bio-Rad, Hercules, CA, U.S.A.). Transformants were screened for orientation and single insertion by PCR. DNA extracted from the positive clones was sequenced to verify the identity of the expression construct. The plasmids with the correct insert sequence were linearized with SalI (MBI Fermentas) and transformed into P. pastoris cells (strain GS115; Invitrogen) by electroporation as described in the Invitrogen Pichia Expression kit manual, version L. The transformants were selectively grown on Regeneration Dextrose (−His) plates. The PCR, as well as colony blotting [33] with antibodies generated towards recombinant PttXET16A expressed in E. coli [7], was used to screen for colonies actively secreting the recombinant enzyme. The pPIC9 vector without the PttXET16A insert was linearized and transformed into P. pastoris to serve as a negative control for expression analysis.
Site-directed mutagenesis of the predicted N-glycosylation site in PttXET16A
Asn93, the predicted site of N-glycan attachment in PttXET16A, was mutated to a serine residue by site-directed mutagenesis using the Transformer™ Site-Directed Mutagenesis kit (Clontech, Stockholm, Sweden). A mutation primer (5′-GAGTTCTTAGGAAGCAGGACTGGCCAG-3′) introduced the desired mutation (underlined sequence) and a selection primer (5′-CCATCTCCTTGCCTGCACCATTCC-3′) was used to remove the unique SphI site from the vector, thus allowing the selection of mutated plasmids by restriction. Plasmids with the correct sequence were transformed into P. pastoris cells as described above for the wild-type gene.
Analysis of the secretion efficiency of PttXET16A and PttXET16A N93S
The ability of P. pastoris to secrete PttXET16A and PttXET16A N93S was analysed by the method described in the Invitrogen Pichia Expression kit manual as follows. Cells were separated from 1 ml samples of PttXET16A, PttXET16A N93S and negative control cultivations. The ‘breaking buffer’ (50 mM sodium phosphate, pH 7.4, 1 mM PMSF, 1 mM EDTA and 5%, v/v, glycerol) and glass beads were then added and, after eight cycles of vortex-mixing (30 s) and incubations on ice (30 s), the cell lysates were centrifuged at 16000 g for 10 min at 4 °C. The radioactive XET assay was performed in duplicate on both the soluble fraction and the culture supernatant for each culture. Cell lysis and subsequent analysis was also performed in duplicate.
Production and purification of the recombinant PttXET16A and PttXET16A N93S
Expression of the recombinant PttXET16A and PttXET16A N93S was performed essentially as described in the Invitrogen Pichia Expression kit manual. Briefly, 5 ml of an overnight culture (30 °C) in BMGY (buffered glycerol/complex medium) was used to inoculate 100 ml of the same medium. This culture was grown overnight in a 1 litre flask at 30 °C and 220 rev./min. To induce the expression, the yeast cells were harvested by centrifugation at 500 g for 15 min and resuspended in BMMY (buffered methanol/complex medium) in non-baffled 5 litre flasks to a final attenuance D600∼1. The PttXET16A culture was incubated for 3 days at 22 °C and 220 rev./min, whereas, for the PttXET16A N93S mutant, the incubation parameters were changed to 4 days at 16 °C and 180 rev./min. Methanol was added to a final concentration of 1% every 24 h. Expression of the active enzyme was confirmed by assaying the culture supernatant with the colorimetric XET assay.
The supernatant was recovered from a methanol-induced PttXET16A cultivation (2.6 litres) by centrifugation at 1200 g for 20 min. After filtration through a 0.45 μm Versapore membrane (Mini Capsule; Pall, Ann Arbor, MI, U.S.A.), the supernatant was concentrated and buffer-exchanged to 100 mM ammonium acetate (pH 5.5) in an ultrafiltration unit (Pall). The sample was then loaded on to 130 ml of SP Trisacryl gel (Sigma) packed in an XK26/40 column (Amersham Biosciences). Proteins were eluted from the cation-exchange column by a linear 0–1 M NaCl gradient in 100 mM ammonium acetate (pH 5.5) over 10 column volumes. Fractions containing active PttXET16A were pooled, dialysed against 100 mM ammonium acetate (pH 5.5) using Spectra/Por 2 membrane tubing (molecular mass cut-off 12–14000; Spectrum Medical Laboratories, Houston, TX, U.S.A.) and applied to a RESOURCE S 8 ml column (Amersham Biosciences). PttXET16A was eluted by a linear gradient of 0–1 M NaCl (10 column volumes) in 100 mM ammonium acetate (pH 5.5). All chromatographic steps were performed at room temperature (20 °C). A cultivation of the PttXET16A N93S mutant was treated identically, except that buffer exchange with the ultrafiltration unit was replaced by a 3 h dialysis at 4 °C using Spectra/Por 2 membrane tubing (molecular mass cut-off 12000–14000). Virgin SP Trisacryl column matrix (20 ml in an XK16/20 column) was used to avoid potential contamination by wild-type enzyme from previously used material. The second cation-exchange step on RESOURCE S was omitted in the purification of PttXET16A N93S. Purified PttXET16A and PttXET16A N93S were stored at 4 °C in the buffer in which they eluted from the cation-exchange columns [100 mM ammonium acetate buffer (acetic acid at a final concentration of 100 mM, titrated with ammonia to pH 5.5) containing approx. 300 mM NaCl].
pH-rate profile and thermal stability
The pH-rate profiles of wild-type PttXET16A and the N93S mutant were determined in duplicate using a 30 min colorimetric assay. The following 100 mM buffer systems were used: sodium formate, pH 3.0–4.0; sodium citrate, pH 4.0–6.2; and sodium phosphate, pH 6.2–8.0. The optimum temperature of the recombinant PttXET16A was estimated by performing duplicate measurements of the XET activity with a 15 min colorimetric assay in 100 mM sodium citrate (pH 5.5) at temperatures in the range 0–60 °C.
