Abstract
Human AE1 (anion exchanger 1), or Band 3, is an abundant membrane glycoprotein found in the plasma membrane of erythrocytes. The physiological role of the protein is to carry out chloride/bicarbonate exchange across the plasma membrane, a process that increases the carbon-dioxide-carrying capacity of blood. To study the topology of TMs (transmembrane segments) 1–4, a series of scanning N-glycosylation mutants were created spanning the region from EC (extracellular loop) 1 to EC2 in full-length AE1. These constructs were expressed in HEK-293 (human embryonic kidney) cells, and their N-glycosylation efficiencies were determined. Unexpectedly, positions within putative TMs 2 and 3 could be efficiently glycosylated. In contrast, the same positions were very poorly glycosylated when present in mutant AE1 with the SAO (Southeast Asian ovalocytosis) deletion (ΔA400–A408) in TM1. These results suggest that the TM2–3 region of AE1 may become transiently exposed to the endoplasmic reticulum lumen during biosynthesis, and that there is a competition between proper folding of the region into the membrane and N-glycosylation at introduced sites. The SAO deletion disrupts the proper integration of TMs 1–2, probably leaving the region exposed to the cytosol. As a result, engineered N-glycosylation acceptor sites in TM2–3 could not be utilized by the oligosaccharyltransferase in this mutant form of AE1. The properties of TM2–3 suggest that these segments form a re-entrant loop in human AE1.
Keywords: anion exchanger, Band 3, membrane protein, N-glycosylation, ovalocytosis, topology
Abbreviations: AE1, anion exchanger 1; DMEM, Dulbecco's modified Eagle's medium; EC, extracellular loop; ER, endoplasmic reticulum; HEK, human embryonic kidney; OST, oligosaccharyltransferase; SA, signal-anchor; SAO, Southeast Asian ovalocytosis; ST, stop-transfer; TM, transmembrane segment
INTRODUCTION
Human AE1 (anion exchanger 1), an abundant glycoprotein found in the plasma membrane of erythrocytes, carries out the electroneutral exchange of chloride and bicarbonate, a process that enhances the carbon-dioxide-carrying capacity of blood. The transport function of this 911-amino-acid glycoprotein is carried out by its C-terminal membrane-embedded domain [1], whereas the N-terminal cytosolic domain provides binding sites to the red-cell cytoskeleton and cytosolic proteins [2]. Mutations in AE1 have been linked to haematological conditions [3], such as SAO (Southeast Asian ovalocytosis). SAO arises from the deletion of the sequence Ala400–Ala408 in AE1 at the boundary of the cytosolic domain and TM (transmembrane segment) 1 [4–7]. The membrane domain of the SAO protein is misfolded [8], and the protein is inactive in anion transport [6].
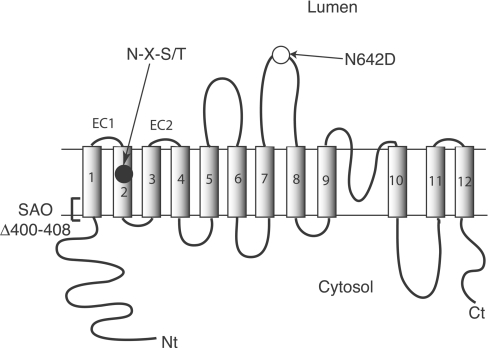
The topology of the membrane domain of human AE1 (Figure 1) has been examined using various techniques [9,10]. Hydropathy analysis suggests that the membrane domain consists of 12–14 TMs [11]. The C-terminal tail of the membrane domain was located to the cytosolic side by antibody binding [12,13]. Proteolytic cleavage experiments localized sensitive sites to the cytosolic or extracytosolic side of the membrane [14–16]. Naturally occurring antigenic sites encompassing Glu658 [17] and Pro854 [18,19] localize these regions to the red-cell exterior. The endogenous N-glycosylation acceptor site at Asn642 positions this region on the extracytosolic side of the membrane. Novel N-glycosylation acceptor sites created in various loops allowed determination of whether the loops are intracellular or extracellular, definition of the limits of ECs (extracellular loops) 3 and 4, as well as identification of a re-entrant T-loop in the C-terminal part of the membrane domain [20,21]. Cysteine-scanning mutagenesis was employed to study the C-terminal part of the membrane domain, revealing possible pore-lining residues in this region [22].
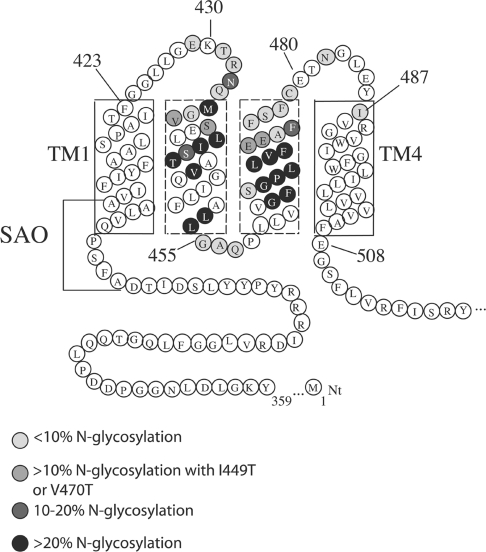
Figure 1. Folding model of human AE1 and N-glycosylation mutants.
Human AE1 consists of an N-terminal cytosolic domain and a membrane domain (12-TM model shown here). The endogenous N-glycosylation acceptor site is located in EC4, at Asn642. Deletion of residues 400–408 in TM1 results in SAO. The N-glycosylation mutants used in the present study were created in a N642D background; individual N-glycosylation acceptor sites (N-X-S/T) were made in the region encompassing EC1–EC2.
Limited information is available concerning the topology of the N-terminal part of the membrane domain of AE1. Hydropathy analysis shows three peaks in the region of residues 400–510 encompassing four TM segments [11]. The first peak is assigned as TM1. The second peak is large enough to accommodate two TMs, assigned as TMs 2 and 3. The third peak is highly hydrophobic and is assigned as TM4. Sensitivity to proteases placed Tyr359 and Lys360 on the cytosolic side of the membrane [15], giving TM1 an NcytCexo orientation. Lys430 in the loop connecting TMs 1 and 2 (EC1) can be labelled with external eosin-5-maleimide [23] and is the site of external reductive methylation [24]. The low-incidence blood group antigen ELO is associated with an R432W mutation in EC1 [25]. These observations indicate that the loop is extracellular. Although a novel N-glycosylation site created at position 429 in EC1 could not be N-glycosylated, this was due to the distance constraint to the OST (oligosaccharyltransferase) active site, which requires the acceptor site to be located a sufficient distance (approx. 12–14 amino acids) away from the end of TMs [20,26,27]. N-Glycosylation sites created in an extended EC1 can be glycosylated [20,21,28], confirming the extracellular disposition of the EC1. The presence of the low-incidence Froese blood group antigen Fra, resulting from the G480K mutation in the loop connecting TMs 3 and 4, confirms the existence of EC2 [29,30]. No internal markers have defined the cytosolic loop linking TMs 2 and 3, although these long hydrophobic segments are connected by a short polar sequence (Gly455-Ala-Pro-Gln).
