Abstract
OPN (osteopontin) is an integrin-binding highly phosphorylated glycoprotein, recognized as a key molecule in a multitude of biological processes such as bone mineralization, cancer metastasis, cell-mediated immune response, inflammation and cell survival. A significant regulation of OPN function is mediated through PTM (post-translational modification). Using a combination of Edman degradation and MS analyses, we have characterized the complete phosphorylation and glycosylation pattern of native human OPN. A total of 36 phosphoresidues have been localized in the sequence of OPN. There are 29 phosphorylations (Ser8, Ser10, Ser11, Ser46, Ser47, Thr50, Ser60, Ser62, Ser65, Ser83, Ser86, Ser89, Ser92, Ser104, Ser110, Ser113, Thr169, Ser179, Ser208, Ser218, Ser238, Ser247, Ser254, Ser259, Ser264, Ser275, Ser287, Ser292 and Ser294) located in the target sequence of MGCK (mammary gland casein kinase) also known as the Golgi kinase (S/T-X-E/S(P)/D). Six phosphorylations (Ser101, Ser107, Ser175, Ser199, Ser212 and Ser251) are located in the target sequence of CKII (casein kinase II) [S-X-X-E/S(P)/D] and a single phosphorylation, Ser203, is not positioned in the motif of either MGCK or CKII. The 36 phosphoresidues represent the maximal degree of modification since variability at many sites was seen. Five threonine residues are O-glycosylated (Thr118, Thr122, Thr127, Thr131 and Thr136) and two potential sites for N-glycosylation (Asn63 and Asn90) are not occupied in human milk OPN. The phosphorylations are arranged in clusters of three to five phosphoresidues and the regions containing the glycosylations and the RGD (Arg-Gly-Asp) integrin-binding sequence are devoid of phosphorylations. Knowledge about the positions and nature of PTMs in OPN will allow a rational experimental design of functional studies aimed at understanding the structural and functional interdependences in diverse biological processes in which OPN is a key molecule.
Keywords: casein kinase II, Golgi kinase, mammary gland casein kinase, O-glycosylation, osteopontin, phosphorylation
Abbreviations: CK, casein kinase; DHB, 2,5-dihydroxybenzoic acid; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MGCK, mammary gland casein kinase; μRPC, μ reverse-phase chromatography; OPN, osteopontin; PNGase F, peptide N-glycosidase F; PTH, phenylthiohydantoin; PTM, post-translational modification; RP, reverse-phase; SA, sinapinic acid; SIBLING, small integrin-binding ligand, N-linked glycoprotein; TFA, trifluoroacetic acid
INTRODUCTION
OPN (osteopontin) is a multifunctional phosphorylated glycoprotein containing an integrin-binding RGD (Arg-Gly-Asp) sequence. OPN was originally isolated from the mineralized matrix of bovine bone [1] and is a member of the SIBLING (small integrin-binding ligand, N-linked glycoprotein) family of glycophosphoproteins expressed in bone and/or teeth [2,3]. However, the expression of OPN is not limited to mineralized tissues but extends to a variety of cells, tissues and body fluids, including blood, urine and milk [4]. OPN is an important regulator of bone remodelling and mineral crystallinity and is associated with pathological events, such as infection, inflammation, wound healing, cancer metastasis and tissue calcification [5–9]. In addition, OPN regulates cytokine production, migration and activation of macrophages [10,11].
OPN is extensively altered through PTM (post-translational modification), such as phosphorylation, glycosylation and proteolytic processing, which have significant effects on the structure and biological properties of the protein [3,4]. The nature of the PTMs decorating OPN is highly tissue- and cell-specific, reflecting the diverse functions of the protein in different physiological systems. Normal rat kidney cells, for example, secrete both phosphorylated and non-phosphorylated variants of OPN that diverge in glycosylation patterns and physiological properties [12]. The level of phosphorylation of OPN is responsive to hormonal levels, e.g. vitamin D [13] and in oncogene-transfected Rat-1 cells, it switches from the synthesis of sialylated to non-sialylated OPN abrogating cell binding between OPN and the cells [14]. Likewise, the regulatory roles of OPN in normal and pathological mineralization are highly dependent on the phosphorylation status of the protein [15,16]. Dephosphorylation of OPN hinders its ability to inhibit hydroxyapatite formation [17], and phosphorylation is necessary for its inhibition of calcium oxalate crystallization in urine [18] and calcification of vascular smooth muscle cells [19]. Phosphorylation of OPN has been shown to be essential for osteoclast attachment, and TRAP (tartrate-resistant acid phosphatase) dephosphorylation of bovine OPN resulted in a partially dephosphorylated protein that could no longer promote RGD-dependent osteoclast attachment [20], and recent data suggest that the phosphorylation of OPN is involved in the stimulation of bone resorption [21]. The interaction between OPN and certain receptors on macrophages is also influenced by the phosphorylation state of the protein. OPN binding to the macrophage β3-integrin receptor is dependent on the phosphorylation of the N-terminal part of the protein, and macrophages are only stimulated to express interleukin-12 if this fragment is phosphorylated [10]. Likewise, phosphorylation is required for the OPN-mediated spreading and activation of macrophages [11] and recently OPN has also been shown to promote trophoblastic cell migration in a process that is dependent on the level of phosphorylation of the protein [22]. Collectively, these studies indicate the crucial role of PTM in the diverse functions of OPN.
Few studies have investigated the PTM patterns of OPN. The initial characterization of rat bone OPN estimated the presence of 12 phosphoserines and one phosphothreonine, plus an additional five or six O-linked and one N-linked oligosaccharides [23]. In a later study, 11 variable phosphorylated serines were localized in OPN from rat bone [24]. Previously, we have described the in vivo PTM pattern of bovine milk OPN containing 28 phosphorylations and three O-linked glycosylations [25]. Human milk OPN has recently been estimated to contain 32 mol phosphate/mol OPN [26]. Other studies have localized ten phosphoserines in OPN from cultured chicken osteoblasts [27] and estimated the presence of eight phosphorylations in bovine bone OPN [28]. Phosphorylations in bovine milk OPN [25,29] are located in recognition motifs of the MGCK (mammary gland casein kinase) [S/T-X-E/S(P)/D] [30,31] and CKII (casein kinase II) [S-X-X-E/S(P)/D] [30,32]. In vitro studies have shown that MGCK, CKI and CKII [33,34] are able to phosphorylate OPN. Minor phosphorylation of OPN can also be mediated by cGMP-dependent kinase and protein kinase C in vitro, whereas cAMP-dependent protein kinase is unable to modify the protein [34].