The unfolding temperature (Tm) of wild-type PttXET16A and the N93S mutant was determined by the fluorescence-based thermal shift assay described by Pantoliano et al. [34]. The fluorescent dye ‘SYPRO® Orange protein gel stain’ (supplied as a ‘5000× concentrate in DMSO’ by Molecular Probes, Eugene, OR, U.S.A.; catalogue number S6650) was used to monitor protein unfolding. The protein samples were heated in the iCycler iQ Real-time Detection System (Bio-Rad) with a charge-coupled device detector to monitor the fluorescent changes. The temperature was increased from 20 to 90 °C at a rate of 0.2 °C/min. Fluorescence intensity was measured with excitation and emission wavelengths of 490 and 575 nm respectively. The curve-fitting program XLfit allowed the calculation of Tm by fitting the data to a Boltzmann model. To each well, 7.5 μl of SYPRO® Orange (diluted 1:1000 from the stock solution) and 17.5 μl of a solution of protein (3.5–4.0 μg) in buffer was added. The Tm values of PttXET16A and PttXET16A N93S were measured in three different buffer systems: 70 mM ammonium acetate (pH 5.5); 70 mM ammonium acetate (pH 5.5) with 350 mM NaCl; and 70 mM ammonium acetate (pH 5.5) with 0.5 M urea.
ESI–MS (electrospray ionization MS) analysis
MS analysis was performed on a Q-Tof™ II mass spectrometer operated at a resolution of >10000 full-width at half maximum peak height (Waters; Micromass MS Technologies, Manchester, U.K.). External mass calibration was obtained over the m/z range 50–2000 using a solution of NaI (2 mg/ml) in 2-propanol/water (1:1, v/v). Intact proteins in ultrapure water or dilute volatile buffer solutions were diluted to 1–5 pmol/μl in acetonitrile/water (1:1, v/v) containing 0.1% formic acid and introduced into the mass spectrometer by infusion (syringe pump, 300–500 nl/min) through a nano-Z spray source (source voltage, approx. 3000 V; cone voltage, 30–35 V). Data were typically acquired over the m/z range 500–2000. The quadrupole mass filter of the Q-Tof™ II was operated in a wide band pass (radio frequency only) mode when collecting TOF (time-of-flight) MS data, and Ar was present in the hexapole collision cell to improve resolution (collision energy, 10 V). The raw combined spectral data obtained were background-subtracted and subjected to MaxEnt1 (Maximum Entropy™ 1 algorithm) deconvolution (Waters; Micromass MS Technologies) to produce reconstructed zero-charge spectra.
Protein deglycosylation by endo H (endoglycosidase H)
Before treatment of recombinant PttXET16A with Streptomyces plicatus endo H (Roche, Mannheim, Germany), both enzymes were dialysed against 10 mM ammonium acetate (pH 5.5, adjusted with acetic acid) using Pierce Slide-A-Lyzer™ MINI flotation dialysis units (10–100 μl size; Boule Nordic AB, Huddinge, Sweden). Protein deglycosylation was performed by adding endo H (0.55 unit, 110 μl) to PttXET16A (55 μl, 1 mg/ml) in 10 mM ammonium acetate buffer (pH 5.5), followed by incubation at 25 °C. Samples for ESI–MS analysis and activity assays were withdrawn at appropriate time intervals. In anticipation of a potentially lower stability of the deglycosylated enzyme, the colorimetric XET activity assay was performed with a reduced assay time (15 min) and temperature (25 °C). In control samples, 10 mM ammonium acetate (pH 5.5) replaced the endo H solution. Background-subtracted, combined spectra over the m/z range 870–1340 were used to generate zero-charge spectra by MaxEnt1 deconvolution (28000–35000 output mass, 1 Da resolution and 50% minimum left and right intensity ratios). This m/z range was chosen to include all major multiple-charged peaks of endo H, deglycosylated PttXET16A and PttXET16A glycoforms. Calculation of the percentage of deglycosylated enzyme was based on the peak-area fraction of the deglycosylated PttXET16A after conversion of MaxEnt1 spectra into centroid spectra [21]: deglycosylation (%)=100(area of peak 32349)/(sum of the areas of peaks 32349, 33849, 34011 and 34173).
RESULTS
Heterologous expression and purification of PttXET16A and PttXET16A N93S
To characterize the enzyme and to study the influence of glycosylation on its catalytic activity, wild-type PttXET16A and the corresponding N93S N-glycosylation site mutant were expressed in the methylotrophic yeast P. pastoris. Screening of the positive transformants by colony blotting [33] using specific antibodies raised against PttXET16A revealed that only 10% were able to secrete satisfactory levels of the recombinant proteins. However, the level of XET secretion from the clones identified by the antibody selection was stably maintained in repeated rounds of cultivation (results not shown).