Assessing the topogenic function of AE1 TMs in a cell-free translation system, Ota et al. [31] found that TMs 1 and 4 act as efficient SA (signal-anchor) and ST (stop-transfer) elements respectively, while isolated TMs 2 and 3 are poor ST and SA elements. The presence of TM1 located close to TM2, however, was able to rescue membrane integration of TM2, as determined using constructs of TM1–2 with a downstream glycosylation reporter domain [31,32]. On the other hand, the presence of TMs 1 and 2 upstream of TM3 did not improve membrane integration of TM3 [31], suggesting that TM3 has no internal SA function, possibly requiring the strong ST character of TM4 for membrane integration [31]. Hamasaki et al. [33,34] proposed that the membrane domain in membrane proteins such as AE1 be divided into three categories. Category 1 consists of hydrophilic connecting loops. Category 2, to which TMs 1–3 belong, are membrane-embedded owing to peptide–peptide interactions and can be extracted by alkaline treatment. Category 3 peptides, such as TM4, are embedded by strong peptide–lipid interactions and are resistant to alkaline extraction.
Complementary fragments of AE1 have been co-expressed in Xenopus oocytes and their ability to be expressed at the cell surface and carry out anion transport has been examined [35,36]. Most combinations of fragments produced anion transport activity. However, co-expressed fragments that were separated in either EC1 [36] or EC2 [35,36] were not functional. Interactions between TMs 2–3 and both TM1 and 4 are likely to be essential for correct membrane integration of TMs 2 and 3. It was also suggested that TMs 1–5 form a folded subdomain [36], proposed to be situated at the dimeric interface of the membrane domain [37].
In the present paper, we describe experiments to examine the folding of the TM1–4 region in AE1 using scanning N-glycosylation mutagenesis. Novel N-glycosylation acceptor sites were created at over 30 different positions in the region encompassing EC1–EC2 in AE1 N642D or SAO N642D background. These constructs were expressed in transiently transfected HEK-293 (human embryonic kidney) cells in the presence of [35S]methionine and were immunoprecipitated with an anti-AE1 antibody. The results that we obtained were surprising, with N-glycosylation observed throughout TM2–3. These sites were not N-glycosylated efficiently in AE1 SAO constructs. These results were interpreted in conjunction with previous studies, providing insight into the folding of TM2–3 in both AE1 and AE1 SAO, and they suggest that the hydrophobic region corresponding to TM2–3 form a re-entrant loop structure in Band 3. The introduction of polar residues into this region may promote its exposure to the ER (endoplasmic reticulum) lumen, allowing N-glycosylation at introduced sites. Similar mutations that occur in membrane proteins that are linked to diseases such as haemolytic anaemia in AE1 and cystic fibrosis in CFTR (cystic fibrosis transmembrane conductance regulator) could result in severe protein misfolding, impaired trafficking and loss of function.
EXPERIMENTAL
Materials
Materials used and their suppliers are as follows: pcDNA3 (Invitrogen, San Diego, CA, U.S.A.); QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.); DMEM (Dulbecco's modified Eagle's medium), calf serum, penicillin and streptomycin (Gibco BRL, Burlington, ON, Canada); EasyTag™ EXPRE35S35S protein labelling mix (PerkinElmer Life Sciences, Mississauga, ON, Canada); concanavalin A (Sigma–Aldrich Canada, Oakville, ON, Canada); Protein G–Sepharose (Amersham Biosciences, Baie d'Urfé, QC, Canada); dropping bottle with 20–30 μm filter unit (Wheaton, Millville, NJ, U.S.A.); and mutagenic primers (ACGT Corp., Toronto, ON, Canada).
Site-directed mutagenesis
The coding sequence for human AE1 was inserted into the XhoI and BamHI sites of pcDNA3 [21]. The construction of AE1 SAO from pcDNA3·AE1 has been described in [21]. To remove the single endogenous N-glycosylation site (N642D constructs), an N642D mutation was carried out with AE1 or AE1 SAO as templates. Single N-glycosylation acceptor sites were created in the region from EC1 to EC2, inclusive, on an AE1 N642D or SAO N642D background in pcDNA3 using the QuikChange™ mutagenesis kit. Sequencing of the constructs was carried out by the ACGT Corp. Figure 1 shows an illustration of the N-glycosylation mutants, listed in Table 1.
Table 1. Constructs used for AE1 scanning N-glycosylation mutagenesis.