It is clear that the expression of OPN in tissues and body fluids differs not only in protein levels but also in the phosphorylation and glycosylation state of the protein. Elucidation of the PTMs of OPN is essential for the understanding of many diverse functions in which the protein participates. Here, we report the complete phosphorylation and glycosylation pattern of native human OPN, containing up to 34 phosphoserines, two phosphothreonines, five O-glycosylated threonines and no apparent N-glycosylation.
EXPERIMENTAL
Materials
Bovine pancreas trypsin (EC 3.4.21.4) and Staphylococcus aureus V8 proteinase (EC 3.4.21.19) were obtained from Worthington Biochemical (Freehold, NJ, U.S.A.). Thermolysin (EC 3.4.24.27) from Bacillus thermoproteolyticus rokko and O-glycosidase (endo-α-N-acetylgalactosaminidase; EC 3.2.1.97) were obtained from Sigma (St. Louis, MO, U.S.A.). The μRPC (μ reverse-phase chromatography) C2/C18 PC 3.2/3 and the Superdex peptide HR 10/30 columns were purchased from Amersham Biosciences (Uppsala, Sweden). Vydac C18 RP (reverse-phase) resin was obtained from The Separations Group (Hesperia, CA, U.S.A.). Reagents used for sequencing were purchased from Applied Biosystems (Warrington, Cheshire, U.K.). SA (sinapinic acid) and DHB (2,5-dihydroxybenzoic acid) were purchased from LaserBio Labs (Sophia-Antipolis Cedex, France). PNGase F (peptide N-glycosidase F; EC 3.5.1.52) kit was obtained from New England Biolabs (Beverly, MA, U.S.A.). Sialidase (neuraminidase; EC 3.2.1.18) was obtained from Roche (Mannheim, Germany). All other chemicals used were of analytical grade.
Purification of human OPN
OPN was purified from human milk from a pool of donors essentially as described in [35]. However, instead of the final RP-HPLC step, the supernatant resulting from the barium chloride and sodium citrate precipitation was dialysed against 0.1% formic acid overnight, centrifuged (31000 g, 25 min) and the resulting supernatant was freeze-dried. SDS/PAGE and N-terminal sequencing verified the very high purity (+98%) of the resulting OPN.
Generation and separation of peptides
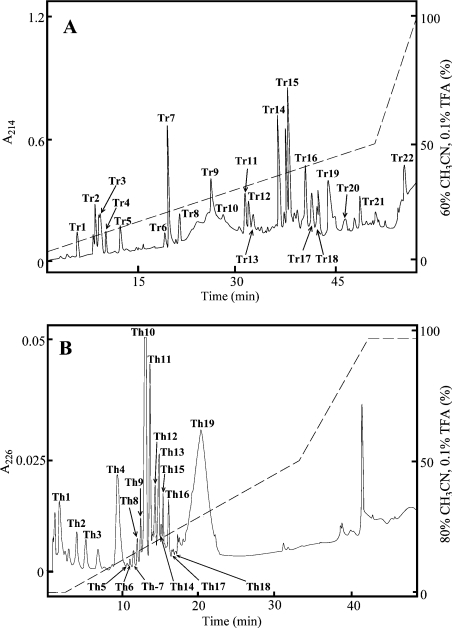
OPN was digested with trypsin using an enzyme-to-substrate ratio of 1:30 (w/w), in 50 mM ammonium bicarbonate (pH 8.1), at 37 °C for 6 h. Tryptic peptides were separated by RP-HPLC on a μRPC C2/C18 PC 3.2/3 column connected to a Pharmacia SMART system (Pharmacia, Uppsala, Sweden). Separation was carried out in 0.1% TFA (trifluoroacetic acid; buffer A) and eluted with a gradient of 60% (v/v) acetonitrile in 0.1% TFA (buffer B) developed for 54 min (0–9 min, 0% buffer B; 9–49 min, 0–50% buffer B; 49–54 min, 50–100% buffer B) at a flow rate of 0.15 ml/min. The peptides were detected in the effluent by measuring the absorbance at 214 nm. Fractions were stored at −20 °C until analysis. For analysis of a large fragment not cleaved with trypsin (∼residues 36–152), some of the tryptic digests were separated by gel filtration on a Superdex Peptide HR 10/30 column. The column was equilibrated with 0.1 M ammonium bicarbonate (pH 8.1) and operated at a flow rate of 0.3 ml/min. The fractions containing the fragment in question were pooled, freeze-dried and further digested with thermolysin using an enzyme-to-substrate ratio of 1:10 (w/w) in 0.1 M pyridine-acetate (pH 6.5) at 56 °C for 6 h. The resulting peptides were separated by RP-HPLC on a Vydac C18 column connected to a Pharmacia LKB system (Pharmacia). Separation was carried out in 0.1% TFA (buffer A) and eluted with a gradient of 80% acetonitrile in 0.1% TFA (buffer B) developed over 50 min (0–5 min, 0% buffer B; 5–40 min, 0–50% buffer B; 40–50 min, 50–95% buffer B) at a flow rate of 0.85 ml/min. Fraction Th19 (Figure 1B) from the thermolysin digest was further digested with S. aureus V8 proteinase at an enzyme concentration of 30 μg/ml in 0.1 M ammonium bicarbonate (pH 8.1) at 37 °C for 3.5 h.
Figure 1. RP-HPLC separation of trypsin and thermolysin digests of OPN.
(A) RP-HPLC of tryptic peptides. Peptides were separated on a μRPC C2/C18 PC 3.2/3 column operated by a Pharmacia SMART system. Separation was carried out in 0.1% TFA and peptides were eluted with a gradient of 60% acetonitrile in 0.1% TFA (- - -) at a flow rate of 0.15 ml/min. The peptides were detected in the effluent by measuring the absorbance at 214 nm (——). (B) RP-HPLC of thermolytic peptides. Peptides were separated by RP-HPLC on a Vydac C18 column operated by a Pharmacia LKB system. Separation was carried out in 0.1% TFA and eluted with a gradient of 80% acetonitrile in 0.1% TFA (- - -) at a flow rate of 0.85 ml/min. The peptides were detected in the effluent by measuring the absorbance at 226 nm (——).