During scale-up of the cultivation under standard conditions at 30 °C, the enzyme activity in the culture medium was found to be unstable, although the total amount of extracellular protein continued to increase (results not shown). Greatly improved yields of active XET were obtained by lowering the cultivation temperature to 22 and 16 °C for PttXET16A and PttXET16A N93S respectively, as well as lowering the incubator shaking speed to 180–220 rev./min.
Determination of the XET activity in 1 ml of culture medium by the radioactive assay revealed that the activity of wild-type PttXET16A was 0.55 and 0.014 units in the extracellular and intracellular fractions respectively. For the PttXET16A N93S mutant, the extracellular and intracellular activities were 0.063 and 0.0065 unit respectively, which indicated that the mutant enzyme was not secreted as efficiently as wild-type XET16A. No XET activity was detected in either fraction from a negative control of P. pastoris harbouring a pPIC9 vector lacking the XET gene.
Optimized methods were developed to obtain highly purified preparations of the wild-type enzyme and the glycosylation-deficient mutant. The wild-type enzyme was purified by employing two sequential cation-exchange chromatography steps (the purification Table is available as online information; see Supplementary Table 1 at http://www.BiochemJ.org/bj/390/bj3900105add.htm). MS analysis of the purified PttXET16A demonstrated N-terminal heterogeneity caused by differential cleavage of the α-factor signal peptide in addition to variable N-glycosylation (Table 1). Analysis of individual column fractions showed that the different variants had identical specific activities (results not shown); subsequently, they were treated as a single protein. A slightly modified version of the purification method was used for the PttXET16A N93S mutant protein (Supplementary Table 1; http://www.BiochemJ.org/bj/390/bj3900105add.htm). The combination of poor secretion efficiency and enzyme instability gave rise to a much lower yield for the PttXET16A N93S enzyme (0.4 mg/l cultivation medium) compared with the wild-type enzyme (12.4 mg/l). Unlike the wild-type enzyme, only one signal peptide cleavage variant was observed, which was identified by MS analysis as YV-PttXET16A N93S (calculated mass 32119, observed mass 32116) lacking glycosylation.
Table 1. Observed polypeptide and glycosylation variants of PttXET16A.
| Polypeptide* | Calculated pI | N-linked glycan | Calculated mass (Da) | Observed mass (Da)† |
|---|---|---|---|---|
| YV-PttXET16A | 7.75 | GlcNAc2Man8 | 33848 | 33849 |
| GlcNAc2Man9 | 34010 | 34011 | ||
| GlcNAc2Man10 | 34172 | 34173 | ||
| EAEAYV-PttXET16A | 6.68 | GlcNAc2Man8 | 34249 | 34250 |
| GlcNAc2Man9 | 34411 | 34412 | ||
| YV-PttXET16A+endo H | 7.75 | GlcNAc | 32349 | 32349 |
| EAEAYV-PttXET16A+endo H | 6.68 | GlcNAc | 32750 | 32750 |
* The polypeptide sequence refers to the complete polypeptide sequence of PttXET16A+additional amino acids from the α-factor secretion signal. pI values were calculated using the Compute pI/Mw tool on the ExPASy Molecular Biology server (http://www.expasy.org/).
† See Figure 2(a).
Properties of the recombinant PttXET16A and PttXET16A N93S
The specific activities of the purified proteins, determined using the radioactive assay and tamarind xyloglucan (1 g/l) as the donor substrate, were 42.5 units/mg for the PttXET16A and 34.5 units/mg for PttXET16A N93S (see also Supplementary Table 1; http://www.BiochemJ.org/bj/390/bj3900105add.htm). During purification and storage, the PttXET16A N93S protein quickly precipitated from the buffer solution; 50% of the enzyme was lost after storage at 4 °C for 5 days in 100 mM ammonium acetate (pH 5.5) with 250 mM NaCl, whereas the specific activity remained unaltered. In contrast, wild-type PttXET16A is stable over several months under the same conditions. Thus, due to uncertainties in the concentration of active PttXET16A N93S in the solutions, highly accurate values of specific activity could not be obtained. Nevertheless, throughout all the experiments, the mutant exhibited a slightly lower specific activity (factor 1.5–3) compared with wild-type PttXET16A. Due to the difficulty in handling highly viscous, concentrated xyloglucan solutions, the maximum donor substrate concentration was limited to 5 g/l for the determination of Michaelis–Menten parameters with the radioactive assay. Nonetheless, it was demonstrated that PttXET16A and PttXET16A N93S have comparable apparent Km values for the donor substrate: 2.2±0.3 and 1.4±0.3 g/l respectively. PttXET16A had an apparent kcat value of 180±10 nmol·min−1·mg−1 and PttXET16A N93S had an apparent kcat value of 60±6 nmol·min−1·mg−1. For the acceptor substrate XLLGol, Km=75±4 μM for PttXET16A and Km=62±5 μM for PttXET16A N93S were deduced by substrate competition [35,36].
Using the BCA reducing sugar assay, no endo-xyloglucanase (EC 3.2.1.151) activity was detected for either wild-type PttXET16A or the N93S mutant at a xyloglucan concentration of 1 g/l. On the basis of the detection limit of the BCA assay, an upper limit can be placed on the hydrolytic rate of 230 pmol·min−1·mg−1, which is 180-fold lower than the observed transglycosylation rate of the wild-type PttXET16A (42.5 nmol·min−1·mg−1; xyloglucan concentration, 1 g/l). Furthermore, calculation of the transglycosylation rate is based on a published tritium counting efficiency of xyloglucan bound to cellulose filter papers of 44% [3]. However, tritium counting efficiency from cellulose filters is generally low (1–4%) and difficult to determine precisely [30,31]. Therefore the observed transglycosylase activity may be underestimated (discussed in [21]), which would make the ratio transglycosylation/hydrolytic activity of PttXET16A even higher.
