Abstract
The Lass (longevity-assurance homologue) family members, which are highly conserved among eukaryotes, function in ceramide synthesis. In the mouse, there are at least five Lass family members, Lass1, Lass2, Lass4, Lass5 and the hitherto uncharacterized Lass6. To investigate specific roles for each Lass member in ceramide synthesis, we cloned these five mouse proteins. Overproduction of any Lass protein in cultured cells resulted in an increase in cellular ceramide, but the ceramide species produced varied. Overproduction of Lass1 increased C18:0-ceramide levels preferentially, and overproduction of Lass2 and Lass4 increased levels of longer ceramides such as C22:0- and C24:0-ceramides. Lass5 and Lass6 produced shorter ceramide species (C14:0- and C16:0-ceramides); however, their substrate preferences towards saturated/unsaturated fatty acyl-CoA differed. In addition to differences in substrate preferences, we also demonstrated by Northern blotting that Lass family members are differentially expressed among tissues. Additionally, we found that Lass proteins differ with regard to glycosylation. Of the five members, only Lass2, Lass5 and Lass6 were N-glycosylated, each at their N-terminal Asn residue. The occurrence of N-glycosylation of some Lass proteins provides topological insight, indicating that the N-termini of Lass family members probably face the luminal side of the endoplasmic reticulum membrane. Furthermore, based on a proteinase K digestion assay, we demonstrated that the C-terminus of Lass6 faces the cytosolic side of the membrane. From these data we propose topology for the conserved Lag1 motif in Lass family members, namely that the N-terminal region faces the luminal side and the C-terminal region the cytosolic side of the endoplasmic reticulum membrane.
Keywords: ceramide synthase, sphingolipid, longevity assurance homologue (Lass), N-glycosylation, topology
Abbreviations: CHO, Chinese hamster ovary; Endo Hf, endoglycosidase H; ER, endoplasmic reticulum; EST, expressed sequence tag; FB1, fumonisin B1; GFP, green fluorescent protein; HA, haemagglutinin; HEK, human embryonic kidney; HOX, homeobox; LAG/Lag, longevity-assurance gene; Lass, longevity-assurance homologue; Lip1p, Lag1p/Lac1p-interacting protein; ORF, open reading frame; PNGase F, N-glycosidase F; TLC domain, TRAM/Lag1p/CLN8 (ceroid-lipofuscinoses, neuronal 8) domain; TM1 (etc.), transmembrane segment 1 (etc.); TRAM, translocating chain-associating membrane; TRH, TRAM homologue
INTRODUCTION
Sphingolipids are essential components of the eukaryotic plasma membrane. Ceramide is the precursor for all sphingolipids, and is generated from a sphingoid long-chain base and an amide-linked fatty acid. In mammalian cells, the ceramides incorporate sphingoid bases, mainly sphingosine (for ceramide) or dihydrosphingosine (for dihydroceramide), although phytosphingosine (forming phytoceramide) has been found in mammalian tissues [1,2]. Furthermore, ceramides exhibit a tissue-dependent bias for amide-linked fatty acids, characterized by chain length, degree of saturation and degree of hydroxylation [3–6]. For example, in brain, C18:0, C24:0 and C24:1 are the major fatty acids found in sphingolipids [3,5], with C18:0-ceramide as the primary short-chain ceramide. In contrast, C16:0 is the most abundant fatty acid in short-chain ceramides found in other cells, such as fibroblasts, endothelial cells and cells of the immune system [4,5].
Ceramide functions as a second messenger in a variety of cellular events, including apoptosis and differentiation [7,8]. Endogenous ceramide levels can be tightly controlled by various mechanisms, particularly activation of sphingomyelinase and de novo ceramide synthesis [9,10]. One of the key enzymes in the de novo pathway is dihydrosphingosine:N-acyltransferase (dihydroceramide synthase). LAG1 (longevity-assurance gene 1) and its homologue LAC1 have been identified in the yeast Saccharomyces cerevisiae as genes that are essential for ceramide synthesis [11,12]. Cells carrying a single deletion in lag1 or lac1 have no remarkable phenotype; however, lag1Δ lac1Δ double mutant cells have drastically reduced sphingolipid levels. In vitro assays using extracts from these cells demonstrated that LAG1 and LAC1 are essential key genes for the acyl-CoA-dependent ceramide synthesis reaction. However, at present it is not clear whether Lag1p and Lac1p are part of the catalytically active enzyme or are essential regulators of the ceramide synthase.
Recent studies have suggested that in mammalian cells there are four different Lag1p/Lac1p homologues: UOG1 (upstream of growth and differentiation factor 1)/Lass1 (longevity-assurance homologue 1), TRH3 [TRAM (translocating chain-associating membrane) homologue 3]/Lass2, TRH1/Lass4 and TRH4/Lass5 [designated hereafter by the MGD (mouse genome database) nomenclature Lass; see Table 1] [13–15]. Mouse Lass1, Lass4 and Lass5 were analysed after being expressed in cultured cells [13,15], whereas human Lass1, Lass2 and Lass4 were expressed in yeast and then characterized [14]. These studies revealed that each Lass member possesses a characteristic substrate preference for a particular fatty acyl-CoA, although the two different assay systems did not always produce accordant results (Table 1). For example, the Lass1 protein selectively regulates the synthesis of C18:0-containing sphingolipids in cultured cells [13], while microsomes made from Lass1-overexpressing lag1Δ lac1Δ yeast cells exhibited mainly C26:0-CoA-dependent ceramide synthase activity [14] (Table 1). Moreover, Lass4 expressed in mammalian cells increased mainly C18:0- and C20:0-CoA-dependent ceramide synthase activity [15], whereas Lass4 expressed in yeast cells showed a substrate preference for C20:0- and C24:0-CoA [14] (Table 1).
Table 1. Reported fatty acyl-CoA preferences of mouse Lass family members.
The Lass name is used in accordance with the mouse genome database (MGD) (http://www.informatics.jax.org/mgihome/submissions/submissions_menu.shtml). Preferred fatty acyl-CoAs have been reported for overexpressing cells [13,15] (*) and for lag1Δ lac1Δ yeast cells [14] (†). Percentage sequence identity and similarity are relative to Lass6. n.d., not determined.
| Current name | Old name | Accession no. | Identity/similarity (%) | Preference |
|---|---|---|---|---|
| Lass1 | UOG1*† | NM_138647 | 16.0/27.4 | C18:0*, C26:0† |
| Lass2‡ | trh3, clone 1† | BC006847 | 35.6/50.9 | C24:0† |
| Lass4 | trh1*, clone 4† | BC003946 | 37.2/51.2 | C18:0*, C20:0*†, C24:0† |
| Lass5 | trh4* | BC046797 | 61.7/68.2 | C16:0* |
| Lass6 | trh1-like | BC057629 | 100/100 | n.d. |
‡ We found that the human Lass2 reported previously [34] was truncated at its N-terminus. Since the truncated N-terminus was essential for its activity (results not shown), the mouse Lass2 used here contained an additional 150 amino acids compared with the previously reported human protein.
In the present study, we have cloned the four known mouse Lass family members, Lass1, Lass2, Lass4 and Lass5, as well as the new, previously uncharacterized Lass6, and characterized them using a uniform set of assays. Each protein exhibited a unique substrate preference for chain length and/or saturation of fatty acyl-CoAs. Additionally, tissue expression patterns differed for the proteins. We also found that Lass2, Lass5 and Lass6 were N-glycosylated on their N-terminal Asn residues. Using these results, together with a proteinase K digestion assay used to investigate the C-terminal orientation of the Lass6 protein, we propose a topology for the conserved Lag1 motif in Lass family members.