Characterization of peptides
Peptides were characterized by MS and amino acid sequence analyses. Amino acid sequence analyses were performed on an Applied Biosystems PROCISE HT protein sequencer with on-line identification of PTH (phenylthiohydantoin) derivatives. Modified serine/threonine residues were identified by the lack of a PTH derivative in the cycles where these amino acids are modified. MS analyses of the peptides were performed using a Voyager DE-PRO MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) mass spectrometer (Applied Biosystems). Samples for MS analyses were prepared by mixing 1 μl of the fraction to be analysed with DHB (peptides) or SA (native OPN; 10 g/l) in a 1:1 ratio directly on the MS-target probe. All spectra were obtained in positive reflector-ion or positive linear-ion mode using a nitrogen laser at 337 nm and an acceleration voltage of 20 kV. Typically, 50–100 laser shots were added per spectrum and calibrated with external standards. The theoretical peptide masses and sequence coverage were calculated using the GPMAW program (Lighthouse Data, Odense, Denmark).
Enzymatic deglycosylation
N-linked deglycosylation was performed as described by the manufacturer using the PNGase F deglycosylation kit. OPN (15 μg) was incubated with 500 units of PNGase F at 37 °C for 20 h. For deglycosylation of O-linked glycans, 100 μg of OPN was incubated with 6 m-units of O-glycosidase in 50 mM tri-sodium citrate dihydrate (pH 6) at 37 °C for 21 h. For the removal of terminal sialic acid, 100 μg of OPN was incubated with 1 m-unit of sialidase, which cleaves terminal α-(2–3), α-(2–6) and α-(2–8) linkages of sialic acid in glycoconjugates, in 50 mM ammonium acetate (pH 5) at 37 °C for 18 h. The reaction products from the enzymatic deglycosylations were visualized by SDS/PAGE.
Amino sugar analysis
Peptide Th19 was subjected to amino acid and amino sugar analyses. The peptide was hydrolysed for 3 h under vacuum in the presence of 6 M HCl, 0.05% phenol and 1% thioglycolic acid at 110 °C and the released amino acids and amino sugars were analysed essentially as described in [36].
RESULTS
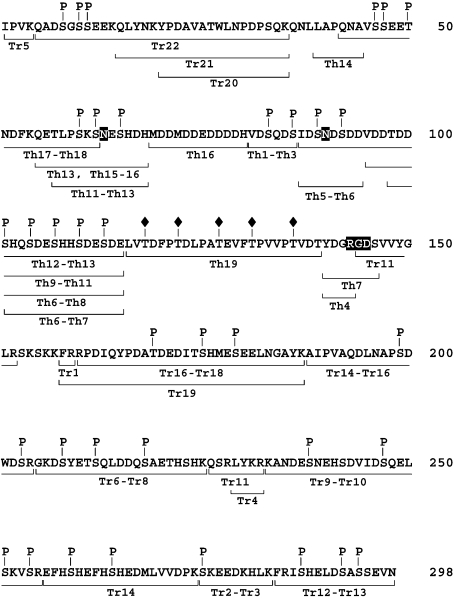
OPN was digested with trypsin and the resulting peptides were separated by RP-HPLC (Figure 1A). A large extremely acidic part of the protein representing residues 36–152 was isolated from the tryptic digest by gel filtration (results not shown) and subsequently digested with thermolysin and the resulting peptides were separated by RP-HPLC (Figure 1B). All fractions were collected and the sites and nature of the PTMs were characterized by a combination of MS and N-terminal Edman sequence analyses (Figure 2).
Figure 2. Localization of PTMs in human OPN.
Solid lines indicate characterized peptides. P denotes identified phosphorylation. ◆, glycosylations. The two putative N-glycosylation sites and the integrin receptor-binding RGD sequence are highlighted. Peptides are named with reference to the elution profiles in Figure 1.
Phosphorylations
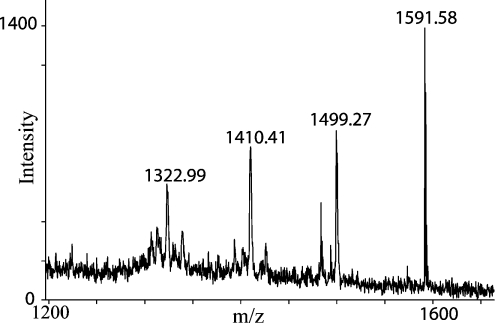
Human OPN contains 42 serine and 14 threonine residues, and 32 of these residues are located in the recognition sequence of the MGCK [30,31] and seven match the recognition sequence for CKII [30,32]. To identify the specific phosphorylated serines and threonines, we analysed all fractions from the RP-HPLC separations by MALDI–TOF-MS and some were also subjected to N-terminal Edman sequence analysis. Identification of phosphorylated peptides by MALDI–TOF-MS was accomplished by detection of the characteristic spectra of phosphorylated serines and threonines. These spectra present a characteristic series of peaks separated by approx. 90–93 Da (Figure 3), which are formed by metastable decomposition of the phosphoserine or phosphothreonine [37]. The mass of an O-linked phosphate group is 80 Da. Peptides with masses differing by 80, 160, 240 etc. from the theoretically calculated mass were identified as being phosphorylated. When more potential sites of phosphorylation were present in a given peptide, the exact location was determined by N-terminal sequence analysis. Modified serine/threonine residues were identified by the lack of a PTH derivative in the cycles where these amino acids are modified.
Figure 3. MALDI–TOF-MS analysis of Th11 from Figure 1(B).
The protonated mass at m/z 1591.58 corresponds to the triple phosphorylated peptide T57LPSKSNESHDH68. The characteristic fragmentation pattern confirms the presence of three phosphorylations in the peptide.
Data from the MALDI–TOF-MS and Edman sequence analyses and the resulting map of PTMs are shown in Table 1 and Figure 2 respectively. In total, we have identified 36 phosphorylation sites in human milk OPN. A total of 29 phosphorylations divided among 27 serines (Ser8, Ser10, Ser11, Ser46, Ser47, Ser60, Ser62, Ser65, Ser83, Ser86, Ser89, Ser92, Ser104, Ser110, Ser113, Ser179, Ser208, Ser218, Ser238, Ser247, Ser254, Ser259, Ser264, Ser275, Ser287, Ser292 and Ser294) and two threonines (Thr50 and Thr169) were located in MGCK recognition sequences. Six serines (Ser101, Ser107, Ser175, Ser199, Ser212 and Ser251) in CKII motifs were phosphorylated and finally a single phosphorylation at Ser203 did not fit the recognition motifs of any of the two kinase. Phosphorylated residues located in overlapping MGCK and CKII motifs are considered to be phosphorylated by MGCK, because of the marginal action of CKII in the mammary gland and in other tissues when its consensus sequence is overlapping that of MGCK [33].
Table 1. Characterization of peptides from human milk OPN.