| (a) AE1 Constructs | ||
|---|---|---|
| Construct* | N-Glycosylation site | Mutation(s) |
| AE1 | N642 (EC4) | None |
| AE1 N642D | None | N642D |
| AE1 N431 | N431 (EC1) | T431N, N433S, N642D |
| AE1 N432 | N432 (EC1) | R432N, Q434S, N642D |
| AE1 N433 | N433 (EC1) | M435S, N642D |
| AE1 N434 | N434 (EC1) | Q434N, G436S, N642D |
| AE1 N435 | N435 (EC1) | M435N, V437T, N642D |
| AE1 N436 | N436 (EC1) | G436N, N642D |
| AE1 N437 | N437 (TM2) | V437N, E439T, N642D |
| AE1 N437/I449T | N437 (TM2) | V437N, E439T, I449T, N642D |
| AE1 N438 | N438 (TM2) | S438N, E439G, L440S, N642D |
| AE1 N438/I449T | N438 (TM2) | S438N, E439G, L440S, I449T, N642D |
| AE1 N441 | N441 (TM2) | L441N, N642D |
| AE1 N442 | N442 (TM2) | I442N, N642D |
| AE1 N443 | N443 (TM2) | S443N, A445T, N642D |
| AE1 N444 | N444 (TM2) | T444N, V446T, N642D |
| AE1 N446 | N446 (TM2) | V446N, G448S, N642D |
| AE1 N453 | N453 (TM2) | L453N, G455S, N642D |
| AE1 N454 | N454 (TM2) | L454N, A456S, N642D |
| AE1 N455 | N455 (TM2) | G455N, Q457S, N642D |
| AE1 N455, P458A | N455 (TM2) | G455N, Q457S, P458A, N642D |
| AE1 N456 | N456 (TM2) | A456N, P458S, N642D |
| AE1 N457 | N457 (TM2) | Q457N, P458A, L459S, N642D |
| AE1 N463 | N463 (TM3) | G463N, F464S, N642D |
| AE1 N464 | N464 (TM3) | F464N, G466S, P467A, N642D |
| AE1 N465 | N465 (TM3) | S465N, P467S, N642D |
| AE1 N466 | N466 (TM3) | G466N, P467A, L468T, N642D |
| AE1 N467 | N467 (TM3) | P467N, L469T, N642D |
| AE1 N468 | N468 (TM3) | L468N, V470T, N642D |
| AE1 N469 | N469 (TM3) | L469N, F471S, N642D |
| AE1 N470 | N470 (TM3) | V470N, F471S, E472T, N642D |
| AE1 N471 | N471 (TM3) | F471N, E472G, E473T, N642D |
| AE1 N472 | N472 (TM3) | E472N, E473G, A474T, N642D |
| AE1 N472/V470T | N472 (TM3) | V470T, E472N, E473G, A474T, N642D |
| AE1 N473 | N473 (TM3) | E473N, F475S, N642D |
| AE1 N473/V470T | N473 (TM3) | V470T, E473N, F475S, N642D |
| AE1 N474 | N474 (TM3) | A474N, F475S, F476S, N642D |
| AE1 N475 | N475 (TM3) | F475N, F476S, N642D |
| AE1 N476 | N476 (TM3) | F476N, F478S, N642D |
| AE1 N477 | N477 (TM3) | S477N, F478S, C479S, N642D |
| AE1 N478 | N478 (TM3) | F478N, E480T, N642D |
| AE1 N479 | N479 (TM3) | C479N, E480G, N642D |
| AE1 N482 | N482 (TM3/EC2) | L484S, N642D |
| AE1 N487 | N487 (EC2) | I487N, G489S, N642D |
| (b) SAO constructs | ||
| Construct* | N-Glycosylation site | Mutation(s) |
| AE1 SAO | N642 (EC4) | Δ400–408 |
| SAO N642D | None | Δ400–408, N642D |
| SAO N431 | N431 (EC1) | Δ400–408, T431N, N433S, N642D |
| SAO N433 | N433 (EC1) | Δ400–408, M435S, N642D |
| SAO N434 | N434 (EC1) | Δ400–408, Q434N, G436S, N642D |
| SAO N441 | N441 (TM2) | Δ400–408, L441N, N642D |
| SAO N442 | N442 (TM2) | Δ400–408, I442N, N642D |
| SAO N446 | N446 (TM2) | Δ400–408, V446N, G448S, N642D |
| SAO N454 | N454 (TM2) | Δ400–408, L454N, A456S, N642D |
| SAO N455 | N455 (TM2) | Δ400–408, G455N, Q457S, N642D |
| SAO N456 | N456 (TM2) | Δ400–408, A456N, P458S, N642D |
| SAO N463 | N463 (TM3) | Δ400–408, G463N, F464S, N642D |
| SAO N464 | N464 (TM3) | Δ400–408, F464N, G466S, P467A, N642D |
| SAO N465 | N465 (TM3) | Δ400–408, S465N, P467S, N642D |
| SAO N475 | N475 (TM3) | Δ400–408, F475N, F476S, N642D |
| SAO N479 | N479 (TM3) | Δ400–408, C479N, E480G, N642D |
| SAO N482 | N482 (TM3/EC2) | Δ400–408, L484S, N642D |
* Constructs refer to the name given to each mutant in the present paper.
Transient transfection and expression of AE1 mutants
HEK-293 cells were transfected using the calcium phosphate precipitation method with 1 μg of plasmid DNA per well of a six-well plate. Cells were grown in DMEM supplemented with 10% (v/v) calf serum, 0.5% penicillin and 0.5% streptomycin under air/CO2 (19:1) at 37 °C [21]. At 1 day post-transfection, cells were labelled with EasyTag™ EXPRE35S35S protein labelling mix for 2 h at 37 °C, then washed with PBS and lysed with RIPA buffer (1% deoxycholate, 1% Triton X-100, 0.1% SDS, 0.15 M NaCl, 10 mM Tris/HCl, pH 7.5, and 1 mM EDTA). A rabbit polyclonal antibody directed against a synthetic peptide corresponding to the C-terminal 16 residues of human AE1 [21] and Protein G–Sepharose were used to immunoprecipitate AE1 proteins. The samples were analysed by concanavalin A lectin-shift SDS/PAGE [38] in order to resolve N-glycosylated (high mannose) from unglycosylated protein, followed by autoradiography. Multiple exposures on film were taken to ensure the signals were within a linear range of sensitivity. Signals were scanned and quantified by ImageJ software. The percentage N-glycosylation was calculated as the intensity of upper (glycosylated) band divided by the total intensity of upper and lower (unglycosylated) bands, in samples prepared from multiple independent transfections.
RESULTS
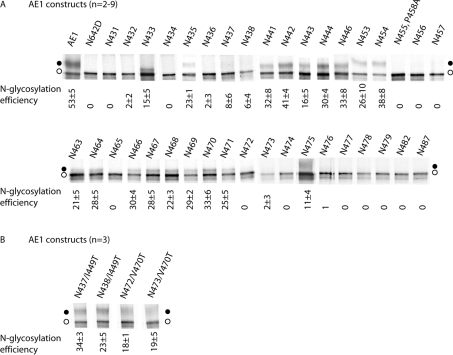
Single N-glycosylation acceptor sites were created in the region from EC1 to EC2 in AE1 (Figure 1 and Table 1). The endogenous N-glycosylation site at Asn642 in these constructs was mutated to Asp642 (Figure 1 and Table 1). To simplify the nomenclature, the mutants named below all contain the N642D mutation unless otherwise specified. The N-glycosylation mutants were transiently expressed in HEK-293 cells. AE1 expressed in transiently transfected HEK-293 cells retains a high-mannose oligosaccharide structure, although it can traffic to the plasma membrane [39]. At 1 day post-transfection, the cells were labelled with [35S]methionine for 2 h. AE1 exits the ER with a half-time of approx. 4 h [39], thus most of the protein produced in the 2-h labelling period represents protein still in the ER. The cells were then lysed with detergent, and AE1 proteins were immunoprecipitated using an antibody directed against the C-terminus. Samples were resolved by concanavalin A lectin-shift gel electrophoresis to separate the high-mannose glycosylated species from the unglycosylated species [38], and radiolabelled AE1 was visualized by autoradiography. Results are shown in Figure 2(A) and are summarized in a histogram in Figure 3. AE1 N642D was not glycosylated as expected, and marks the position of the non-glycosylated AE1 on the SDS gels (Figure 2A). AE1 was present as two bands. The upper band was susceptible to endoglycosidase H digestion [28], indicating that the protein contains high-mannose oligosaccharide. The lower band runs at the same position as AE1 N642D and represents non-glycosylated AE1. Note that the upper bands in the various lanes were shifted to different heights relative to the lower bands, since the samples were run on different lectin shift gels. The scanning N-glycosylation mapping results for AE1 constructs are summarized in Figure 3.