EXPERIMENTAL
Cell culture and transfection
HEK (human embryonic kidney) 293T cells were grown in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum, in a humidified atmosphere of 5% CO2 at 37 °C. CHO (Chinese hamster ovary) cells were similarly grown in Ham's F-12 medium containing 10% (v/v) fetal bovine serum. Cells were transfected with the indicated cDNA using Lipofectamine Plus® (Invitrogen, Carlsbad, CA, U.S.A.), according to the manufacturer's directions. Lysates of transiently transfected cells were prepared 24 h after transfection.
Cloning and plasmid construction
An EST (expressed sequence tag) clone (GenBank® accession number BC057629) containing a Lass6 ORF (open reading frame) was used to amplify Lass6 cDNA by PCR with the primers 5′-CGGATCCGCCACCATGGCAGGGATCTTAGCCTG-3′ and 5′-CTTCCAGGAACAAACAGTTCC-3′ (Lass6-L). The amplified DNA fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI, U.S.A.) to generate pGEM-Lass6A. pcDNA3-HA-Lass6 was then constructed by cloning a 1.2 kb BamHI/EcoRI fragment from pGEM-Lass6A into the BamHI/EcoRI site of pcDNA3-HA, which had been constructed to create an N-terminally HA (haemagglutinin)-tagged gene from pcDNA3 (Invitrogen). The Lass6 fragment used to construct the C-terminally enhanced GFP (green fluorescent protein)-fused gene was amplified using the primers 5′-GGCCACCATGGCAGGGATCTTAGCCTG-3′ and 5′-GGGATCCGAATCATCCACGGAACAAGGAC-3′, and the amplified DNA fragment was then cloned into pGEM-T Easy vector to generate pGEM-Lass6B. Next, pEGFP-Lass6 was constructed by cloning the 1.2 kb EcoRI/BamHI fragment pGEM-Lass6B into the EcoRI/BamHI site of pEGFP-N1 (Clontech, BD Biosciences, Palo Alto, CA, U.S.A.).
The cDNAs containing the other Lass family ORFs were cloned similarly into pcDNA3-HA using the appropriate EST clones and primers. Specifically, for pcDNA3-HA-Lass1, we used the EST clone AW106349 and primers 5′-CGGATCCGATAGCATGGCTGCTGCCG-3′ and 5′-AAGAGACTCAGAAGAGCTTGTCCTTC-3′; for pCDNA3-HA-Lass2, we used the EST clone BI329498 and primers 5′-CGTCGACATGCTCCAGACCTTGTATGAC-3′ and 5′-GTCAGTCATTCTTAGGATGATTG-3′; for pCDNA3-HA-Lass4, we used the EST clone AA672434 and primers 5′-CGTCGACATGTCGTTCAGCTTGAGTGAG-3′ and 5′-GTCTATGTGGCCCGGGTGTGC-3′; and for pCDNA3-HA-Lass5, we used the EST clone BF785764 and primers 5′-CGGATCCCGTAAGATGGCGACTGCAGCAGCG-3′ and 5′-TCTAGTCACAGGAGTGTAGATGTG-3′. Although we also tried to find the mouse homologue of human Lass3 (GenBank accession no. BC010032), we could not find it in any mouse EST databases.
Site-directed mutagenesis
Oligonucleotide-mediated site-directed mutagenesis was performed in vitro using the QuikChange™ kit (Stratagene, La Jolla, CA, U.S.A.) according to the manufacturer's instructions. Asn residues at positions 18 and 285 of Lass6, 26 of Lass5 and 19 of Lass2 were replaced with Gln. The following primers were used in preparing HA–Lass constructs for each mutated gene: for pcDNA3-HA-Lass6-N18Q, 5′-CGGTTTTGGCTTCCGCACCAAGTCACCTGGGCAG-3′ and 5′-CTGCCCAGGTGACTTGGTGCGGAAGCCAAAACCG-3′; for pcDNA3-HA-Lass6-N285Q, 5′-CTCTCTGGGTGTTGCAAACCACATTATTTGAAAG-3′ and 5′-CTTTCAAATAATGTGGTTTGCAACACCCAGAGAG-3′; for pcDNA3-HA-Lass5-N26Q, 5′-CTGGCTGCCTCAGCAAGTGAGCTGGGCGGAC-3′ and 5′-GTCCGCCCAGCTCACTTGCTGAGGCAGCCAG-3′; and for pcDNA3-HA-Lass2-N19Q, 5′-CTATGGCTGCCTGTGCAATTAACCTGGGCTG-3′ and 5′-CAGCCCAGGTTAATTGCACAGGCAGCCATAG-3′.
Ceramide synthase assay
In vitro dihydroceramide synthase assays were performed essentially as described previously [16] with minor modifications. Briefly, HEK 293T cells transfected with the indicated plasmid were suspended in buffer A [50 mM Hepes/NaOH, pH 7.5, 1×protease inhibitor mixture (Complete™; Roche Molecular Biochemicals, Indianapolis, IN, U.S.A.) and 0.5 mM dithiothreitol], and lysed by sonication. After removal of cell debris by centrifugation, the resulting total cell lysates (20–40 μg of protein) were mixed with 5 μM dihydrosphingosine (Biomol, Plymouth Meeting, PA, U.S.A.), 0.2 μCi of [4,5-3H]D-erythro-dihydrosphingosine (50 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO, U.S.A.) and 25 μM fatty acyl-CoA in buffer B (50 mM Hepes/NaOH, pH 7.5, 0.5 μM dithiothreitol and 1 mM MgCl2) in a volume of 100 μl. Digitonin (final concentration 0.1%) was added to those assays using a long-chain acyl-CoA (C20:0–C26:0). After incubation for 15 min at 37 °C, the reaction was stopped with 100 μl of chloroform/methanol (1:1, v/v), and the lipids were extracted by the method of Bligh and Dyer [17]. The dried lipids were resuspended in 30 μl of chloroform/methanol (2:1, v/v), and separated on Silica Gel 60 TLC plates (Merck, Whitestation, NJ, U.S.A.), using chloroform/methanol/2 M ammonium hydroxide (40:10:1, by vol.). The TLC plates were sprayed with the fluorographic reagent EN3HANCE™ (PerkinElmer Life Sciences, Ontario, Canada), dried, and exposed to X-ray films at −80 °C. For quantification, the silica was scraped from the TLC plates, and the radioactivity was measured using a liquid scintillation counter (LSC-3600; Aloka, Tokyo, Japan). The following fatty acyl-CoAs were used for this assay: myristoyl (C14:0)-CoA, palmitoyl (C16:0)-CoA, stearoyl (C18:0)-CoA, oleoyl (C18:1)-CoA, arachidoyl (C20:0)-CoA (all from Sigma, St. Louis, MO, U.S.A.), behenoyl (C22:0)-CoA, lignoceroyl (C24:0)-CoA and hexacosanoyl (C26:0)-CoA (all from American Radiolabeled Chemicals).