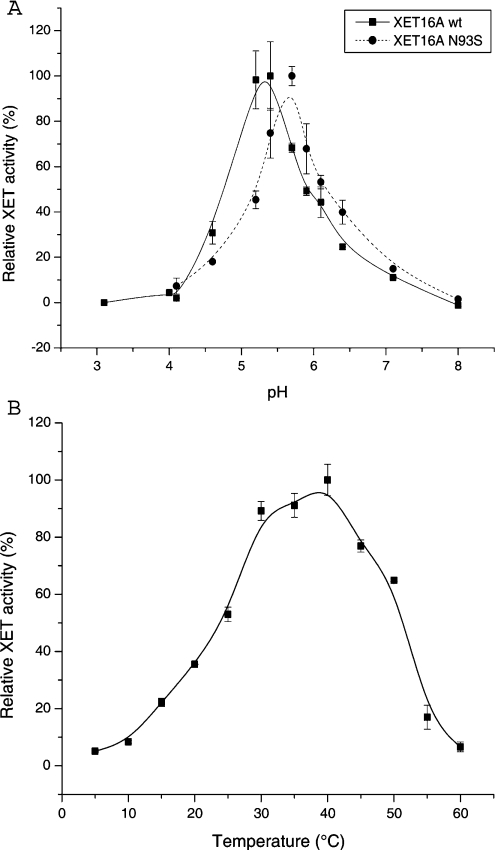
The pH-rate profiles of the recombinant PttXET16A and PttXET16A N93S are shown in Figure 1(A). PttXET16A has a slightly acidic optimum pH and demonstrates a rapid loss of activity in the pH range 4–5, which is typical for XET isoenzymes from various sources [21,23,35]. The basic limb of the pH-rate profile of PttXET16A N93S is slightly shifted towards higher pH values, and a slightly more pronounced loss in activity was observed at pH values below 5.5. The activity profile obtained for PttXET16A by varying the temperature of the colorimetric assay indicates that the optimum temperature for the enzyme is in the range 30–40 °C (Figure 1B).
Figure 1. Environmental effects on PttXET16A catalysis.
(A) pH-rate profile of the PttXET16A and PttXET16A N93S. (B) Temperature-rate profile. B-spline curves were drawn through the data points in each panel using Microcal™ Origin® v.6.0 to serve as ocular guides. Error bars represent S.D. values calculated from duplicate determinations.
The unfolding temperatures (Tm) of PttXET16A and PttXET16A N93S in 70 mM ammonium acetate (pH 5.5) were 52.9 and 52.3 °C respectively (Table 2). Addition of 350 mM NaCl in this buffer (analogous to the buffer conditions under which the enzyme elutes from the cation-exchange columns) decreased the Tm for both enzymes by approx. 4 °C; the addition of urea to 0.5 M had a similar effect (Table 2). An extensive range of solvent conditions was examined to find improved storage conditions for the enzymes (see Supplementary Tables 2–4; http://www.BiochemJ.org/bj/390/bj3900105add.htm). Additives such as glycerol and sucrose (20%) stabilized the wild-type enzyme by up to 5 °C, whereas high salt concentrations (up to 1 M NaCl) decreased the Tm of the wild-type by up to 10 °C.
Table 2. Unfolding temperature of PttXET16A and the N93S mutant.
AA, 70 mM ammonium acetate (pH 5.5).
| Protein | Final buffer concentration | Tm (°C) |
|---|---|---|
| PttXET16A wild-type | AA | 52.9 |
| AA with 350 mM NaCl | 48.4 | |
| AA with 0.5 M urea | 49.4 | |
| PttXET16A N93S | AA | 52.3 |
| AA with 350 mM NaCl | 49.2 | |
| AA with 0.5 M urea | 49.0 |
Effect of endo H-catalysed N-deglycosylation of PttXET16A on enzyme activity
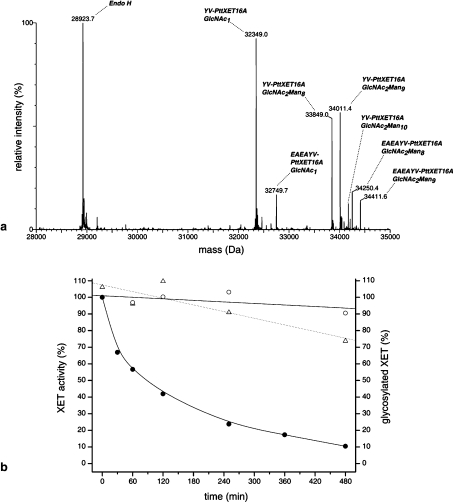
As shown in Table 1, P. pastoris N-glycosylates recombinant PttXET16A with high-mannose-type glycans typically bearing 8–10 mannose units, although glycoforms bearing up to 12 mannose units have been observed in some cultivations (results not shown). To complement mutagenesis studies on the N-glycosylation of PttXET16A, the enzyme was treated with endo H, which cleaves the chitobiose core of high-mannose-type N-glycans to leave a single asparagine-linked GlcNAc residue. The deglycosylation was performed under relatively gentle conditions to maximize PttXET16A enzyme stability without decreasing the rate of deglycosylation [21]. A time-course experiment was then performed in which activity was measured as a function of the glycosylation state of the enzyme. ESI–MS analysis of endo H-treated PttXET16A allowed the simultaneous mass determination and quantification of the total protein in the reaction mixture, including endo H, deglycosylated PttXET16A and the major glycoforms of heterologously expressed PttXET16A (Figure 2a). This, in turn, allowed the extent of deglycosylation at each time point to be calculated directly as a percentage on the basis of the peak-area fraction [defined as (peak area)/(sum of all peak areas considered)] of the deglycosylated enzyme (Figure 2b). Peak areas for the major glycoforms of the YV-PttXET16A cleavage variant at 32349, 33849, 34011 and 34173 Da were included in the analysis. Lower abundance peaks due to glycoforms of the EAEAYV-PttXET16A cleavage variant were omitted for simplicity. As shown in Figure 2(b), PttXET16A is 90% deglycosylated after 480 min, yet it retained approx. 