Figure 2. Scanning N-glycosylation mapping results for AE1 constructs.
HEK-293 cells transiently transfected with N-glycosylation mutants were labelled with [35S]methionine for 2 h. This represents proteins localized mainly to the ER. The cells were then lysed, and the cell lysate was subjected to immunoprecipitation with anti-AE1 antibody. The samples were separated by lectin-shift SDS/PAGE and analysed by autoradiography. (A) AE1 constructs with TM2/3 glycosylation acceptor site mutation. (B) AE1 constructs with TM2/3 glycosylation acceptor site and I449T or V470T mutations. N-Glycosylation efficiencies were calculated by dividing the upper band (glycosylated) intensity by the total intensity of upper and lower (non-glycosylated) bands, and listed beneath each lane (±S.D.). ●, Glycosylated proteins; ○, non-glycosylated proteins.
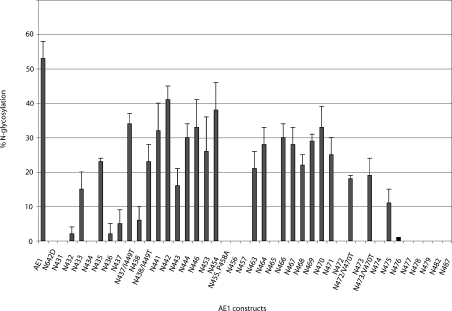
Figure 3. N-Glycosylation efficiencies of AE1 constructs.
The N-glycosylation efficiencies of various AE1 constructs expressed in HEK-293 cells are shown (±S.D.).
AE1 was glycosylated at Asn642 at 53±5% (Figure 2A), consistent with previous observations [28]. The presence of unglycosylated AE1 was probably due to overexpression of the protein in the transiently transfected HEK-293 cells. Previous experiments showed that an N-glycosylation site created at position 429 could not be glycosylated in an AE1 construct [20]. Our results show that AE1 N431 and N432 were also not glycosylated, probably because they were too close to the end of TM1 (Figure 2A). The downstream positions showed discontinuous stretches of glycosylation in HEK-293 cells (Figure 3). AE1 N433 was glycosylated, although not efficiently, at 15±5% (Figure 2A). No glycosylation was seen in AE1 N434, but AE1 N435 was glycosylated (23±1%). A very low level of glycosylation was again observed for AE1 N436, N437 and N438. Efficient N-glycosylation was observed in AE1 N441, N442, N443, N444, N446, N453 and N454: in most cases, more than 30%.
N-Glycosylation efficiencies abruptly dropped in positions immediately downstream of N454: AE1 N455 (results not shown), N456 and N457 were not glycosylated. To rule out the possibility that Pro458 may interfere with recognition of the N-glycosylation sequon by OST, the mutation P458A was introduced into AE1 N455, producing the construct AE1 N455, P458A. The P458A mutation did not improve the glycosylation efficiency of AE1 N455 (Figure 2A). N-Glycosylation was again observed in AE1 N463 (21±5%) and N464 (28±5%), but was absent in AE1 N465 (Figure 2A). The stretch of positions AE1 N466, N467, N468, N469, N470 and N471 were more than 20% glycosylated (Figure 2A). AE1 N472, N473 and N474 were not glycosylated, whereas AE1 N475 was weakly glycosylated (11±4%). Positions close to TM4, AE1 N476, N477, N478, N479, N482 and N487 were not N-glycosylated.
The discontinuous pattern of N-glycosylation was unexpected. Introduced sites that were a sufficient distance from the ends of adjacent TMs were expected to be N-glycosylated if the TM2-3 region is exposed to the ER lumen during biosynthesis. The introduction of novel N-glycosylation acceptor sites often results in a change in the polarity of the region. An increase in polarity in the region may enhance the exposure of the region to the lumen, allowing N-glycosylation to occur. In contrast, changes that do not increase polarity significantly may not alter the integration of TM2–3 into the membrane. To investigate the relationship of hydrophobicity of the TM (Table 2) and utilization of N-glycosylation acceptor sites created within the TM, additional mutants were created. These mutations were designed to decrease the hydrophobicity of TM2 or 3 (I449T or V470T) in select TM2 or TM3 glycosylation mutants (Tables 1 and 2) that showed poor glycosylation (Figure 2A). Hydrophobic residues were mutated to threonine, similar to mutations to create novel N-glycosylation sites, resulting in similar changes in polarity. These mutants were expressed in HEK-293 cells and were radiolabelled, and the immunoprecipitated AE1 proteins were resolved by lectin-shift SDS/PAGE (Figure 2B). While AE1 N437, N438, N472 and N473 were poorly or not glycosylated (Figure 2A), the glycosylation efficiencies of the same sites in N437/I449T, N438/I449T, N472/V470T and N473/V470T were greatly increased (Figure 2B) by the introduction of a polar residue elsewhere in the region.