Peak numbers correspond to those of Figure 1(A) (Tr), Figure 1(B) (Th) and Figure 2. Peptide amino acid sequences were identified by sequence and/or MALDI–TOF-MS analyses. ‘Sequence’ data are the result of N-terminal sequence analyses, where ‘–’ denotes that the peptide has not been sequenced to the end and ‘*’ denotes that no PTH derivative was identified, which indicates the presence of a modified residue. The observed molecular masses were determined by MALDI–TOF-MS. Unless mentioned otherwise, all masses are monoisotopic. Mass difference is that between the observed and calculated masses. The type of modification corresponding to the mass difference is given in parentheses: phosphorylation (P), oxidation of methionine (Mox), oxidation of tryptophan (Wox) and pyroglutamic acid formed from N-terminal glutamine (pGlu).
| Peak | Peptide | Sequence | Observed mass | Calculated mass | Mass difference |
|---|---|---|---|---|---|
| Tr1 | F158–R159 | FR | 322.13 | 322.18 | |
| Tr2 | S275–K283 | 1193.55 | 1113.58 | 79.97 (1P) | |
| Tr3 | S275–K283 | 1113.53 | 1113.58 | ||
| Tr4 | L229–K232 | LYKR | 579.29 | 579.35 | |
| Tr5 | I1–K4 | IPVK | 456.23 | 456.31 | |
| Tr6 | G205–K225 | 2603.01 | 2363.03 | 239.98 (3P) | |
| Tr7 | G205–K225 | 2523.05 | 2363.03 | 160.02 (2P) | |
| Tr8 | G205–K225 | 2443.09 | 2363.03 | 80.06 (1P) | |
| Tr9 | K233–R255 | 2827.20/2907.19 | 2587.22 | 239.98 (3P)/319.97 (4P) | |
| Tr10 | K233–R255 | 2747.25 | 2587.22 | 160.03 (2P) | |
| Tr11 | Q226–R232 | 950.52 | 950.55 | ||
| G144–R152 | 965.55 | 965.50 | |||
| Tr12 | F284–N298 | 1930.67 | 1690.80 | 239.87 (3P) | |
| Tr13 | F284–N298 | 1850.72 | 1690.80 | 159.92 (2P) | |
| Tr14 | A188–R204 | 2014.80/2018.79/2046.80 | 1854.89 | 159.9 (2P)/163.9(2P,Wox)/191.91 (2P, Wox) | |
| E256–K274 | 2480.09/2496.05 | 2320.04 | 160.05 (2P)/176.01 (2P, Mox) | ||
| Tr15 | A188–R204 | AIPVAQDL- | 1934.85/1938.84/1950.83 | 1854.89 | 79.96 (1P)/83.95 (1P, Wox)/95.94 (1P, Wox) |
| Tr16 | A188–R204 | AIPV- | 1854.89/1858.95/1886.88 | 1854.89 | Not modified/4.06(Wox)/31.99(Wox) |
| R160–K187 | RPDI- | 3464.30/3480.21 | 3224.43 | 239.87 (3P)/255.78 (3P, Mox) | |
| Tr17 | R160–K187 | 3384.48/3400.49 | 3224.43 | 160.05 (2P)/176.06 (2P, Mox) | |
| Tr18 | R160–K187 | 3304.52/3320.54 | 3224.43 | 80.09 (1P)/96.11 (1P, Mox) | |
| Tr19 | F158–K187 | 3687.57/3703.53/3768.93† | 3527.59 | 159.98 (2P)/175.94(2P, Mox)/239.16‡ (3P) | |
| Tr20 | Y20–K35 | 1801.85/1805.84/1833.84 | 1801.87 | Not modified/3.97(Wox)/31.97(Wox) | |
| Tr21 | Q15–K35 | 2435.19/2448.20/2452.23 | 2448.21 | −13.02 (pGlu, Wox)/not modified/4.02 (Wox) | |
| Tr22 | Q5–K35 | QAD*G**E- | 3690.92†/3708.42† | 3466.63 | 222.21‡ (3P, pGlu)/239.71‡ (3P) |
| Th1 | V81–S86 | VD*QD* | 810.19 | 650.26 | 159.93 (2P) |
| Th2 | V81–S86 | VD*QDS | 730.22 | 650.26 | 79.96 (1P) |
| Th3 | V81–S86 | 650.24 | 650.26 | ||
| Th4 | Y140–R143 | YDGR | 510.19 | 510.22 | |
| Th5 | I87–D94 | 1040.26 | 880.31 | 159.95 (2P) | |
| Th6 | I87–D94 | IDSND- | 960.23 | 880.31 | 79.92 (1P) |
| S101–E115 | SHQ*D- | 2045.47 | 1725.61 | 319.86 (4P) | |
| T98–E115 | TDD*H- | 2456.44 | 2056.72 | 399.72 (5P) | |
| Th7 | Y140–S146 | YDGR- | 769.29 | 769.30 | |
| S101–E115 | SHQ*- | 1965.64 | 1725.61 | 240.03 (3P) | |
| T98–E115 | TDD*- | 2376.61 | 2056.72 | 319.89 (4P) | |
| Th8 | T98–E115 | TDD*- | 2296.55 | 2056.72 | 239.83 (3P) |
| Th9 | V95–E115 | VDDTD- | 2785.87 | 2385.84 | 400.03 (5P) |
| Th10 | V95–E115 | 2625.70/2705.80 | 2385.84 | 239.86 (3P)/319.96 (4P) | |
| Th11 | T57–H68 | TLP*K*N- | 1591.58 | 1351.62 | 239.96 (3P) |
| V95–E115 | VDDTDDS- | 2545.91 | 2385.84 | 160.07 (2P) | |
| Th12 | T57–H68 | TLPSK*N- | 1511.57 | 1351.62 | 159.95 (2P) |
| I87–E115 | IS*ND*D- | 3726.97/3809.30† | 3247.14 | 479.83 (6P)/560.36‡ (7P) | |
| Th13 | T57–H68 | 1431.60 | 1351.62 | 79.98 (1P) | |
| Q55–H68 | 1848.70 | 1608.72 | 239.98 (3P) | ||
| I87–E115 | 3647.08 | 3247.14 | 399.94 (5P) | ||
| Th14 | L39–A44 | 613.28 | 613.32 | ||
| Th15 | Q55–H68 | 1831.67 | 1608.72 | 222.95 (3P, pGlu) | |
| Th16 | M69–H80 | 1467.45/1483.44 | 1467.41 | Not modified/16.03 (Mox) | |
| Q55–H68 | 1751.65 | 1608.72 | 142.93 (2P, pGlu) | ||
| Th17 | Q42–S62 | 2738.88 | 2339.09 | 399.79 (5P) | |
| Th18 | Q42–S62 | 2659.11 | 2339.09 | 320.02 (4P) | |
| Th19 | L116–T139 | L116–T139 | –§ |
† Average molecular masses determined by MALDI–TOF-MS in a linear mode.