[3H]Dihydrosphingosine labelling assay
HEK 293T cells transfected with the indicated plasmid were incubated with or without 20 μM FB1 (fumonisin B1) for 6 h, then metabolically labelled with 1.0 μCi of [4,5-3H]D-erythro-dihydrosphingosine by incubation at 37 °C for 4 h. The cells were washed with PBS, followed by lipid extraction as described previously [18]. The labelled lipids were separated on Silica Gel 60 HPTLC plates (Merck) using chloroform/methanol/acetic acid (190:9:1, by vol.). After drying, the lipids were separated again by HPTLC with the same solvent system.
Northern blot analysis
Analysis was performed on mouse poly(A)+ blots containing 2 μg of poly(A)+ RNA from adult mouse tissues (Origene, Rockville, MD, U.S.A.). The Lass6 probe was prepared from a DNA fragment corresponding to the N-terminal 600 bp of the Lass6 ORF, which had been amplified from pGEM-Lass6A using the primers 5′-ATGGCAGGGATCTTAGCCTG-3′ and 5′-GTCCTTCCTTTTGATATCTG-3′. The amplified fragment was then labelled with [32P]dCTP using a Random Primer DNA Labeling Kit (TAKARA, Shiga, Japan). The other Lass probes were prepared similarly using DNA fragments that included the N-terminal 600 bp of the BamHI/KpnI fragment of pGEM-Lass1, the N-terminal 500 bp of the EcoRV fragment of pcDNA3-HA-Lass2, the C-terminal 700 bp of the KpnI/PstI fragment of pGEM-Lass4, and the C-terminal 700 bp of the XhoI fragment of pcDNA3-HA-Lass5. The β-actin cDNA control probe was purchased from Origene. Hybridization was carried out in ExpressHyb buffer (Clontech, BD Biosciences) for 5 h at 68 °C.
Deglycosylation of Lass2, Lass5 and Lass6 proteins
HEK 293T cells, transfected with the indicated plasmids, were washed with cold PBS, then suspended in buffer A and lysed by sonication. For deglycosylation, the cell lysates were denatured with 10×denaturing buffer (5% SDS and 10% β-mercaptoethanol) at 37 °C for 10 min. Each denatured sample was further treated with 1000 units of Endo Hf (endoglycosidase H; BioLabs Inc., Beverly, MA, U.S.A.) or 500 units of PNGase F (N-glycosidase F; BioLabs Inc.) for 1 h at 37 °C according to the manufacturer's instructions.
Proteinase K digestion
Digestion with proteinase K was performed as described previously [19].
Immunoblotting
Total cell lysates were denatured in SDS sample buffer (final: 10 mM Tris/HCl, pH 6.8, 0.4% SDS, 2% glycerol and 5% β-mercaptoethanol) for 10 min at 37 °C. Samples were separated on an SDS/10%-polyacrylamide gel and transferred to PVDF microporous membranes, which were then blocked with 5% (w/v) skimmed milk in TBS-T (20 mM Tris/HCl, pH 7.5, 137 mM NaCl and 0.05% Tween 20). The membranes were incubated with the anti-HA antibody HA7 (1:2000 dilution; Sigma), anti-GFP antibodies (0.8 μg/ml; Roche Diagnostics) or anti-calnexin antibodies (0.1 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) for 60 min at room temperature. The membranes were then washed, and incubated with the appropriate horseradish peroxidase-coupled secondary antibody (1:5000 dilution; Amersham Biosciences, Piscataway, NJ, U.S.A.) for 60 min at room temperature. After a wash with TBS-T, the membranes were developed using an enhanced chemiluminescence system (ECL® kit; Amersham Biosciences) and visualized using X-ray film.
Immunofluorescence microscopy
Immunofluorescence microscopy was performed as described previously [19]. The anti-HA antibody HA7 (1:200 dilution; Sigma) and an anti-KDEL antibody (1 μg/ml; Molecular Probes, Inc., Eugene, OR, U.S.A.) were used as primary antibodies. Alexa 488-conjugated anti-rabbit (5 μg/ml; Molecular Probes) and Alexa 594-conjugated anti-mouse (5 μg/ml; Molecular Probes) antibodies were used as secondary antibodies.
RESULTS
Mouse Lass6 is a member of the mouse Lass family
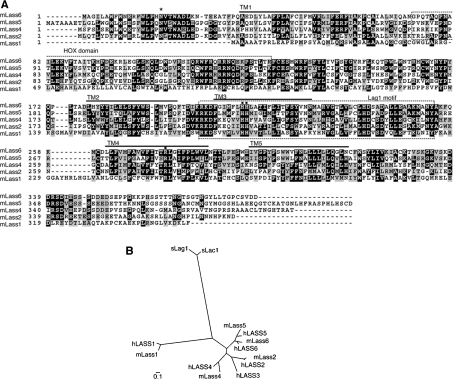
To understand the contribution of each Lass protein to the diversity of the total cellular ceramide composition, we first cloned all available mouse Lass members (Lass1, Lass2, Lass4 and Lass5), as well as the previously uncharacterized member Lass6. The Lass6 gene, which had been registered previously with GenBank® (accession number BC057629), encodes a 384-amino-acid protein with a predicted molecular mass of 44.8 kDa. The Lass6 protein shares significantly high identity with other Lass family members (Figure 1A). It exhibits the highest identity with Lass5 (61.7% identity and 68.2% similarity) and the lowest identity with Lass1 (16.0% identity and 27.4% similarity) (Table 1).
Figure 1. Sequence comparison of five mouse Lass family members.
(A) Amino acid sequence alignment for mouse proteins Lass1, Lass2, Lass4, Lass5 and Lass6. GenBank® accession numbers are shown in Table 1. The alignment was generated using the ClustalW (http://www.ch.embnet.org/software/ClustalW.html) and BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html) programs. Black boxes indicate identical residues, and grey boxes show amino acid similarity. Lass6 transmembrane segments (TM1–TM5) predicted by TopPredII are underlined [23]. The HOX domain and Lag1 motif are indicated and enclosed in boxes formed by broken lines. The conserved His and Arg residues (■) and the conserved N-glycosylation site (*) are indicated. (B) Dendrogram of the known mouse (m) and human (h) Lass family members and yeast (s) Lag1p/Lac1p. Sequences from the Lass family members of mouse, human and yeast were aligned using ClustalW. GenBank® accession numbers of the mouse family members are shown in Table 1. The GenBank accession numbers for human Lass family members are M62302 (hLASS1), BC010032 (hLASS2), BC010032 (hLASS3), AK022151 (hLASS4), BC032565 (hLASS5) and BC030800 (hLASS6), and those for yeast Lag1p/Lac1p members are NP_011860 (sLag1p) and NP_012917(sLac1p). The scale bar indicates the genetic distance for 0.1 amino acid substitution per site.
Lass proteins are known to contain a TLC [TRAM/Lag1p/CLN8 (ceroid-lipofuscinoses, neuronal 8)] homology domain (SMART accession no. SM00724) of approx. 200 amino acid residues [20,21]. This domain contains a single Arg residue that is strictly conserved, and, often, two consecutive His residues. In Lass6 the TLC domain encompasses the region from Thr-130 to Ser-331, with conserved residues at Arg-275, His-211 and His-212 aligning with those of the other Lass family members (Figure 1A). Within the TLC domain, there is a region called the Lag1 motif (Figure 1A, boxed), which exhibits high identity among Lass family members and includes 17 conserved residues. Additionally, some Lass family members (Lass2, Lass4 and Lass5, but not Lass1 or yeast Lag1p or Lac1p) also contain a HOX (homeobox) domain (SMART accession no. SM00389), which is conserved in some transcription factors involved in developmental regulation [21,22]. Lass6 also contains a HOX domain, from Gly-73 to Arg-131 (Figure 1A, boxed).