Table 2. TM2 and 3 sequences and Kyte–Doolittle (KD) values of TM2 and TM3 mutants.
| Construct* | TM2 sequence amino acids 437–458† | TM3 sequence amino acids 459–481† | Average KD of TM2‡ | Average KD of TM3‡ |
|---|---|---|---|---|
| AE1 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.34 | 1.34 |
| AE1 N642D | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N431 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N432 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N433 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N434 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N435 | TSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.11 | 1.34 |
| AE1 N436 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N437 | NSTLLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.11 | 1.34 |
| AE1 N437/I449T | NSTLLISTAVQGTLFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.87 | 1.34 |
| AE1 N438 | VNGSLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.14 | 1.34 |
| AE1 N438/I449T | VNGSLISTAVQGTLFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.9 | 1.34 |
| AE1 N441 | VSELNISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.00 | 1.34 |
| AE1 N442 | VSELLNSTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.97 | 1.34 |
| AE1 N443 | VSELLINTTVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 1.1 | 1.34 |
| AE1 N444 | VSELLISNATQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.99 | 1.34 |
| AE1 N446 | VSELLISTANQSILFALLGAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.97 | 1.34 |
| AE1 N453 | VSELLISTAVQGILFANLSAQP | LLVVGFSGPLLVFEEAFFSFCET | 0.99 | 1.34 |
| AE1 N454 | VSELLISTAVQGILFALNGSQP | LLVVGFSGPLLVFEEAFFSFCET | 0.89 | 1.34 |
| AE1 N455 | VSELLISTAVQGILFALLNASP | LLVVGFSGPLLVFEEAFFSFCET | 1.32 | 1.34 |
| AE1 N455, P458A | VSELLISTAVQGILFALLNASA | LLVVGFSGPLLVFEEAFFSFCET | 1.47 | 1.34 |
| AE1 N456 | VSELLISTAVQGILFALLGNQS | LLVVGFSGPLLVFEEAFFSFCET | 1.13 | 1.34 |
| AE1 N457 | VSELLISTAVQGILFALLGANA | SLVVGFSGPLLVFEEAFFSFCET | 1.49 | 1.14 |
| AE1 N463 | VSELLISTAVQGILFALLGAQP | LLVVNSSGPLLVFEEAFFSFCET | 1.34 | 1.05 |
| AE1 N464 | VSELLISTAVQGILFALLGAQP | LLVVGNSSALLVFEEAFFSFCET | 1.34 | 1.2 |
| AE1 N465 | VSELLISTAVQGILFALLGAQP | LLVVGFNGSLLVFEEAFFSFCET | 1.34 | 1.26 |
| AE1 N466 | VSELLISTAVQGILFALLGAQP | LLVVGFSNATLVFEEAFFSFCET | 1.34 | 1.16 |
| AE1 N467 | VSELLISTAVQGILFALLGAQP | LLVVGFSGNLTVFEEAFFSFCET | 1.34 | 1.06 |
| AE1 N468 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPNLTFEEAFFSFCET | 1.34 | 0.83 |
| AE1 N469 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLNVSTEAFFSFCET | 1.34 | 0.99 |
| AE1 N470 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLNSTEAFFSFCET | 1.34 | 0.97 |
| AE1 N471 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVNGTAFFSFCET | 1.34 | 1.33 |
| AE1 N472 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFNGTFFSFCET | 1.34 | 1.37 |
| AE1 N472/V470T | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLTFNGTFFSFCET | 1.34 | 1.16 |
| AE1 N473 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFENASFSFCET | 1.34 | 1.25 |
| AE1 N473/V470T | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLTFENASFSFCET | 1.34 | 1.04 |
| AE1 N474 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEENSSSFCET | 1.34 | 0.8 |
| AE1 N475 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEANSSFCET | 1.34 | 0.91 |
| AE1 N476 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFNSSCET | 1.34 | 0.91 |
| AE1 N477 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFNSSET | 1.34 | 0.93 |
| AE1 N478 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSNCTT | 1.34 | 1.19 |
| AE1 N479 | VSELLISTAVQGILFALLGAQP | LLVVGFSGPLLVFEEAFFSFNGT | 1.34 | 1.22 |
| AE1 N482 | Unaltered | Unaltered | 1.34 | 1.34 |
| AE1 N487 | Unaltered | Unaltered | 1.34 | 1.34 |
* Constructs refer to the name given each mutant in the paper.
† Mutated sites are in bold.
‡ Altered Kyte–Doolittle scores are in bold.
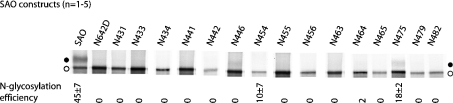
N-Glycosylation mutations within the EC1–2 region were also created in an AE1 SAO N642D background (Figure 1 and Table 1). [35S]Methionine-labelling of transiently transfected HEK-293 cells showed a different glycosylation pattern from that of the AE1 mutants (Figure 4). AE1 SAO had an N-glycosylation efficiency of 45±7% at the endogenous acceptor site, similar to that of AE1 (53±5%). The control AE1 SAO N642D was not glycosylated. Whereas AE1 constructs showed N-glycosylation at stretches of positions from EC1 to EC2, no detectable N-glycosylation was seen in the corresponding positions in AE1 SAO constructs (Figure 4). Faint upper bands that may be visible to the eye were present in some lanes (e.g. SAO N446), but these were not quantifiable owing to the high intensity of the lower band and probably represented less than 5% glycosylation. The first acceptor site to be utilized by OST in the SAO mutants was N475 (18±2%), near the end of TM3. This site was also N-glycosylated to a similar level (11±4%) in wild-type AE1.
Figure 4. Scanning N-glycosylation mapping results for AE1 SAO constructs.
Transiently transfected HEK-293 cells were radiolabelled with [35S]methionine and lysed, and AE1 was immunoprecipitated. The samples were analysed by lectin-shift SDS/PAGE and autoradiography. N-Glycosylation efficiencies were calculated by dividing the upper band (glycosylated) intensity by the total intensity of upper and lower (non-glycosylated) bands, and listed beneath each lane (±S.D.). ●, Glycosylated proteins; ○, non-glycosylated proteins.
DISCUSSION
In the present study, we performed scanning N-glycosylation mutagenesis on the region encompassing EC1–EC2 in AE1, and observed unexpected results that positions within TM2 and TM3 could be glycosylated. The corresponding region was not glycosylated in AE1 SAO, suggesting a disruption of membrane integration extending to TM3 due to the deletion ΔA400–A408 in TM1.
At least three factors contributed to determining whether a particular N-glycosylation acceptor site was utilized: (i) distance of the site to an adjacent TM; (ii) hydrophobicity of the segment involved; and (iii) protein–protein interactions requiring specific residues. The region close to the lumenal end of TM1 (N429 [20], N431, N432 and N434) was not glycosylated, since it could not reach the active site of the OST, which requires 12–14 residues from the ends of TMs to reach it [26]. Within TM2–3 region, efficient glycosylation was observed from N441 to N471, punctuated by positions that were not glycosylated (N455–N457 and N465).