‡ Difference between the observed average mass and the theoretical calculated average mass (mass value not shown).
§ No ions were detected by MALDI–TOF-MS.
In a few cases, the mass of an observed phosphopeptide was assigned to a sequence of amino acids containing more potential phosphor-accepting residues than the observed amount of phosphate groups, e.g. the mass analysis of Tr6 (Figure 1A and Table 1) gave a mass of 2603.01 Da and a characteristic fragmentation pattern indicating three phosphorylations. The observed mass correlates with the calculated protonated mass (2602.94 Da) of the peptide G205KDSYETSQLDDQSAETHSHK225 containing three phosphate groups. The peptide contains four serine and two threonine residues but only three of the serine residues are positioned in the recognition motifs of MGCK or CKII and therefore these serines are considered to be the most likely phosphoacceptor sites. This conclusion is supported by the presence of phosphorylations at homologous positions in bovine milk OPN [25,29]. The same approach was used when assigning the phosphorylations in Tr9, Tr12 and Tr16.
Digestion of OPN with trypsin and thermolysin gives rise to complex peptide mixtures and hence the RP separation results in chromatograms with some unresolved peaks. Accordingly, N-terminal sequence analyses showed the presence of several peptides in some fractions. Likewise, peptides with identical N-termini but with mass differences of 80, 160 Da etc. appeared at several different positions in the chromatograms, indicating heterogeneous phosphorylation in many peptides. A notable example is the peptide V95DDTDDSHQSDESHHSDESDE115 (Th9–Th11 in Table 1 and Figure 1B). This peptide was observed in four phosphorylation variants ranging from two to five modified residues.
Phosphorylations were assigned to residues in motifs for MGCK or CKII in all but one case. Mass analysis of Tr14 (Figure 1A) showed a mass (2014.80 Da) correlating with the calculated protonated mass (2014.83 Da) of the tryptic peptide A188IPVAQDLNAPSDWDSR204 containing two phosphorylations. This peptide contains exactly two potential phosphor-accepting residues (Ser199 and Ser203). Ser199 is located in a CKII motif, but Ser203 is not located in the recognition sequence of either of the kinases MGCK or CKII. In summary, MALDI–TOF-MS analyses and N-terminal sequencing of human milk OPN revealed 36 phosphorylated amino acids divided among 34 serine and two threonine residues. All residues except one were located in recognition motifs for either MGCK or CKII (Figure 2).
Glycosylations
MS analysis of Th19 (Figure 1B) revealed no distinct peaks, suggesting that the peptide considered is heavily modified, most likely by oligosaccharides. N-terminal sequencing showed that this fraction contained a peptide starting at Leu116. The region in bovine milk corresponding to this peptide contains three O-linked oligosaccharides [25]. The peptide in Th19 was too large for a trustworthy assignment of O-linked glycosylation sites by N-terminal sequencing due to decreasing yield of PTH derivatives during the Edman degradation, a phenomenon that was intensified by the presence of many proline residues present in the sequence. Thermolysin failed to cleave this peptide and as a consequence, the peptide was digested with S. aureus V8 proteinase. This resulted in the peptides L116VTDFPTDLPATE128 and V129FTPVVPTVDT139 that were separated by RP-HPLC (results not shown). Amino acid sequencing of these peptides gave no identifiable PTH-amino acids in the cycles corresponding to Thr118, Thr122, Thr127, Thr131 and Thr136, suggesting the presence of O-linked glycosylations at these five threonine residues. Short-time hydrolysis of the peptide Th19 followed by amino acid sugar analysis showed the presence of high amounts of the amino sugars, galactosamine and glucosamine, strongly indicating O-linked glycosylation at the five threonine residues. No trace of PTH-Thr was ever observed at any of these positions during this study, indicating full glycosylation. The yield of PTH-Thr in the cycle corresponding to Thr139 showed that this threonine is not modified in human OPN. The presence of O-linked glycans was further investigated by enzymatic deglycosylation with sialidase and O-glycosidase. The effect of each enzyme was tested by incubation of OPN with each enzyme alone, and the combined effect was tested by incubation with sialidase and O-glycosidase in succession. Neither of these enzymes alone nor in combination had any effect on OPN migration in SDS/PAGE (results not shown), indicating that the O-glycans in human OPN do not contain terminal sialic residues and are not of the unsubstituted core 1 type.
Two asparagine residues in human OPN (Asn63 and Asn90) are located in the putative N-glycosylation motif Asn-X-Ser/Thr. MS and amino acid sequence analyses of the peptides from the thermolysin digest containing these asparagines showed that none of these are glycosylated in human milk OPN. Furthermore, incubation of OPN with PNGase F did not result in changes in SDS/PAGE migration.
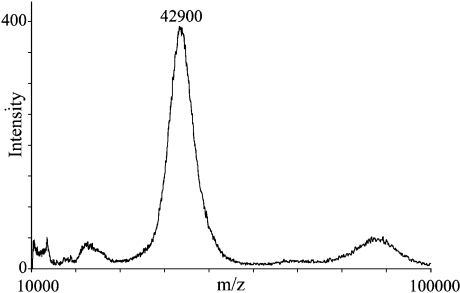
MS analysis of native human milk OPN showed a mass of approx. 42.9 kDa (Figure 4). Subtraction of the calculated mass of the amino acids and the 32 phosphate groups found to be the average phosphorylation level of human milk OPN [26] leaves approx. 6.6 kDa for the glycosylations. Likewise, MS analysis of the fragment representing residues 36–152, in which all the glycosylations are accounted for (results not shown), also showed an excess mass of approx. 6.6 kDa, corresponding to roughly 30–32 monosaccharides to be divided among the five O-glycans.
Figure 4. MALDI–TOF-MS analysis of intact native human OPN.
The average mass peak at 42.9 kDa represents native human OPN. The peaks at approx. 22 and 85 kDa represent the derived (M2H+) and (2MH+) protonated species respectively.
Oxidations
In this study, several peptides containing oxidized methionine and tryptophan residues were observed (Table 1). Oxidized tryptophans can give rise to +4, +16 and +32 Da mass increases [38] and can thereby support the identification of peptides containing tryptophan. Methionines are often observed with a 16 Da mass increase due to oxidation of the sulphur-containing side chain. Although it cannot be demonstrated unequivocally whether oxidation occurred during sample handling or in vivo, it is most likely that the oxidations are artifacts of sample handling and MS analysis. This assumption is supported by MS analysis of methionine- and tryptophan-containing peptides from other proteins, which resulted in similar characteristic mass increases (results not shown).