The TopPredII program [23] predicted that Lass6 comprises up to five transmembrane-spanning domains (TM1, amino acid residues 36–56; TM2, 178–198; TM3, 204–224; TM4, 262–282; TM5, 294–314) (Figure 1A, underlined). Four of these (TM1, TM3, TM4 and TM5) exhibited high probability scores for being membrane-spanning regions (TM1, 1.2417; TM3, 1.7417; TM4, 1.5760; TM5, 1.3135), whereas the score for TM2 was relatively low (0.8010).
Characterization of Lass6
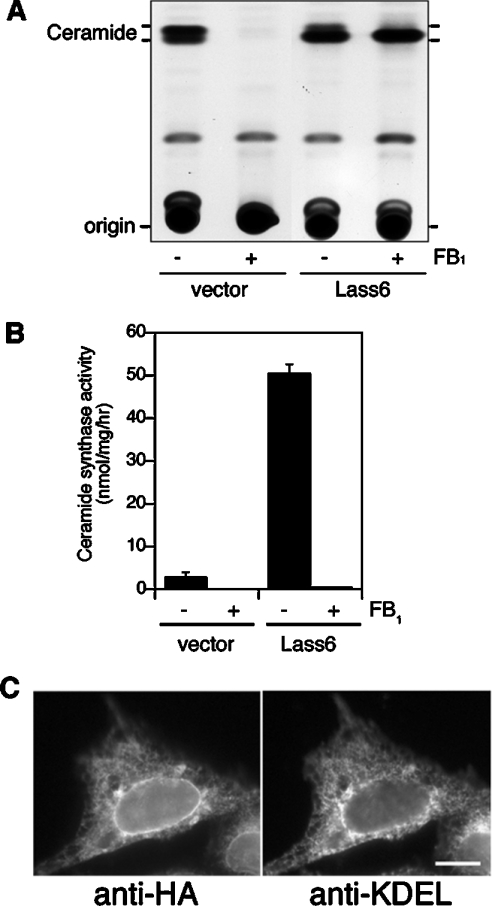
On beginning to characterize the Lass6 protein, we investigated its possible involvement in ceramide synthesis. HEK 293T cells were transiently transfected with control vector or pcDNA3-HA-Lass6 (encoding N-terminally HA-tagged Lass6), then metabolically labelled with [3H]dihydrosphingosine. The cells transfected with the control vector produced two lipid bands (Figure 2A). The upper and lower bands exhibited migration identical to that of unlabelled C16:0-ceramide and C24:0-ceramide respectively (results not shown), so correspond to long-chain and short-chain ceramides. In HEK 293T cells, sphingolipids are composed mainly of long-chain ceramides such as C24:0- and C24:1-ceramides, as well as short-chain ceramides such as C16:0-ceramide [13,15]. In vector-transfected cells, the amount of long-chain ceramide was higher than that of short-chain ceramide, whereas short-chain ceramide was prominent in the HA–Lass6-overproducing cells (Figure 2A). Treatment with 20 μM FB1, a well-known ceramide synthase inhibitor, completely inhibited ceramide synthesis in HEK 293T cells transfected with the control vector. In contrast, in the presence of FB1, HA-Lass6-overexpressing cells specifically produced ceramide with short-chain fatty acids. These results suggest that the Lass6 protein is involved in the synthesis of short-chain (dihydro)ceramides.
Figure 2. Overproduction of Lass6 induces ceramide synthesis in mammalian cells.
(A) Lass6 enhances ceramide synthesis in cultured cells. HEK 293T cells were transfected with pcDNA3 (vector) or pcDNA3-HA-Lass6 (Lass6) and incubated for 4 h in medium containing 1 μCi of [3H]dihydrosphingosine in the presence or absence of FB1 (20 μM). Lipids were extracted and separated by TLC. (B) Sensitivity to FB1 of Lass6 activity in vitro. Total cell lysates were prepared from HA–Lass6-overproducing HEK 293T cells or from cells transfected with control vector, and preincubated for 1 h on ice with (+) or without (−) FB1 (20 μM). A dihydroceramide synthase assay was performed by incubating 25 μM C16:0-CoA, 5 μM dihydrosphingosine (including [3H]dihydrosphingosine) and cell lysates from control (40 μg of protein) or HA–Lass6-overproducing (20 μg of protein) cells for 15 min at 37 °C. Results shown are means±S.D. from three independent experiments. (C) Lass6 is localized in the ER. CHO cells transfected with pcDNA3-HA-Lass6 were immunostained with anti-HA (left panel) and anti-KDEL (right panel) antibodies. Bar, 10 μm.
We next examined the activity of Lass6 in dihydroceramide synthesis in vitro. Total lysates prepared from HEK 293T cells transfected with control vector or pcDNA3-HA-Lass6 were incubated with [3H]dihydrosphingosine and C16:0-CoA. Dihydroceramide synthase activity in lysates from HA–Lass6-overproducing cells was ∼18.5-fold higher than the activity in control cells (Figure 2B). Treatment with 20 μM FB1 completely inhibited this activity. Such a discrepancy in sensitivity to FB1 between in vitro and in vivo assays was also reported for cells overproducing Lass1, Lass4 and Lass5 [13,15]. Thus Lass6 appears to share similar enzymatic characteristics with other Lass members.
In previous studies, Lass1, Lass4 and Lass5 were shown to be localized in the ER (endoplasmic reticulum) [13,15]. To investigate whether Lass6 is also localized in this compartment, indirect immunofluorescence microscopy was performed on CHO cells transfected for 24 h with a pcDNA3-HA-Lass6 plasmid. HA–Lass6 was detected on the nuclear envelope and as a reticular structure in the perinuclear and cytoplasmic regions (Figure 2C, left panel). This staining pattern was similar to that of the ER marker KDEL (Figure 2C, right panel) and their patterns did indeed overlap (results not shown). The signal for HA–Lass6 was high at the nuclear envelope and the perinuclear region of the reticular structure, but weaker in the cytoplasmic region. In contrast, the anti-KDEL antibodies stained the nuclear envelope only weakly. Similar results were observed for the ER localization of Lass2 in CHO cells transfected with a pcDNA3-HA-Lass2 plasmid (results not shown). These results indicate that Lass6 is an ER-resident protein, similar to other Lass members, including Lass2.