Mutations that were required to generate the glycosylation acceptor sites in TMs 2 or 3 altered the average hydrophobicity [40] of the TM (Table 2). In all positions that were glycosylated, at least one hydrophobic residue had been mutated to the hydrophilic amino acids asparagine and/or serine/threonine (Table 1). Mutations in TM2–3 to introduce novel N-glycosylation sites that did not decrease hydrophobicity (Gly455-Ala-Gln-Pro sequence) did not result in N-glycosylation. We hypothesized the reduction in average hydrophobicity of the TM played a role in causing the normally embedded positions of TM2–3 to become lumenally exposed and accessible to the OST. N-Glycosylation of these sites would profoundly affect the structure of AE1, preventing them from integrating into the protein and trapping the segment in the ER lumen. Work on aquaporin-1 [41] has shown that regions that are membrane-embedded in membrane proteins may be transiently exposed to the ER lumen or cytosol during biosynthesis, and subsequently fold back into the protein. The trapped form of the N-glycosylation mutants may represent a folding intermediate of AE1 during biosynthesis, where TM2–3 becomes transiently exposed to the ER lumen.
We found a good correlation between altered hydrophobicity of the TM segments and utilization of the acceptor sites in the mutants: mutants with TMs having lower hydrophobicity were more likely to be glycosylated. The mutations may have also caused disruption of specific protein–protein interactions leading to exposure of TM2–3 to the ER lumen and subsequent N-glycosylation. It is conceivable that naturally occurring mutations that introduce a polar residue within a TM segment with marginal hydrophobicity may give rise to disease by severely disrupting the folding of the membrane domain in a similar manner to what was observed in the present study.
A refined folding model of AE1 TM1–4 from the present study is shown in Figure 5. As an estimate to the ends of TMs1 and 4, we took positions 435 (23±1% glycosylation) and 473 (19±5% for N473/V470T) as the glycosylation boundaries of the region encompassing EC1–EC2. Introduced N-glycosylation sites between these residues were efficiently N-glycosylated if the segment hydrophobicity was lowered as a result of mutagenesis. Applying the ‘12+14 rule’ puts the C-terminal end of TM1 at around Phe423 and the N-terminal end of TM4 at around Ile487. A previous N-glycosylation mapping study from our laboratory using a construct with an extended EC1 (1+/4− construct) suggested that the C-terminal end of TM1 in wild-type AE1 is around Pro419 [28]. The extension of EC1 may have an effect on the positioning of TM1 in the membrane. Phe423 is a more likely end of TM1, as it is followed by a pair of glycine residues that could serve as helix-terminators [42]. Ile487 marks the beginning of a hydrophobic TM4 segment that includes one arginine residue within the first turn of the helix (Figure 5). These estimates agree with existing topological data on EC1 [23–25], and are consistent with the region surrounding Tyr486 being extracellular [29,30]. As well, our new model (Figure 5) puts Glu480 in EC2, rather than within TM3 [21], and accounts for its association with the Fra blood group antigen [29,30]. The polar sequences Gly455-Ala-Pro-Gln linking TMs 2 and 3, and Glu508-Gly-Ser are placed on the cytosolic side of the membrane in our new model.
Figure 5. Refined model of the topology of TM1–4 in human AE1.
The first four TMs of human AE1 are shown, with residues involved in SAO deletion. The results of scanning N-glycosylation mapping are summarized. Position 429 cannot be N-glycosylated, as shown previously [20]. Residues are shaded according to their N-glycosylation efficiencies. Using the ‘12+14 rule’ places the lumenal end of TM1 at Phe423 and the beginning of TM4 at Ile487.
The TM 2 and 3 positions that were glycosylated in AE1 constructs (Figure 2A) did not show detectable glycosylation in SAO constructs (Figure 4). The first tested position to be glycosylated in SAO constructs was SAO N475, near the end of the proposed TM3. This site was also glycosylated in the wild-type AE1 construct. We had proposed that the population of membrane domain of AE1 SAO is probably a mixture of three possible configurations [28]: (i) SAO TMs 1–3 are membrane-integrated, with the C-terminus of SAO TM1 positioned similarly in the membrane as wild-type TM1; (ii) owing to altered interaction with SAO TM1, TMs 2 and 3 are extruded into the ER lumen; and (iii) SAO TMs 1 and 2 fail to insert into the membrane and are exposed to the cytosol. The present study indicates that the population of AE1 SAO protein predominantly exhibits scenario (iii). This is supported by the lack of glycosylation in TM2 positions and positions in the N-terminus of TM3 (N463 and N464) in the SAO constructs, suggesting that they were not lumenally exposed. Only the C-terminal part of TM3 (N475) in the N-glycosylation mutants was exposed to the ER lumen and OST.
We propose that TMs 2 and 3 in AE1 form a re-entrant loop (P-loop) structure similar to that found in other membrane proteins. The crystal structure of the Streptomyces lividans KcsA K+ channel [43] showed a P-loop that plays a key role in ion selectivity of the channel. Studies on the GAT-1 γ-aminobutyric acid transporter [44], the potassium channel ROMK1 [45], the Na+/K+-ATPase [46] and the human transient receptor potential 3 [47] have shown that N-glycosylation acceptor sites engineered into the P-loop region can become glycosylated. A possible P-loop has been identified in the membrane domain of human AE1, between TMs 9 and 10, via scanning N-glycosylation mutagenesis studies [20,21]. TMs 2 and 3 have been shown to be susceptible to proteolysis after alkaline treatment [33,34]. These category 2 peptides are not necessarily α-helical, but may be flexible [48]. Assessing the topogenic functions of each TM by protease-protection assay, TM2 showed a markedly lower ST function than most other TMs in AE1 [31]; TM3 was a poor SA and was unable to initiate translocation of a downstream reporter domain [31]. Gtag(2:3), where a glycosylation tag was present upstream of a construct of TM2–3, was not glycosylated, indicating that TM2 failed to integrate into the membrane in this construct [36]. Gtag(2:4), on the other hand, was glycosylated, suggesting that the presence of TM4 promotes membrane integration of TM2 [36]. In addition, proper membrane integration of TM2 requires the presence of TM1 in close proximity [32]. It appears that the membrane integration of TM2–3 requires the presence of TMs 1 and 4.
There is some evidence that the TM2–3 region may be involved in transport function of AE1. Wang et al. [35] expressed complementary fragments of the AE1 membrane domain in Xenopus oocytes and assayed for transport function. They found that the integrity of EC2 was essential for anion transport. We speculate that TM2–3 may form a flexible pore-lining structure, which normally folds into the protein, but is transiently exposed to the ER lumen during biosynthesis. N-Glycosylation of such a loop would trap it in the ER lumen [49], resulting in protein misfolding and likely loss of function.