DISCUSSION
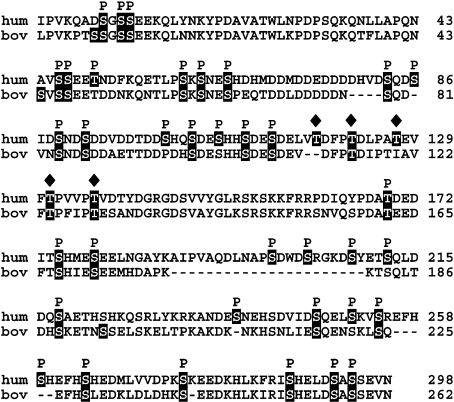
The complete phosphorylation and glycosylation pattern of native human OPN has been determined, showing the presence of up to 34 phosphoserine, two phosphothreonine and five O-glycosylated threonine residues. The degree of phosphorylation of human milk OPN is significantly higher than that reported for rat bone OPN [23], chicken osteoblast OPN [27] and the in vitro estimated phosphorylation in bovine bone [28], but only slightly higher than the 28 phosphate groups decorating the bovine milk OPN molecule [25]. An alignment of the human and bovine OPN sequences including the sites of PTM is shown in Figure 5. It seems evident that milk OPN contains significantly more phosphorylations than OPN in mineralized tissues. This suggests a tissue-specific phosphorylation and/or dephosphorylation, which is in line with the multitude of biological functions of OPN in different tissues and body fluids.
Figure 5. Alignment of human and bovine OPN sequences.
Modified residues are highlighted and introduced gaps are indicated by broken lines. P denotes identified phosphorylation and filled diamonds symbolize glycosylations.
The phosphorylation motifs in human OPN show that the primary requirement for serine and threonine phosphorylation is a negatively charged residue in the n+2 or n+3 positions relative to the phosphoacceptor residue. These motifs are identical with the recognition sequences of MGCK [30,31] and CKII [30,32] respectively. This implies that both kinases modify OPN in the human mammary gland; however, the situation is probably more complicated. It has long been recognized that MGCK is the most active kinase in the mammary gland and that the major milk proteins, the caseins [31], as well as component PP3 [39], all have their phosphate groups attached in MGCK recognizable sequences. In addition, minor phosphorylation by MGCK on serine residues located in CKII motifs [30] and in sequences with negatively charged residues in the n+5 to n+7 positions has been observed [40]. Altogether, this could suggest a situation where MGCK is solely responsible for the phosphorylation of OPN in the mammary gland. On the other hand, serines, and threonines in particular, with an aspartic acid in the n+2 position are phosphorylated much less efficiently by MGCK compared with residues with a glutamic acid or phosphoserine at the equivalent positions [30,41], making the modifications of Ser65, Ser83, Ser86, Ser89, Ser92, Thr50 and Thr169 by MGCK atypical. The specificity of CKII may in some instances overlap the specificity of MGCK, as negatively charged residues at positions n+1, n+2 and/or n+3 can potentially confer phosphorylation by CKII [32]. In addition, 11 phosphoserines are also positioned in sequences that potentially could be phosphorylated by CKI. Ten of them overlie MGCK motifs and a single phosphoserine overlies a CKII recognition sequence. In fact, in vitro experiments with OPN as substrate for these kinases have shown that CKI is a better OPN kinase than CKII, but both are inferior to MGCK [33]. So the question whether MGCK acts alone or in concert with CKI/CKII in the phosphorylation of OPN in vivo remains to be elucidated. However, MGCK is present within the Golgi compartments whereas CKI and CKII are cytosolic enzymes and the in vivo phosphorylation of OPN is therefore most probably mediated by the MGCK. The ‘cryptic’ phosphorylation at Ser203 could be a product of the above-mentioned phosphorylation of serine residues located in sequences atypical of those normally recognized by either MGCK or CKII. In vitro experiments have shown that minor phosphorylation of OPN can be mediated by cGMP-dependent protein kinase and protein kinase C [34]. The cGMP-dependent protein kinase favours phosphorylation when an arginine residue is located C-terminal to the phosphate-accepting residue [41], which makes it possible that this kinase is responsible for the phosphorylation of Ser203 in human OPN.
The phosphorylation motifs reported in the present study are in agreement with the patterns described for OPN from rat bone [24] and chicken osteoblasts [27], where almost all phosphorylations are located in the recognition sequence of MGCK and a few in motifs indicating CKII-mediated phosphorylation. Likewise, in the related SIBLINGs, BSP (bone sialoprotein) [42] and DSP (dentin sialoprotein) [43], almost all phosphorylations are located in motifs with acidic residues in the n+2 and/or n+3 position, indicating that these are phosphorylated by identical or analogous kinases.
In the present study, we showed that human milk OPN contains 36 potential sites of phosphorylation. This number represents the maximal level of phosphorylation, as a number of sites are only partially occupied. The average level of phosphorylation is approx. 32 phosphorylations/OPN molecule [26]. In a screening of cDNA libraries from human mammary gland and milk cells only full-length OPN mRNA was detected (M. S. Nielsen and E. S. Sørensen, unpublished work), indicating that the observed heterogeneity is not a result of alternative splicing of the OPN transcript. The phenomenon of partial phosphorylation reported in this study is also observed for rat bone OPN [24] and for chicken osteoblast OPN [27], whereas no variation has been reported in the phosphorylation of bovine milk OPN [25]. The biological significance of this variation in utilization of potential phosphorylation sites as well as a possible variation in the rate of phosphorylation and dephosphorylation within tissues, such as the mammary gland and bone, is not yet known. The heterogeneity seen in bone OPN could be explained by the continuous formation and aging of the bone matrix and the action of phosphatases [44,45]. The partially phosphorylated sites observed in this study are distributed over most of the proteins and are located in both MGCK and CKII motifs.
The arrangement of phosphorylations in clusters of three to five, spanned by stretches of unmodified amino acids, is consistent with the pattern described for bovine OPN [25] (Figure 5). Regions with high concentrations of phosphorylations and acidic amino acids constitute potential binding sites for minerals and calcium salts. These highly anionic regions could enable OPN to form soluble complexes with calcium ions and thereby inhibit unintentional calcium crystallization and precipitation, e.g. in milk and urine [15,18]. Likewise, the phosphorylation clusters could be responsible for induction of structures and scaffolds that mediate OPN binding to hydroxyapatite in mineralized tissues. During bone resorption, osteoclasts bind to OPN via the RGD integrin-binding sequence and OPN anchors the osteoclasts to the hydroxyapatite via phosphorylations [5]. Cluster phosphorylation of OPN could play an essential role in these biological scenarios.