Lass2, Lass5 and Lass6 are glycosylated
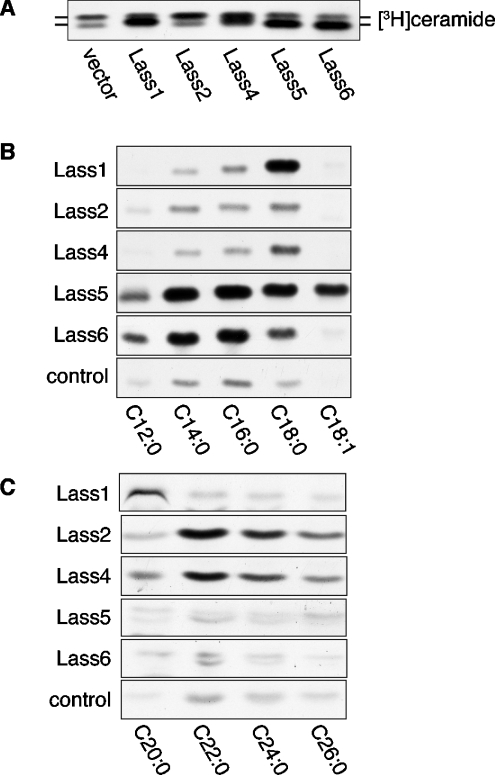
We expressed mouse Lass1, Lass2, Lass4, Lass5 and Lass6 in HEK 293T cells as HA-tagged proteins and detected them by immunoblotting (Figure 3A). Although HA–Lass1 and HA–Lass4 were each detected as a single band, three bands were observed for HA–Lass2 and two bands each for HA–Lass5 and HA–Lass6. Since these proteins contain predicted N-glycosylation motifs (NXT, NXS), we investigated the possibility that Lass2, Lass5 and Lass6 are modified by N-glycosylation. We incubated lysates from HA–Lass6-overproducing cells with PNGase F, which removes the entire carbohydrate chain from the Asn residue of N-linked glycoproteins. On PNGase F treatment the upper band disappeared, so that only the lower band was apparent (Figure 3B). Furthermore, treatment with Endo Hf, which cleaves within the sugar core of high-mannose and some hybrid oligosaccharides of N-linked glycoproteins, also resulted in a single, lower band. These results confirm that Lass6 is modified by N-glycosylation of a high-mannose and/or hybrid type.
Figure 3. Lass2, Lass5 and Lass6 are N-glycosylated.
(A) Confirmation of the overexpression of mouse Lass family members in HEK 293T cells by immunoblotting. HEK 293T cells were transfected with pcDNA3-HA (vector), pcDNA3-HA-Lass1, pcDNA3-HA-Lass2, pcDNA3-HA-Lass4, pcDNA3-HA-Lass5 or pcDNA3-HA-Lass6. Total cell lysates (5 μg of protein) were separated on an SDS/10%-polyacrylamide gel and then transferred to membranes. Immunoblotting was then performed using an anti-HA antibody. (B) PNGase F and Endo Hf digestion assay. Denatured samples were prepared from HEK 293T cells transfected with pcDNA3-HA-Lass6, pcDNA3-HA-Lass5 or pcDNA3-HA-Lass2. After incubating with buffer alone, PNGase F or Endo Hf, the cell lysates (5 μg of protein) were separated and subjected to immunoblotting as in (A). An asterisk (*) indicates a glycosylated protein.
We also examined the N-glycosylation of Lass5, which appears as two bands, and Lass2, which has three bands (Figure 3A). After treatment with PNGase F and Endo Hf, the upper band of HA–Lass5 shifted to the lower position (Figure 3B). Similarly, the upper band of Lass2 disappeared and the two lower bands remained after PNGase F and Endo Hf treatment. This indicates that both Lass5 and Lass2 are also N-glycosylated (Figure 3B). The reasons behind the unglycosylated forms of Lass2 appearing as two bands are unclear. It is possible that the lower band represents a proteolytically truncated form of the protein.
Comparison of the (dihydro)ceramide synthase activities of mouse Lass family members
Next, we examined the in vivo (dihydro)ceramide synthase activity of Lass family members using a metabolic labelling assay (Figure 4A). Control HEK 293T cells produced two bands corresponding to long-chain ceramide (upper band) and short-chain ceramide (lower band). Overproduction of either Lass member resulted in an increase in ceramide levels, although the ceramide species varied. Overproduction of HA–Lass1, HA–Lass5 or HA–Lass6 increased the levels of short-chain ceramide. On the other hand, only long-chain ceramide was increased in the HA–Lass2-overproducing cells. When HA–Lass4 was expressed, a ceramide exhibiting an intermediate mobility on TLC appeared, in addition to the increase in long-chain ceramide.
Figure 4. Comparison of ceramide synthesis activity of Lass family members.
(A) Lass proteins increase ceramide synthesis in cultured cells. HEK 293T cells transfected with plasmids as in Figure 3(A) were incubated for 4 h in medium with 1 μCi of [3H]dihydrosphingosine. Lipids were extracted and separated by TLC. (B) Comparison of in vitro ceramide synthase activities of Lass family members on fatty acyl-CoAs of various lengths and saturation. Samples (20 μg of protein) from the same cell lysates as used in Figure 3(A) were incubated with [3H]dihydrosphingosine/5 μM dihydrosphingosine and the indicated fatty acyl-CoA (25 μM) at 37 °C for 15 min. This experiment was performed three times with identical results. (C) Comparison of long-chain ceramide synthase activities of Lass proteins. Samples (40 μg of protein) from the cell lysates were incubated with [3H]dihydrosphingosine/5 μM dihydrosphingosine, the indicated fatty acyl-CoA (25 μM) and 0.1% digitonin (final concentration) at 37 °C for 15 min. The experiment was performed three times with identical results.
To compare the substrate specificity of Lass family members more precisely, we examined dihydroceramide synthase activities in vitro using various fatty acyl-CoAs (25 μM) and [3H]dihydrosphingosine (Figures 4B and 4C). Overproduction of Lass1 induced C18:0-CoA-specific ceramide synthesis, similar to a previous report [13], and the cells also exhibited weak C20:0-ceramide synthesis. The detected levels of C12:0-, C14:0- and C16:0-ceramides were the same as for the control vector-transfected cells. Interestingly, while C18:0-CoA was the best substrate for Lass1-dependent ceramide synthesis, C18:1-CoA, which has the same chain length as C18:0, was not a good substrate at all. Lass2 and Lass4 preferred fatty acyl-CoAs with longer chain lengths than those preferred by Lass1. Both C22:0- and C24:0-CoAs were used effectively for Lass2- and Lass4-dependent ceramide synthesis, and C26:0-, C20:0- and C18:0-CoAs also acted as substrates, albeit weakly. Lass5, which presented the highest identity to Lass6 (Figure 1), also had the most similar substrate preference. Both Lass5- and Lass6-overproducing cells exhibited high dihydroceramide synthesis activity using C16:0- and C14:0-CoAs and low activity with C12:0- and C18:0-CoAs. On the other hand, whereas the Lass5-overproducing cells demonstrated significant C18:1-ceramide synthesis, the Lass6-overproducing cells did not show any such increase. Thus Lass5 and Lass6 have similar preferences for fatty acyl-CoA length, yet different activities towards unsaturated fatty acyl-CoA. In summary, Lass family members each have distinct preferences for fatty acyl-CoAs, although with some overlap.
Tissue distribution of the mRNA expression of mouse Lass family members
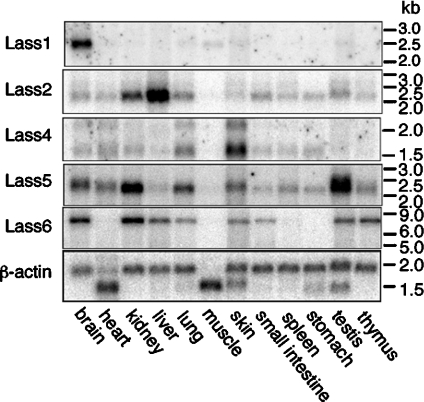
To compare the distributions of the five mouse Lass family members, we performed Northern blot analyses of poly(A)+ RNAs from 12 different mouse tissues (Figure 5). Lass1 was detected in brain as a 2.6 kb band and just weakly in muscle, indicating that expression is mostly brain-specific. In contrast, mRNAs for Lass2, Lass4, Lass5 and Lass6 were found to be mostly ubiquitous, although no expression was detected in muscle for any of these. Lass2 was detected as a 2.2 kb band, with the highest expression in liver, but also high expression in kidney. A major band at 1.6 kb and a minor band at 2.2 kb were observed for Lass4, and were quite strong in skin, with no apparent expression in muscle, testis or thymus. The highest expression of the 2.2 kb major band of Lass5 was in testis, and the expression in kidney was also high. A 7.0 kb band of Lass6 was most highly expressed in kidney, followed by brain. No expression of Lass6 was observed in heart, muscle, spleen or stomach. These results clearly demonstrate that Lass family members are differentially expressed among tissues.