References
- 1.Grinstein S., Ship S., Rothstein A. Anion transport in relation to proteolytic dissection of band 3 protein. Biochim. Biophys. Acta. 1978;507:294–304. doi: 10.1016/0005-2736(78)90424-8. [DOI] [PubMed] [Google Scholar]
- 2.Low P. S. Structure and function of the cytoplasmic domain of band 3: center of erythrocyte membrane–peripheral protein interactions. Biochim. Biophys. Acta. 1986;864:145–167. doi: 10.1016/0304-4157(86)90009-2. [DOI] [PubMed] [Google Scholar]
- 3.Tanner M. J. A. Band 3 anion exchanger and its involvement in erythrocyte and kidney disorders. Curr. Opin. Hematol. 2002;9:133–139. doi: 10.1097/00062752-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Jarolim P., Palek J., Amato D., Hassan K., Sapak P., Nurse G. T., Rubin H. L., Zhai S., Sahr K. E., Liu S. C. Deletion in erythrocyte band 3 gene in malaria-resistant Southeast Asian ovalocytosis. Proc. Natl. Acad. Sci. U.S.A. 1991;88:11022–11026. doi: 10.1073/pnas.88.24.11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohandas N., Winardi R., Knowles D., Leung A., Parra M., George E., Conboy J., Chasis J. Molecular basis for membrane rigidity of hereditary ovalocytosis: a novel mechanism involving the cytoplasmic domain of band 3. J. Clin. Invest. 1992;89:686–692. doi: 10.1172/JCI115636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schofield A. E., Reardon D. M., Tanner M. J. A. Defective anion transport activity of the abnormal band 3 in hereditary ovalocytic red blood cells. Nature (London) 1992;355:836–838. doi: 10.1038/355836a0. [DOI] [PubMed] [Google Scholar]
- 7.Takeshima Y., Sofro A. S., Suryantoro P., Narita N., Matsuo M. Twenty seven nucleotide deletion within exon 11 of the erythrocyte band 3 gene in Indonesian ovalocytosis. Jpn. J. Hum. Genet. 1994;39:181–185. doi: 10.1007/BF01915953. [DOI] [PubMed] [Google Scholar]
- 8.Moriyama R., Ideguchi H., Lombardo C. R., Van Dort H. M., Low P. S. Structural and functional characterization of band 3 from Southeast Asian ovalocytes. J. Biol. Chem. 1992;267:25792–25797. [PubMed] [Google Scholar]
- 9.Kopito R. R., Lodish H. F. Primary structure and transmembrane orientation of the murine anion exchange protein. Nature (London) 1985;316:234–238. doi: 10.1038/316234a0. [DOI] [PubMed] [Google Scholar]
- 10.Jennings M. L. Topography of membrane proteins. Annu. Rev. Biochem. 1989;58:999–1027. doi: 10.1146/annurev.bi.58.070189.005031. [DOI] [PubMed] [Google Scholar]
- 11.Reithmeier R. A. F., Chan S. L., Popov M. Structure of the erythrocyte band 3 anion exchanger. In: Konings W. N., Kaback H. R., Lolkema J. S., editors. Transport Processes in Eukaryotic and Prokaryotic Organisms, vol. 2. Amsterdam: Elsevier Science; 1996. pp. 281–309. [Google Scholar]
- 12.Wainwright S. D., Tanner M. J. A., Martin G. E., Yendle J. E., Holmes C. Monoclonal antibodies to the membrane domain of the human erythrocyte anion transport protein: localization of the C-terminus of the protein to the cytoplasmic side of the red cell membrane and distribution of the protein in some human tissues. Biochem. J. 1989;258:211–220. doi: 10.1042/bj2580211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman D. M., Reithmeier R. A. F. Localization of the carboxyl terminus of Band 3 to the cytoplasmic side of the erythrocyte membrane using antibodies raised against a synthetic peptide. J. Biol. Chem. 1988;263:10022–10028. [PubMed] [Google Scholar]
- 14.Jennings M. L. Anion transport proteins. In: Seldin D. W., Giebisch G., editors. The Kidney: Physiology and Pathophysiology. New York: Raven Press, Ltd; 1992. pp. 503–535. [Google Scholar]
- 15.Steck T. L., Ramos B., Strapazon E. Proteolytic dissection of band 3, the predominant transmembrane polypeptide of the human erythrocyte membrane. Biochemistry. 1976;15:1153–1161. doi: 10.1021/bi00650a030. [DOI] [PubMed] [Google Scholar]
- 16.Jennings M. L., Anderson M. P., Monaghan R. Monoclonal antibodies against human erythrocyte band 3 protein: localization of proteolytic cleavage sites and stilbenedisulfonate-binding lysine residues. J. Biol. Chem. 1986;261:9002–9010. [PubMed] [Google Scholar]
- 17.Bruce L. J., Ring S. M., Anstee D. J., Reid M. E., Wilkinson S., Tanner M. J. A. Changes in the blood group Wright antigens are associated with a mutation at amino acid 658 in human erythrocyte band 3: a site of interaction between band 3 and glycophorin A under certain conditions. Blood. 1995;85:541–547. [PubMed] [Google Scholar]
- 18.Zelinski T., Coghlan G., White L., Philipps S. The Diego blood group locus is located on chromosome 17q. Genomics. 1993;17:665–666. doi: 10.1006/geno.1993.1387. [DOI] [PubMed] [Google Scholar]
- 19.Bruce L. J., Anstee D. J., Spring F. A., Tanner M. J. A. Band 3 Memphis variant II. Altered stilbene disulfonate binding and the Diego (Dia) blood group antigen are associated with the human erythrocyte band 3 mutation Pro854→Leu. J. Biol. Chem. 1994;269:16155–16158. [PubMed] [Google Scholar]
- 20.Popov M., Tam L. Y., Li J., Reithmeier R. A. F. Mapping the ends of transmembrane segments in a polytopic membrane protein: scanning N-glycosylation mutagenesis of extracytosolic loops in the anion exchanger, band 3. J. Biol. Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 21.Popov M., Li J., Reithmeier R. A. F. Transmembrane folding of the human erythrocyte anion exchanger (AE1, Band 3) determined by scanning and insertional N-glycosylation mutagenesis. Biochem. J. 1999;339:269–279. [PMC free article] [PubMed] [Google Scholar]
- 22.Fujinaga J., Tang X. B., Casey J. R. Topology of the membrane domain of human erythrocyte anion exchange protein, AE1. J. Biol. Chem. 1999;274:6626–6633. doi: 10.1074/jbc.274.10.6626. [DOI] [PubMed] [Google Scholar]
- 23.Cobb C. E., Beth A. H. Identification of the eosinyl-5-maleimide reaction site on the human erythrocyte anion-exchange protein: overlap with the reaction sites of other chemical probes. Biochemistry. 1990;29:8283–8290. doi: 10.1021/bi00488a012. [DOI] [PubMed] [Google Scholar]