80% of the activity of the control sample without endo H. [The time values indicated in Figure 2(b) represent the times at which samples were withdrawn for each respective analysis. The rate of deglycosylation during the 15 min XET assay would have been greatly reduced by dilution of XET and endo H by an overall factor of 1:20 in the assay mixture (no direct inhibition of endo H activity by the XET assay conditions is assumed). In comparison, samples for MS analysis were immediately diluted in acetonitrile/water (1:1, v/v) containing 0.1% formic acid (pH<4) and spectral acquisition was completed in less than 3 min. Endo H has no detectable XET or endo-xyloglucanase activity at the concentrations used in this experiment (results not shown)]. Based on experience with the N-glycosylation mutant, this loss of activity is attributed to slow precipitation of the deglycosylated enzyme from solution. The lack of a strict correlation between the extent of enzymatic deglycosylation and the loss of activity (Figure 2b) supports the kinetic results obtained for the PttXET16 N93S glycosylation mutant; the N-glycan is not catalytically important in the XET reaction. The enzymatic removal of the N-glycan of PttXET16A does not increase the hydrolytic activity of the enzyme to a detectable level.
Figure 2. Endo H-catalysed deglycosylation of recombinant PttXET16A.
(a) Reconstructed zero-charge spectrum of a deglycosylation reaction containing PttXET16A and endo H at 60 min. (b) Progress curve of the deglycosylation reaction. ●, percentage of glycosylated PttXET16A calculated from MS peak-area fractions; △, relative activity of endo H-treated sample; ○, relative activity of control (no endo H) sample. Linear least-squares fits to the activity data on the deglycosylated and control samples are shown with dotted and solid lines respectively; a B-spline curve is drawn through the MS data.
DISCUSSION
We have characterized an enzyme from the XTH gene family of Populus. To avoid potential contamination by other cell-wall enzymes, we expressed the enzyme in a non-native host, P. pastoris. The recombinant enzyme purified from the yeast culture medium exhibited a clear transglycosylating activity, but no detectable rate of hydrolytic activity, and is thus a true XET (EC 2.4.1.207). This makes PttXET16A the second enzyme in the XTH gene family that has been unequivocally demonstrated to be a strict transglycosylase when acting on tamarind xyloglucan [21].
Most XTH genes described so far have a conserved N-glycosylation site located within 5–15 residues towards the C-terminus from the conserved active-site residues. In spite of its location very close to the catalytic site, removal of the N-glycan by site-directed mutagenesis did not significantly reduce the transglycosylating activity of PttXET16A, nor did it increase the hydrolytic rate of the enzyme to a detectable level. However, the secretion of the PttXET16A N93S mutant in P. pastoris was noticeably impaired relative to wild-type PttXET16A, perhaps due to incorrect folding or limited solubility of the protein lacking an appendant N-glycan [37,38]. The purified PttXET16A N93S protein exhibited thermal stability similar to wild-type PttXET16A, but was highly susceptible to precipitation, which may be attributable to increased hydrophobicity of the non-glycosylated protein. In spite of the low yield and reduced solubility of PttXET16A N93S, the enzyme could be kinetically characterized. The results show that it exhibits similar reaction kinetics compared with the wild-type enzyme and previously described XETs [14,35,36]. A time-course experiment, in which the enzymatic deglycosylation of the wild-type enzyme was followed by MS, corroborated the observation that the lack of an N-glycan did not affect the intrinsic enzyme activity.