Figure 5. Tissue distribution of the mRNA expression of Lass family members.
32P-labelled Lass1, Lass2, Lass4, Lass5, Lass6 and β-actin probes were hybridized to 2 μg of poly(A)+ RNA from the indicated adult mouse tissues immobilized on a positively charged nylon membrane. All Lass family members and the β-actin control were detected using the same membrane.
Asn-18 of Lass6 is glycosylated
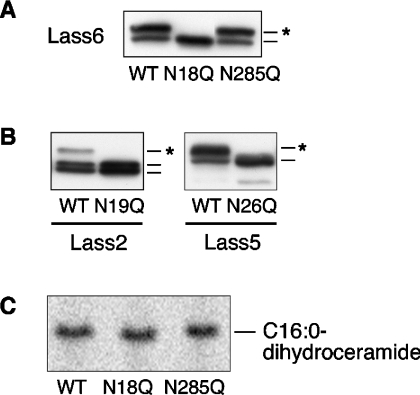
As shown in Figure 3, we observed glycosylated forms of Lass6, Lass5 and Lass2. To identify the specific N-glycosylation site in Lass6, we constructed two Asn→Gln mutants for the predicted glycosylation sites Asn-18 and Asn-285 (Lass6-N18Q and Lass6-N285Q) and performed an immunoblotting assay (Figure 6A). The mutant HA–Lass6-N18Q appeared as a single band with the same mobility as the unglycosylated band of the wild-type form. On the other hand, the mutant HA–Lass6-N285Q exhibited two bands with the same mobilities as observed for the untreated wild-type form of the protein. Thus the Lass6 protein is N-glycosylated at only the Asn-18 position.
Figure 6. The N-glycosylation site is conserved in Lass2, Lass5 and Lass6.
(A) Identification of the N-glycosylation site of Lass6. HEK 293T cells were transfected with pcDNA3-HA-Lass6 (wild type; WT), pcDNA3-HA-Lass6-N18Q (N18Q) or pcDNA3-HA-Lass6-N285Q (N285Q). Extracts (5 μg of protein) from transfected cells were separated on SDS/PAGE. An asterisk (*) indicates a glycosylated protein. (B) Identification of the conserved N-glycosylation site in Lass2 and Lass5. Extracts (5 μg of protein) for immunoblotting were prepared from HEK 293T cells transfected with each corresponding wild-type (WT) pcDNA3-HA-Lass, or with pcDNA3-HA-Lass2-N19Q (N19Q) or pcDNA3-HA-Lass5-N26Q (N26Q). An asterisk (*) indicates a glycosylated protein. (C) In vitro dihydroceramide synthesis activity of Lass6 mutants. Samples (40 μg of protein) from the same lysates as used in (A) were incubated at 37 °C for 15 min with [3H]dihydrosphingosine/5 μM dihydrosphingosine and 25 μM C16:0-CoA.
Since Asn-18 of Lass6 is conserved in Lass2 and Lass5, we next investigated the possible N-glycosylation of the respective Asn residues (Asn-19 of Lass2 and Asn-26 of Lass5) using the Lass2-N19Q and Lass5-N26Q mutants. As shown in Figure 6(B), the mutant proteins had the same mobilities as the unglycosylated forms of their wild-type proteins, indicating that Asn-19 of Lass2 and Asn-26 of Lass5 are also glycosylated. N-glycosylation can take place only on the luminal side of the ER. Therefore the N-termini of Lass2, Lass5 and Lass6 are exposed to the ER lumen.
Next, we examined whether glycosylation of the Lass6 protein is essential for in vitro dihydroceramide synthesis using cell lysates of the two Lass6 mutants (HA–Lass6-N18Q and HA–Lass6-N285Q) and C16:0-CoA (Figure 6C). No loss of activity was observed for the unglycosylated mutant (HA–Lass6-N18Q) when compared with either the glycosylated mutant HA–Lass6-N285Q or the glycosylated wild-type Lass6. Thus N-glycosylation is not essential for the dihydroceramide synthase activity of Lass6.
The C-terminus of mouse Lass6 faces the cytosolic side of the ER
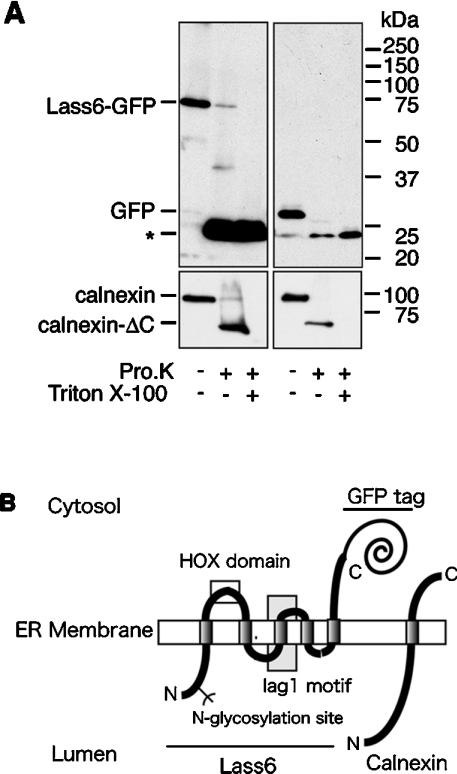
To examine the topology of the C-terminus of Lass6, we performed a proteinase K digestion assay using C-terminally GFP-tagged Lass6 (Lass6–GFP) (Figure 7A). Lass6–GFP demonstrated dihydroceramide synthesis activity in vitro (results not shown), indicating that the GFP fusion protein probably exhibits normal topology. Intact organelles were prepared by disrupting cells under isotonic buffer conditions, and were treated with proteinase K. To assess the isolation of ER, the type I ER membrane protein calnexin, which has a large N-terminal domain that is exposed to the lumen of the ER, was detected by immunoblotting with an antibody against its N-terminal region (amino acid residues 1–70). Upon treatment with proteinase K, calnexin was converted into a C-terminally truncated form of calnexin (calnexin-ΔC), as reported previously [19,24]. The production of calnexin-ΔC only in the absence of Triton X-100 confirmed the preparation of intact ER that was properly sealed and oriented. Using the same samples, we also detected Lass6–GFP by immunoblotting with an anti-GFP antibody. Proteinase K treatment converted Lass6–GFP into a 24 kDa band. Since this band was also produced in the presence of Triton X-100, it must represent a proteinase K-resistant GFP fragment. When free GFP was expressed and subjected to the same assay, a 29 kDa band was also converted into a 24 kDa band by proteinase K. In a topology model in which the C-terminus of Lass6 faces the ER lumen, the production of a fragment larger than free GFP would be expected, since GFP plus the C-terminus of Lass6 (at least the C-terminal transmembrane-to-tail portion) would be protected from proteinase K digestion by the ER membrane. However, the GFP portion of Lass6–GFP was digested similarly to the cytosolic GFP, indicating that the C-terminus of Lass6 is exposed to the cytosolic side of the ER membrane.