Human OPN contains a threonine-proline-rich region (residues 118–139). This region contains six evenly distributed threonine residues (Thr118, Thr122, Thr127, Thr131, Thr136 and Thr139), of which all but Thr139 were found to be O-glycosylated. The threonine-proline-rich region in OPN is well conserved and the three threonine residues corresponding to Thr122, Thr131 and Thr136 in human OPN are strictly conserved in all known mammalian sequences [4] and also glycosylated in bovine milk OPN [25] (Figure 5). The five O-linked glycans identified in human milk OPN are equivalent to the five or six O-linked carbohydrates estimated to be present in rat bone OPN [23]. The migration of OPN in SDS/PAGE did not change after sialidase and/or O-glycosidase treatment. This indicates that the O-glycans of OPN are not sialylated at their termini and nor do they consist of unsubstituted core 1 structures only. The assay employed here will not reveal the presence of sialic acid residues if monosaccharides other than this make up the terminal carbohydrate residue. Hence, these studies do not exclude the possibility that sialic acid is a part of the carbohydrate chains associated with human OPN.
Human OPN contains two putative N-glycosylation sites, Asn63 and Asn90. Characterization of the peptides containing these asparagine residues shows that none of these are glycosylated in human milk OPN. In support of this, incubation of human OPN with PNGase F revealed no changes in SDS/PAGE migration, which would have been expected if N-glycans were present. Furthermore, MS analyses showed that the mass of all glycosylations in OPN is approx. 6.6 kDa, which is unlikely to comprise more than the five identified O-glycosylations. The absence of N-linked glycans is consistent with the results from bovine milk OPN, which is also devoid of modified asparagine residues [25]. Identification of mannose in a crude carbohydrate extract from rat bone has led to the estimation that one, or less than one, N-linked oligosaccharide is associated with rat bone OPN [23], and both neutral and sialylated N-linked glycans have been described in human bone [46]. Collectively, the present and other studies show that N-linked oligosaccharides are preferentially associated with bone OPN, whereas this kind of modification is absent from milk OPN, indicating tissue-specific glycosylation.
In summary, 36 phosphorylation sites and five sites of O-linked glycosylation have been localized in human milk OPN. The phosphorylations are predominantly located in clusters of three to five modified residues spanned by stretches of unmodified amino acids. The regions containing the carbohydrates and the RGD cell-adhesion site are devoid of phosphorylations. This specific arrangement of phosphoresidues and carbohydrates may play a critical role in orchestrating the many diverse functions of OPN. The tissue-specific PTM leading to different levels of phosphorylation and glycosylation among bone and milk OPN probably reflects different biological functions of the protein. The nature and extent of PTM is essential for the functioning of this key molecule in a multitude of biological processes. The present study describes the in vivo phosphorylation and glycosylation patterns of human OPN; a basis that will allow rational experimental design of functional studies aimed at understanding the structure–function relationship of OPN.
Acknowledgments
We thank L. M. Fogh for excellent technical assistance.
References
- 1.Franzén A., Heinegård D. Isolation and characterization of two sialoproteins present only in bone calcified matrix. Biochem. J. 1985;232:715–724. doi: 10.1042/bj2320715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher L. W., Torchia D. A., Fohr B., Young M. F., Fedarko N. S. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem. Biophys. Res. Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 3.Qin C., Baba O., Butler W. T. Post-translational modifications of SIBLING proteins and their roles in osteogenesis and dentinogenesis. Crit. Rev. Oral Biol. Med. 2004;15:126–136. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 4.Sodek J., Ganss B., McKee M. D. Osteopontin. Crit. Rev. Oral Biol. Med. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 5.Giachelli C. M., Steitz S. Osteopontin: a versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19:615–622. doi: 10.1016/s0945-053x(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 6.Denhardt D. T., Noda M., O'Regan A. W., Pavlin D., Berman J. S. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Invest. 2001;107:1055–1061. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boskey A. L., Spevak L., Paschalis E., Doty S. B., McKee M. D. Osteopontin deficiency increases mineral content and mineral crystallinity in mouse bone. Calcif. Tissue Int. 2002;71:145–154. doi: 10.1007/s00223-001-1121-z. [DOI] [PubMed] [Google Scholar]
- 8.Carlinfante G., Vassiliou D., Svensson O., Wendel M., Heinegård D., Andersson G. Differential expression of osteopontin and bone sialoprotein in bone metastasis of breast and prostate carcinoma. Clin. Exp. Metastasis. 2003;20:437–444. doi: 10.1023/a:1025419708343. [DOI] [PubMed] [Google Scholar]
- 9.Rittling S. R., Chambers A. F. Role of osteopontin in tumour progression. Br. J. Cancer. 2004;90:1877–1881. doi: 10.1038/sj.bjc.6601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashkar S., Weber G. F., Panoutsakopoulou V., Sanchirico M. E., Jansson M., Zawaideh S., Rittling S. R., Denhardt D. T., Glimcher M. J., Cantor H. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 11.Weber G. F., Zawaideh S., Hikita S., Kumar V. A., Cantor H., Ashkar S. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J. Leukoc. Biol. 2002;72:752–761. [PubMed] [Google Scholar]
- 12.Singh K., DeVouge M. W., Mukherjee B. B. Physiological properties and differential glycosylation of phosphorylated and nonphosphorylated forms of osteopontin secreted by normal rat kidney cells. J. Biol. Chem. 1990;265:18696–18701. [PubMed] [Google Scholar]