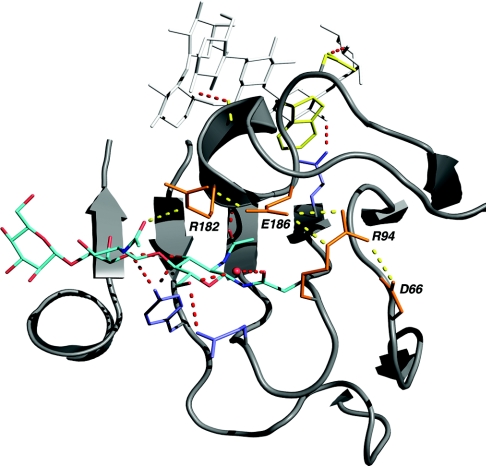
The recent three-dimensional structure of PttXET16A [19] helps to provide a structural interpretation of the enzyme glycosylation data. The PttXET16A polypeptide folds into a curved β-sandwich with a long binding cleft typical of the GH16 family. The glycosylation of Asn93 is clearly detectable in the structure of PttXET16A, with good electron density for two GlcNAc residues and one mannose residue in the vicinity of the active-site cleft [19]. As shown in Figure 3, these three sugar residues have several interactions with the amino acid side chains of PttXET16A. The chitobiose core of the N-glycan is observed within the hydrogen-bonding distance of the side chain of Arg182, which is located on the acceptor-binding loop (Ala176-Trp190; Figure 4) of the enzyme. The importance of this loop for XET activity is illustrated in the PttXET16A structure in complex with a xyloglucan-derived nonasaccharide, XLLG [PDB (Protein Data Bank) code 1UMZ] [19]. The ligand bound in the acceptor sites +1 to +3 has favourable interactions with the side chains of Trp179 and Asp178 and the carbonyl oxygen of Gly183 (Figure 3). Trp179 is strictly conserved in all XETs and is, by way of stacking interactions and hydrogen bond donation, important for the binding of three sugar residues in the acceptor subsites [19]. Therefore the position of the loop with respect to the ligand is likely to have a large contribution to acceptor substrate binding, which is necessary for PttXET16A to function as a transglycosylase.
Figure 3. Structural relationship of the PttXET16A N-glycan to the active-site cleft.
The α-carbon trace is depicted as a grey cartoon. The side chain of Asn93, the N-glycan, and a bound water molecule are coloured by atom type (red, oxygen; blue, nitrogen; cyan, carbon). Side chains of residues involved in an extended electrostatic interaction (Arg182-Glu186-Arg94-Asp66) are shown in orange. Electrostatic and hydrogen-bonding interactions of Arg182, Glu186, Arg94 and Asp66 are represented as dotted yellow lines. Side chains interacting with the N-glycan (Gln97 and Tyr118), as well as the predicted catalytic acid/base residue (Glu89), are shown in blue. The carbonyl group of Gly183 and the side chains of Asp178 and Trp179, which interact with the XGO ligand (white), are shown in yellow. Predicted hydrogen-bonding interactions are shown as dotted red lines. The representation was composed from data in PDB code 1UMZ (chain A) using PyMOL version 0.93 (http://www.pymol.org/) and was rendered using POV-Ray version 3.5 (http://www.povray.org).
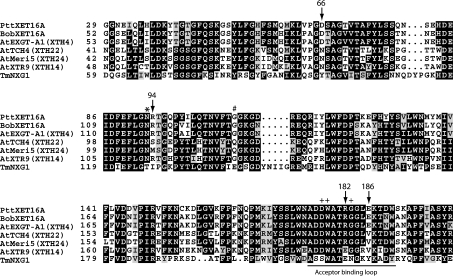
Figure 4. Partial sequence alignment of PttXET16A with XET-like enzymes from other species.
The following protein sequences were used (GenBank® accession numbers in parentheses): PttXET16A, Populus tremula×tremuloides XET16A (AF515607); BobXET16A, Brassica oleracea var. botrytis XET16A (AY156708); AtEXGT-A1 (XTH4), A. thaliana XTH4 (D164541); AtTCH4 (XTH22), A. thaliana XTH22 (AF051338); AtMeri5 (XTH24), A. thaliana XTH24 (D63508); AtXTR9 (XTH14), A. thaliana XTH14 (AF093672); TmNXG1, Tropaeolum majus NXG1 (X68254). The numbering of the PttXET16A sequence is as given in [19]; all other sequences are numbered from the first amino acid of the predicted preprotein. Numbered arrows indicate residues of PttXET16A involved in an extended salt bridge to the acceptor-binding loop. ‘+’ indicates acceptor loop residues involved in binding at the negative (acceptor) subsites. Asterisks indicate the conserved N-glycosylation site observed in many XET-like enzymes proximal to the active site. ‘#’ indicates the location of the N-glycosylation site in ‘subfamily 3’ XETs, which is shifted 15 residues towards the C-terminus [5].
Characterization of the enzymatic properties of genetically deglycosylated PttXET16A revealed decreased solubility, but provided no indication that the N-glycan plays a crucial role in stabilizing the substructures directly involved in catalysis. Further analysis of the structure of the acceptor-binding loop of PttXET16A revealed that residues Glu186 and Arg182 are involved in bidentate electrostatic interactions (Figure 3). Glu186 forms a salt bridge with both Arg182 and Arg94, thus connecting the acceptor-binding loop and the loop at the end of strand β7, which harbours the glycosylation site. Arg94, in turn, forms a salt bridge with Asp66 on the loop between strands β5 and β6. It is therefore possible that these extended electrostatic interactions are more important than the interaction with the N-glycan for the stabilization and positioning of the acceptor-binding loop in PttXET16A.