- 24.Jennings M. L., Nicknish J. S. Erythrocyte band 3 protein: evidence for multiple membrane-crossing segments in the 17 000-dalton chymotryptic fragment. Biochemistry. 1984;23:6432–6436. doi: 10.1021/bi00321a024. [DOI] [PubMed] [Google Scholar]
- 25.Jarolim P., Rubin H. L., Zakova D., Storry J., Reid M. E. Characterization of seven low incidence blood group antigens carried by erythrocyte band 3 protein. Blood. 1998;92:4836–4843. [PubMed] [Google Scholar]
- 26.Nilsson I. M., von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 27.Landolt-Marticorena C., Reithmeier R. A. F. Asparagine-linked oligosaccharides are localized to single extracytosolic segments in multi-span membrane glycoproteins. Biochem. J. 1994;302:253–260. doi: 10.1042/bj3020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung J. C., Reithmeier R. A. F. Membrane integration and topology of the first transmembrane segment in normal and Southeast Asian ovalocytosis human erythrocyte anion exchanger 1. Mol. Membr. Biol. 2005;22:203–214. doi: 10.1080/09687860500093115. [DOI] [PubMed] [Google Scholar]
- 29.McManus K., Lupe K., Coghlan G., Zelinski T. An amino acid substitution in the putative second extracellular loop of RBC band 3 accounts for the Froese blood group polymorphism. Transfusion. 2000;40:1246–1249. doi: 10.1046/j.1537-2995.2000.40101246.x. [DOI] [PubMed] [Google Scholar]
- 30.Jarolim P., Kalabova D., Reid M. E. Substitution Glu480Lys in erythroid band 3 corresponds to the Fra blood group antigen and supports existence of the second ectoplasmic loop of band 3. Transfusion. 2004;44:684–689. doi: 10.1111/j.1537-2995.2004.03291.x. [DOI] [PubMed] [Google Scholar]
- 31.Ota K., Sakaguchi M., Hamasaki N., Mihara K. Assessment of topogenic functions of anticipated transmembrane segments of human band 3. J. Biol. Chem. 1998;273:28286–28291. doi: 10.1074/jbc.273.43.28286. [DOI] [PubMed] [Google Scholar]
- 32.Ota K., Sakaguchi M., Hamasaki N., Mihara K. Membrane integration of the second transmembrane segment of band 3 requires a closely apposed preceding signal-anchor sequence. J. Biol. Chem. 2000;275:29743–29748. doi: 10.1074/jbc.M002468200. [DOI] [PubMed] [Google Scholar]
- 33.Hamasaki N., Kuma H., Ota K., Sakaguchi M., Mihara K. A new concept in polytopic membrane proteins following from the study of band 3 protein. Biochem. Cell Biol. 1998;76:729–733. doi: 10.1139/bcb-76-5-729. [DOI] [PubMed] [Google Scholar]
- 34.Hamasaki N., Okubo K., Kuma H., Kang D., Yae Y. Proteolytic cleavage sites of band 3 protein in alkali-treated membranes: fidelity of hydropathy prediction for band 3 protein. J. Biochem. (Tokyo) 1997;122:577–585. doi: 10.1093/oxfordjournals.jbchem.a021792. [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Groves J. D., Mawby W. J., Tanner M. J. A. Complementation studies with co-expressed fragments of the human red cell anion transporter (Band 3; AE1): the role of some exofacial loops in anion transport. J. Biol. Chem. 1997;272:10631–10638. doi: 10.1074/jbc.272.16.10631. [DOI] [PubMed] [Google Scholar]
- 36.Groves J. D., Tanner M. J. A. Topology studies with biosynthetic fragments identify interacting transmembrane regions of the human red-cell anion exchanger (band 3; AE1) Biochem. J. 1999;344:687–697. [PMC free article] [PubMed] [Google Scholar]
- 37.Groves J. D., Tanner M. J. A. Structural model for the organization of the transmembrane spans of the human red-cell anion exchanger (band 3; AE1) Biochem. J. 1999;344:699–711. [PMC free article] [PubMed] [Google Scholar]
- 38.Popov M., Li J., Reithmeier R. A. F. Resolution of glycoproteins by a lectin gel-shift assay. Anal. Biochem. 2000;279:90–95. doi: 10.1006/abio.1999.4443. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Quilty J., Popov M., Reithmeier R. A. F. Processing of N-linked oligosaccharide depends on its location in the anion exchanger, AE1, membrane glycoprotein. Biochem. J. 2000;349:51–57. doi: 10.1042/0264-6021:3490051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y., Turnbull I. R., Bragin A., Carveth K., Verkman A. S., Skach W. R. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reithmeier R. A. F. Characterization and modeling of membrane proteins using sequence analysis. Curr. Opin. Struct. Biol. 1995;5:491–500. doi: 10.1016/0959-440x(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 43.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 44.Clark J. A. Analysis of the transmembrane topology and membrane assembly of the GAT-1 γ-aminobutyric acid transporter. J. Biol. Chem. 1997;272:14695–14704. doi: 10.1074/jbc.272.23.14695. [DOI] [PubMed] [Google Scholar]
- 45.Schwalbe R. A., Wang Z., Bianchi L., Brown A. M. Novel sites of N-glycosylation in ROMK1 reveal the putative pore-forming segment H5 as extracellular. J. Biol. Chem. 1996;271:24201–24206. doi: 10.1074/jbc.271.39.24201. [DOI] [PubMed] [Google Scholar]
- 46.Schneider H., Scheiner-Bobis G. Involvement of the M7/M8 extracellular loop of the sodium pump alpha subunit in ion transport: structural and functional homology to P-loops of ion channels. J. Biol. Chem. 1997;272:16158–16165. doi: 10.1074/jbc.272.26.16158. [DOI] [PubMed] [Google Scholar]
- 47.Vannier B., Zhu X., Brown D., Birnbaumer L. The membrane topology of human transient receptor potential 3 as inferred from glycosylation-scanning mutagenesis and epitope immunocytochemistry. J. Biol. Chem. 1998;273:8675–8679. doi: 10.1074/jbc.273.15.8675. [DOI] [PubMed] [Google Scholar]
- 48.Hamasaki N., Abe Y., Tanner M. J. A. Flexible regions within the membrane-embedded portions of polytopic membrane proteins. Biochemistry. 2002;41:3852–3854. doi: 10.1021/bi011918l. [DOI] [PubMed] [Google Scholar]
- 49.Goder V., Bieri C., Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]