Figure 7. Membrane topology of the Lass6 protein.
(A) Proteinase K digestion assay. Crude membrane fractions were prepared from HEK 293T cells transfected with pEGFP-Lass6 (Lass6–GFP; left panel) or pEGFP-C1 (control vector; right panel). These were untreated or treated with 0.5 mg/ml proteinase K (Pro.K) at 4 °C for 2 h in the absence or presence of 1% Triton X-100. Proteins were separated on an SDS/10%-polyacrylamide gel, followed by immunoblotting with anti-GFP (upper panel) or anti-calnexin (lower panel) antibodies. Calnexin-ΔC is a C-terminally truncated form of calnexin. (B) A model of the proposed topology of Lass6 relative to the established topology of calnexin. Lass6 is proposed to be an internal membrane protein with five transmembrane domains. The HOX (homeobox) domain (clear box) is localized on the cytosolic side of the ER membrane. The highly conserved Lag1 motif contains the putative third transmembrane domain (grey box). The N-glycosylation site (Asn-18) is localized in the N-terminal hydrophilic domain. The N-terminus is located in the lumen of the ER, whereas the C-terminus is located in the cytosol.
DISCUSSION
At least five Lass family members, including the newly characterized Lass6, exist in mouse. Very recently, purified yeast Lass members (Lac1p and Lag1p) were shown to exhibit (dihydro)ceramide synthase activity, indicating that the Lass members are indeed subunits of ceramide synthases [25]. In that study, Lip1p (Lag1p/Lac1p-interacting protein) was co-purified as an essential component of the ceramide synthase. At present, it remains unclear whether Lass family members and/or some Lip1p/Lip1 homologues act as catalytic subunits of ceramide synthase. Alternatively, Lass family proteins may act as regulatory subunits, functioning in the transport of certain types of fatty acyl-CoA.
In our in vitro assays, Lass5 and Lass6 preferred short-chain acyl-CoAs, whereas Lass2 and Lass4 utilized long-chain acyl-CoAs (Figures 4B and 4C). On the other hand, Lass1 exhibited a high specificity towards C18:0-CoA (Figure 4B). Lass5 and Lass6 share high sequence identity and showed similar preferences for short-chain acyl-CoAs, such as C14:0- and C16:0-CoAs, yet they differed in substrate preference for unsaturated acyl-CoAs such as C18:1-CoA (Figure 4B). Lass2 and Lass4 also exhibited some differences in substrate preferences. Although both preferred long-chain acyl-CoAs such as C22:0- and C24:0-CoAs in vitro, Lass4 could also utilize C18:0- and C20:0-CoAs. Additionally, Lass2 overproduction in vivo resulted in an increase only in long-chain ceramides, whereas Lass4 overproduction caused increases in both middle- and long-chain ceramides (Figure 4A). However, in in vitro assays using similar mammalian expression systems, there were some discrepancies between our results and those of another study on Lass4 [15]. In the earlier report, Lass4 had C18:0- and C20:0-CoA-dependent ceramide synthesis activity, but it could not utilize C22:0- or C24:0-CoAs [15]. In contrast, we detected only weak activity for C18:0- and C20:0-CoA-dependent synthesis, but did observe C22:0- and C24:0-CoA-dependent ceramide synthesis (Figures 4B and 4C). Although the exact reason is unclear, we suppose that a difference in assay conditions caused this discrepancy. We added 0.1% digitonin to the assay mixtures that included long-chain acyl-CoAs to increase their water-solubility, a step that was omitted in our assays for short-chain ceramide synthesis and in the previous study.
In addition to the availability of certain fatty acyl-CoA species, differences in the levels of expression of Lass family members in tissues may produce tissue-specific ceramides. Indeed, we found that each Lass family member exhibits a characteristic tissue distribution pattern (Figure 5). For instance, Lass1, which showed high specificity towards C18:0-CoA, was expressed in brain abundantly and specifically (Figure 5); in brain, C18:0-ceramide is the major short-chain ceramide [3,5]. On the other hand, in most other tissues C16:0 is a more abundant fatty acid in ceramide than C18:0 [4], and Lass5 and Lass6, which exhibited high activities towards C16:0-CoA, were found to be expressed ubiquitously (Figure 5).
We identified an N-glycosylation site at the N-termini of Lass2, Lass5 and Lass6 (Figures 6A and 6B). N-glycosylation occurs only on the luminal side of the ER, indicating that the N-termini of these Lass proteins face the luminal side in the ER membrane. A proteinase K digestion of Lass6 (Figure 7A) provided further evidence that the C-terminus faces the cytosolic side in the ER membrane. These N- and C-terminal topologies of Lass6 revealed that Lass6 spans the ER membrane an odd number of times. The TopPredII program predicted that Lass6 has four or five transmembrane segments. Thus it is most likely that Lass6 spans the membrane five times, as illustrated in Figure 7(B). Lass family members share sequence identity throughout their entire amino acid sequences and have similar hydropathy profiles. Thus the transmembrane organization illustrated in Figure 7(B) may not be restricted to Lass6, but may be applicable to all Lass members. Our topology model predicts that the HOX domain is localized on the cytosolic side in the ER membrane, and that the highly conserved Lag1 motif contains the putative TM3 domain, with its N-terminus facing the lumen of the ER. In this model, then, the highly conserved segments DXXRKD (located near the N-terminus of the Lag1 motif) and LHDXXDXXLE (located near the C-terminus) are oriented on the luminal side and the cytosolic side respectively in the ER membrane.
Previous biochemical analyses using mouse liver tissue suggest that sphingolipid synthesis is initiated in the cytosolic leaflet of the lipid bilayer of the ER membrane, proceeding, at least, to the point of (dihydro)ceramide synthesis [26]. This conclusion is based on the sensitivity of these sphingolipid biosynthetic enzymes to an exogenous proteinase and to membrane-impermeable chemicals. However, our studies suggest that important residues of Lass members are localized on both sides of the ER membrane (Figure 7B). Additionally, the large hydrophilic domain of Lip1p, an essential subunit of yeast ceramide synthase, is localized in the luminal side of ER [25]. Thus it remains unclear on which side of the ER membrane (dihydro)ceramide is synthesized. Several reports have suggested different trans-bilayer orientations for ceramide. The active sites of the relevant galactosyltransferase and glucosyltransferase are located on the luminal side of the ER and the cytosolic side of the Golgi membrane respectively [27–30]. Therefore their substrate, ceramide, must exist on both sides of the ER membrane. The active site of the yeast sphingoid long-chain base-1-phosphate phosphatase, Lcb3p, is proposed to be located on the luminal side of the ER, suggesting that the long-chain base, the substrate for (dihydro)ceramide synthase, is generated on the luminal leaflet [31]. On the other hand, the recently identified ceramide transporter CERT is considered to transfer ceramide on the cytosolic leaflet of the ER membrane to the Golgi apparatus [32]. These results suggest that ceramide is located on both the cytosolic and luminal leaflets. It is most probable that ceramide generated on a specific side of the ER membrane traverses the membrane by spontaneous flip/flop or by the aid of certain translocases.