The effect of deglycosylation on the enzymatic activity has so far been studied with eight different XET isoforms, including PttXET16A. A sequence comparison between these enzymes (Figure 4), based on the three-dimensional structure of PttXET16A, suggests an explanation why some of the enzymes are inactivated by deglycosylation, whereas others are not. In two cases, BobXET16A [21] and AtEXGT (renamed AtXTH4 [16]), all of the residues involved in the acceptor-loop salt bridges are conserved and their enzymatic activity survives deglycosylation [21,23]. TmNXG1 [39], one of the few XET-like enzymes without an N-glycosylation site, does not possess the residues participating in the extended electrostatic interactions observed in PttXET16A. However, the acceptor-binding loop region of TmNXG1 carries a glutamate and lysine residue (corresponding to residues 182 and 185 of PttXET16A), which could be involved in an analogous stabilizing interaction, perhaps together with so far unidentified interactions (Figure 4). AtXTR9 (renamed AtXTH14), which retains full transglycosylase activity after N-deglycosylation, also contains a glutamate at position 182 (PttXET16A numbering), while the lysine residue is conservatively substituted with an arginine [23].
In two cases, AtTCH4 (renamed AtXTH22) and AtMeri5 (renamed AtXTH24), none of the interactions described above and proposed to stabilize the acceptor loop are conserved. The residue corresponding to Arg94 in PttXET16A is replaced with either serine or methionine and the residue corresponding to Glu186 is replaced by valine in both enzymes. However, the residue corresponding to Arg182 in PttXET16A, which is involved in a hydrogen-bonding interaction with the second GlcNAc residue of the N-glycan, is conserved in AtXTH22 and AtXTH24, and the activity of both enzymes is significantly reduced by glycosidase treatment [22,23]. It is thus possible that the N-glycans play a more crucial role in the positioning of the acceptor loop for catalysis in these two enzymes.
Whereas the position of the N-glycosylation site of PttXET16A is conserved in most XETs, in those grouped into subfamily 3 by a phylogenetic analysis, the glycosylation site is shifted 15 residues towards the C-terminus [5]. In these sequences, the N-glycosylation site aligns with Gly108 of PttXET16A (Figure 4), which is positioned directly across the active-site cleft, on the opposite end of strand β8. This implies that the N-glycan has no direct catalytic effect in subfamily 3. Generally, the N-glycosylation of plant proteins is believed to prevent proteolytic degradation, induce correct folding or be involved in protein targeting [40]. Mounting biochemical evidence for members of the XET family indicates that glycosylation does not play a specific role in the catalytic mechanism, but may instead improve enzyme solubility and, in some cases, participate in positioning the acceptor loop.
Online data
Acknowledgments
We thank M. Baumann and W. Lodewyckx for assistance with the preparation of XGOs and U. Ericsson (Department of Structural Biochemistry, Stockholm University, Stockholm, Sweden) for help with the Tm measurements. This work was generously supported by the Knut and Alice Wallenberg Foundation as well as a Marie Curie Individual Fellowship to K.P. (contract no. HPMF-CT-2002-02009). The purchase of the MS equipment was funded by the Wallenberg Consortium North.
References
- 1.Vincken J. P., York W. S., Beldman G., Voragen A. G. J. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol. 1997;114:9–13. doi: 10.1104/pp.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vierhuis E., York W. S., Kolli V. S. K., Vincken J. P., Schols H. A., Van Alebeek G. J. W. M., Voragen A. G. J. Structural analyses of two arabinose containing oligosaccharides derived from olive fruit xyloglucan: XXSG and XLSG. Carbohydr. Res. 2001;332:285–297. doi: 10.1016/s0008-6215(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 3.Fry S. C., Smith R. C., Renwick K. F., Martin D. J., Hodge S. K., Matthews K. J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishitani K., Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- 5.Campbell P., Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- 6.Pauly M., Qin Q., Greene H., Albersheim P., Darvill A., York W. S. Changes in the structure of xyloglucan during cell elongation. Planta. 2001;212:842–850. doi: 10.1007/s004250000448. [DOI] [PubMed] [Google Scholar]
- 7.Bourquin V., Nishikubo N., Abe H., Brumer H., Denman S., Eklund M., Christiernin M., Teeri T. T., Sundberg B., Mellerowicz E. J. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell. 2002;14:3073–3088. doi: 10.1105/tpc.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Q., Schlueter S. D., Brendel V. PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 2004;32:D354–D359. doi: 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vocadlo D. J., Davies G. J., Laine R., Withers S. G. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature (London) 2001;412:835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 10.Sinnott M. L. Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 1990;90:1171–1202. [Google Scholar]
- 11.Lorences E. P., Fry S. C. Xyloglucan oligosaccharides with at least two α-D-xylose residues act as acceptor substrates for xyloglucan endotransglycosylase and promote the depolymerisation of xyloglucan. Physiol. Plant. 1993;88:105–112. [Google Scholar]
- 12.Edwards M., Dea I. C., Bulpin P. V., Reid J. S. Purification and properties of a novel xyloglucan-specific endo-(1->4)-beta-D-glucanase from germinated nasturtium seeds (Tropaeolum majus L.) J. Biol. Chem. 1986;261:9489–9494. [PubMed] [Google Scholar]
- 13.Tabuchi A., Kamisaka S., Hoson T. Purification of xyloglucan hydrolase/endotransferase from cell walls of azuki bean epicotyls. Plant Cell Physiol. 1997;38:653–658. [Google Scholar]
- 14.Schröder R., Atkinson R. G., Langenkamper G., Redgwell R. J. Biochemical and molecular characterisation of xyloglucan endotransglycosylase from ripe kiwifruit. Planta. 1998;204:242–251. doi: 10.1007/s004250050253. [DOI] [PubMed] [Google Scholar]
- 15.Tabuchi A., Mori H., Kamisaka S., Hoson T. A new type of endo-xyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant Cell Physiol. 2001;42:154–161. doi: 10.1093/pcp/pce016. [DOI] [PubMed] [Google Scholar]
- 16.Rose J. K., Braam J., Fry S. C., Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- 17.Sterky F., Regan S., Karlsson J., Hertzberg M., Rohde A., Holmberg A., Amini B., Bhalerao R., Larsson M., Villarroel R., et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertzberg M., Aspeborg H., Schrader J., Andersson A., Erlandsson R., Blomqvist K., Bhalerao R., Uhlén M., Teeri T. T., Lundeberg J., et al. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14732–14737. doi: 10.1073/pnas.261293398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson P., Brumer H., III, Baumann M. J., Kallas Å. M., Henriksson H., Denman S. E., Teeri T. T., Jones T. A. Crystal structures of a xyloglucan endotransglycosylase reveal details of the transglycosylation acceptor binding. Plant Cell. 2004;16:874–886. doi: 10.1105/tpc.020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrissat B., Teeri T. T., Warren R. A. J. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 21.Henriksson H., Denman S. E., Campuzano I. D. G., Ademark P., Master E. R., Teeri T. T., Brumer H. N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem. J. 2003;375:61–73. doi: 10.1042/BJ20030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell P., Braam J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant J. 1998;15:553–561. doi: 10.1046/j.1365-313x.1998.00239.x. [DOI] [PubMed] [Google Scholar]
- 23.Campbell P., Braam J. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 1999;18:371–382. doi: 10.1046/j.1365-313x.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- 24.Arrowsmith D. A., de Silva J. Characterization of two tomato fruit-expressed cDNAs encoding xyloglucan endo-transglycosylase. Plant Mol. Biol. 1995;28:391–403. doi: 10.1007/BF00020389. [DOI] [PubMed] [Google Scholar]
- 25.Oh M.-H., Romanow W. G., Smith R. C., Zamski E., Sasse J., Clouse S. D. Soybean Bru1 encodes a functional xyloglucan endotransglycosylase that is highly expressed in inner epicotyl tissues during brassinosteroid-promoted elongation. Plant Cell Physiol. 1998;39:124–130. [Google Scholar]
- 26.Catalá C., Rose J. K., York W. S., Albersheim P., Darvill A. G., Bennett A. B. Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiol. 2001;127:1180–1192. doi: 10.1104/pp.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrissey J. H. Silver stain for proteins in polyacrylamide gels – a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 28.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Sulová Z., Lednicka M., Farkaš V. A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal. Biochem. 1995;229:80–85. doi: 10.1006/abio.1995.1381. [DOI] [PubMed] [Google Scholar]
- 30.Gill D. M. Liquid scintillation counting of tritiated compounds supported by solid filters. Int. J. Appl. Radiat. Isot. 1967;18:393–398. [Google Scholar]
- 31.Brandsome E. D., Grower M. F. Liquid scintillation counting of H-3 and C-14 on solid supports: a warning. Anal. Biochem. 1970;38:401–408. doi: 10.1016/0003-2697(70)90464-1. [DOI] [PubMed] [Google Scholar]
- 32.Garcia E., Johnston D., Whitaker J. R., Shoemaker S. P. Assessment of endo-1,4-beta-D-glucanase activity by a rapid colorimetric assay using disodium 2,2′-bicinchoninate. J. Food Biochem. 1993;17:135–145. [Google Scholar]
- 33.Holmquist M., Tessier D. C., Cygler M. High-level production of recombinant Geotrichum candidum lipases in yeast Pichia pastoris. Prot. Expr. Purif. 1997;11:35–40. doi: 10.1006/prep.1997.0747. [DOI] [PubMed] [Google Scholar]
- 34.Pantoliano M. W., Petrella E. C., Kwasnoski J. D., Lobanov V. S., Myslik J., Graf E., Carver T., Asel E., Springer B. A., Lane P., et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 35.Steele N. M., Fry S. C. Differences in catalytic properties between native isoenzymes of xyloglucan endotransglycosylase (XET) Phytochemistry. 2000;54:667–680. doi: 10.1016/s0031-9422(00)00203-x. [DOI] [PubMed] [Google Scholar]
- 36.Purugganan M. M., Braam J., Fry S. C. The Arabidopsis TCH4 xyloglucan endotransglycosylase. Substrate specificity, pH optimum, and cold tolerance. Plant Physiol. 1997;115:181–190. doi: 10.1104/pp.115.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu A., Wang Z. K., Beavis R. Structural studies of α-N-acetyl-galactosaminidase: effect of glycosylation on the level of expression, secretion efficiency, and enzyme activity. Arch. Biochem. Biophys. 1998;352:1–8. doi: 10.1006/abbi.1998.0575. [DOI] [PubMed] [Google Scholar]
- 38.Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 39.de Silva J., Jarman C. D., Arrowsmith D. A., Stronach M. S., Chengappa S., Sidebottom C., Reid J. S. Molecular characterization of a xyloglucan-specific endo-(1–>4)-beta-D-glucanase (xyloglucan endo-transglycosylase) from nasturtium seeds. Plant J. 1993;3:701–711. [PubMed] [Google Scholar]
- 40.Rayon C., Lerouge P., Faye L. The protein N-glycosylation in plants. J. Exp. Bot. 1998;49:1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.