A cytosolic orientation has been determined for the active sites of the enzymes catalysing the first and second steps of sphingolipid biosynthesis, i.e. serine palmitoyltransferase and 3-ketodihydrosphingosine reductase respectively [19,33]. To reveal the complete trans-bilayer organization of the sphingolipid biosynthetic pathway, determination of the topology of the active site of (dihydro)ceramide synthase is also required. It will be essential for future studies to determine whether the Lass members are (dihydro)ceramide synthases or regulators.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (B) (12140201) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Okabe K., Keenan R. W., Schmidt G. Phytosphingosine groups as quantitatively significant components of the sphingolipids of the mucosa of the small intestines of some mammalian species. Biochem. Biophys. Res. Commun. 1968;31:137–143. doi: 10.1016/0006-291x(68)90043-0. [DOI] [PubMed] [Google Scholar]
- 2.Coderch L., Lopez O., de la Maza A., Parra J. L. Ceramides and skin function. Am. J. Clin. Dermatol. 2003;4:107–129. doi: 10.2165/00128071-200304020-00004. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien J. S., Rouser G. The fatty acid composition of brain sphingolipids: sphingomyelin, ceramide, cerebroside, and cerebroside sulfate. J. Lipid Res. 1964;5:339–342. [PubMed] [Google Scholar]
- 4.Pettus B. J., Bielawska A., Kroesen B. J., Moeller P. D., Szulc Z. M., Hannun Y. A., Busman M. Observation of different ceramide species from crude cellular extracts by normal-phase high-performance liquid chromatography coupled to atmospheric pressure chemical ionization mass spectrometry. Rapid. Commun. Mass Spectrom. 2003;17:1203–1211. doi: 10.1002/rcm.1043. [DOI] [PubMed] [Google Scholar]
- 5.Pettus B. J., Baes M., Busman M., Hannun Y. A., Van Veldhoven P. P. Mass spectrometric analysis of ceramide perturbations in brain and fibroblasts of mice and human patients with peroxisomal disorders. Rapid Commun. Mass Spectrom. 2004;18:1569–1574. doi: 10.1002/rcm.1520. [DOI] [PubMed] [Google Scholar]
- 6.Breimer M. E., Hansson G. C., Karlsson K. A., Leffler H., Pimlott W., Samuelsson B. E. Selected ion monitoring of glycosphingolipid mixtures. Identification of several blood group type glycolipids in the small intestine of an individual rabbit. Biomed. Mass. Spectrom. 1979;6:231–241. doi: 10.1002/bms.1200060603. [DOI] [PubMed] [Google Scholar]
- 7.Perry D. K., Hannun Y. A. The role of ceramide in cell signaling. Biochim. Biophys. Acta. 1998;1436:233–243. doi: 10.1016/s0005-2760(98)00145-3. [DOI] [PubMed] [Google Scholar]
- 8.Gulbins E. Regulation of death receptor signaling and apoptosis by ceramide. Pharmacol. Res. 2003;47:393–399. doi: 10.1016/s1043-6618(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 9.Perry D. K. Serine palmitoyltransferase: role in apoptotic de novo ceramide synthesis and other stress responses. Biochim. Biophys. Acta. 2002;1585:146–152. doi: 10.1016/s1388-1981(02)00335-9. [DOI] [PubMed] [Google Scholar]
- 10.Marchesini N., Hannun Y. A. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem. Cell Biol. 2004;82:27–44. doi: 10.1139/o03-091. [DOI] [PubMed] [Google Scholar]
- 11.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1pl. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr, Futerman A. H. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 14.Guillas I., Jiang J. C., Vionnet C., Roubaty C., Uldry D., Chuard R., Wang J., Jazwinski S. M., Conzelmann A. Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 15.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr, Futerman A. H. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 2003;278:43452–43459. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 16.Wang E., Merrill A. H., Jr Ceramide synthase. Methods Enzymol. 1999;311:15–21. doi: 10.1016/s0076-6879(00)11062-6. [DOI] [PubMed] [Google Scholar]
- 17.Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Med. Sci. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 18.Mizutani Y., Kihara A., Igarashi Y. Identification of the human sphingolipid C4-hydroxylase, hDES2, and its up-regulation during keratinocyte differentiation. FEBS Lett. 2004;563:93–97. doi: 10.1016/S0014-5793(04)00274-1. [DOI] [PubMed] [Google Scholar]
- 19.Kihara A., Igarashi Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 2004;279:49243–49250. doi: 10.1074/jbc.M405915200. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J. C., Kirchman P. A., Zagulski M., Hunt J., Jazwinski S. M. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 21.Winter E., Ponting C. P. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem. Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- 22.Venkataraman K., Futerman A. H. Do longevity assurance genes containing Hox domains regulate cell development via ceramide synthesis? FEBS Lett. 2002;528:3–4. doi: 10.1016/s0014-5793(02)03248-9. [DOI] [PubMed] [Google Scholar]
- 23.Claros M. G., Heijne G. v. Prediction of transmembrane segments in integral membrane proteins, and the putative topologies, using several algorithms. Comput. Appl. Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 24.Ou W. J., Bergeron J. J., Li Y., Kang C. Y., Thomas D. Y. Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg-ATP and Ca2+ J. Biol. Chem. 1995;270:18051–18059. doi: 10.1074/jbc.270.30.18051. [DOI] [PubMed] [Google Scholar]
- 25.Vallee B., Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschberg K., Rodger J., Futerman A. H. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 1993;290:751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coste H., Martel M. B., Got R. Topology of glucosylceramide synthesis in Golgi membranes from porcine submaxillary glands. Biochim. Biophys. Acta. 1986;858:6–12. doi: 10.1016/0005-2736(86)90285-3. [DOI] [PubMed] [Google Scholar]
- 28.Futerman A. H., Pagano R. E. Determination of the intracellular sites and topology of glucosylceramide synthesis in rat liver. Biochem. J. 1991;280:295–302. doi: 10.1042/bj2800295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeckel D., Karrenbauer A., Burger K. N., van Meer G., Wieland F. Glucosylceramide is synthesized at the cytosolic surface of various Golgi subfractions. J. Cell Biol. 1992;117:259–267. doi: 10.1083/jcb.117.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprong H., Kruithof B., Leijendekker R., Slot J. W., van Meer G., van der Sluijs P. UDP-galactose:ceramide galactosyltransferase is a class I integral membrane protein of the endoplasmic reticulum. J. Biol. Chem. 1998;273:25880–25888. doi: 10.1074/jbc.273.40.25880. [DOI] [PubMed] [Google Scholar]
- 31.Kihara A., Sano T., Iwaki S., Igarashi Y. Transmembrane topology of sphingoid long-chain base-1-phosphate phosphatase, Lcb3p. Genes Cells. 2003;8:525–535. doi: 10.1046/j.1365-2443.2003.00653.x. [DOI] [PubMed] [Google Scholar]
- 32.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature (London) 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda S., Nishijima M., Hanada K. Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J. Biol. Chem. 2003;278:4176–4183. doi: 10.1074/jbc.M209602200. [DOI] [PubMed] [Google Scholar]
- 34.Pan H., Qin W. X., Huo K. K., Wan D. F., Yu Y., Xu Z. G., Hu Q. D., Gu K. T., Zhou X. M., Jiang H. Q., et al. Cloning, mapping, and characterization of a human homologue of the yeast longevity assurance gene LAG1. Genomics. 2001;77:58–64. doi: 10.1006/geno.2001.6614. [DOI] [PubMed] [Google Scholar]