- 13.Safran J. B., Butler W. T., Farach-Carson M. C. Modulation of osteopontin post-translational state by 1,25-(OH)2-vitamin D3. Dependence on Ca2+ influx. J. Biol. Chem. 1998;273:29935–29941. doi: 10.1074/jbc.273.45.29935. [DOI] [PubMed] [Google Scholar]
- 14.Shanmugam V., Chackalaparampil I., Kundu G. C., Mukherjee A. B., Mukherjee B. B. Altered sialylation of osteopontin prevents its receptor-mediated binding on the surface of oncogenically transformed tsB77 cells. Biochemistry. 1997;36:5729–5738. doi: 10.1021/bi961687w. [DOI] [PubMed] [Google Scholar]
- 15.Shiraga H., Min W., VanDusen W. J., Clayman M. D., Miner D., Terrell C. H., Sherbotie J. R., Foreman J. W., Przysiecki C., Neilson E. G., et al. Inhibition of calcium oxalate crystal growth in vitro by uropontin: another member of the aspartic acid-rich protein superfamily. Proc. Natl. Acad. Sci. U.S.A. 1992;89:426–430. doi: 10.1073/pnas.89.1.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wada T., McKee M. D., Steitz S., Giachelli C. M. Calcification of vascular smooth muscle cell cultures: inhibition by osteopontin. Circ. Res. 1999;84:166–178. doi: 10.1161/01.res.84.2.166. [DOI] [PubMed] [Google Scholar]
- 17.Boskey A. L., Maresca M., Ullrich W., Doty S. B., Butler W. T., Prince C. W. Osteopontin-hydroxyapatite interactions in vitro: inhibition of hydroxyapatite formation and growth in a gelatin-gel. Bone Miner. 1993;22:147–159. doi: 10.1016/s0169-6009(08)80225-5. [DOI] [PubMed] [Google Scholar]
- 18.Hoyer J. R., Asplin J. R., Otvos L. Phosphorylated osteopontin peptides suppress crystallization by inhibiting the growth of calcium oxalate crystals. Kidney Int. 2001;60:77–82. doi: 10.1046/j.1523-1755.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- 19.Jono S., Peinado C., Giachelli C. M. Phosphorylation of osteopontin is required for inhibition of vascular smooth muscle cell calcification. J. Biol. Chem. 2000;275:20197–20203. doi: 10.1074/jbc.M909174199. [DOI] [PubMed] [Google Scholar]
- 20.Ek-Rylander B., Flores M., Wendel M., Heinegård D., Andersson G. Dephosphorylation of osteopontin and bone sialoprotein by osteoclastic tartrate-resistant acid phosphatase. Modulation of osteoclast adhesion in vitro. J. Biol. Chem. 1994;269:14853–14856. [PubMed] [Google Scholar]
- 21.Razzouk S., Brunn J. C., Qin C., Tye C. E., Goldberg H. A., Butler W. T. Osteopontin posttranslational modifications, possibly phosphorylation, are required for in vitro bone resorption but not osteoclast adhesion. Bone. 2002;30:40–47. doi: 10.1016/s8756-3282(01)00637-8. [DOI] [PubMed] [Google Scholar]
- 22.Al-Shami R., Sørensen E. S., Ek-Rylander B., Andersson G., Carson D. D., Farach-Carson M. C. Phosphorylated osteopontin promotes migration of human choriocarcinoma cells via a p70 S6 kinase-dependent pathway. J. Cell. Biochem. 2005;94:1218–1233. doi: 10.1002/jcb.20379. [DOI] [PubMed] [Google Scholar]
- 23.Prince C. W., Oosawa T., Butler W. T., Tomana M., Bhown A. S., Bhown M., Schrohenloher R. E. Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J. Biol. Chem. 1987;262:2900–2907. [PubMed] [Google Scholar]
- 24.Neame P. J., Butler W. T. Posttranslational modification in rat bone osteopontin. Connect. Tissue Res. 1996;35:145–150. doi: 10.3109/03008209609029185. [DOI] [PubMed] [Google Scholar]
- 25.Sørensen E. S., Højrup P., Petersen T. E. Posttranslational modifications of bovine osteopontin: identification of twenty-eight phosphorylation and three O-glycosylation sites. Protein Sci. 1995;4:2040–2049. doi: 10.1002/pro.5560041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sørensen S., Justesen S. J., Johnsen A. H. Purification and characterization of osteopontin from human milk. Protein Expr. Purif. 2003;30:238–245. doi: 10.1016/s1046-5928(03)00102-5. [DOI] [PubMed] [Google Scholar]
- 27.Salih E., Ashkar S., Gerstenfeld L. C., Glimcher M. J. Identification of the phosphorylated sites of metabolically 32P-labeled osteopontin from cultured chicken osteoblasts. J. Biol. Chem. 1997;272:13966–13973. doi: 10.1074/jbc.272.21.13966. [DOI] [PubMed] [Google Scholar]
- 28.Salih E., Zhou H. Y., Glimcher M. J. Phosphorylation of purified bovine bone sialoprotein and osteopontin by protein kinases. J. Biol. Chem. 1996;271:16897–16905. doi: 10.1074/jbc.271.28.16897. [DOI] [PubMed] [Google Scholar]
- 29.Sørensen E. S., Petersen T. E. Identification of two phosphorylation motifs in bovine osteopontin. Biochem. Biophys. Res. Commun. 1994;198:200–205. doi: 10.1006/bbrc.1994.1028. [DOI] [PubMed] [Google Scholar]
- 30.Lasa-Benito M., Marin O., Meggio F., Pinna L. A. Golgi apparatus mammary gland casein kinase: monitoring by a specific peptide substrate and definition of specificity determinants. FEBS Lett. 1996;382:149–152. doi: 10.1016/0014-5793(96)00136-6. [DOI] [PubMed] [Google Scholar]
- 31.Mercier J. C. Phosphorylation of caseins, present evidence for an amino acid triplet code posttranslationally recognized by specific kinases. Biochimie. 1981;63:1–17. doi: 10.1016/s0300-9084(81)80141-1. [DOI] [PubMed] [Google Scholar]
- 32.Meggio F., Pinna L. A. One-thousand-and-one substrates of protein kinase CK2? Biochimie. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 33.Lasa M., Chang P. L., Prince C. W., Pinna L. A. Phosphorylation of osteopontin by Golgi apparatus casein kinase. Biochem. Biophys. Res. Commun. 1997;240:602–605. doi: 10.1006/bbrc.1997.7702. [DOI] [PubMed] [Google Scholar]
- 34.Salih E., Ashkar S., Gerstenfeld L. C., Glimcher M. J. Protein kinases of cultured osteoblasts: selectivity for the extracellular matrix proteins of bone and their catalytic competence for osteopontin. J. Bone Miner. Res. 1996;11:1461–1473. doi: 10.1002/jbmr.5650111013. [DOI] [PubMed] [Google Scholar]
- 35.Senger D. R., Perruzzi C. A., Papadopoulos A., Tenen D. G. Purification of a human milk protein closely similar to tumor-secreted phosphoproteins and osteopontin. Biochim. Biophys. Acta. 1989;996:43–48. doi: 10.1016/0167-4838(89)90092-7. [DOI] [PubMed] [Google Scholar]
- 36.Barkholt V., Jensen A. L. Amino acid analysis: determination of cysteine plus half-cystine in proteins after hydrochloric acid hydrolysis with a disulfide compound as additive. Anal. Biochem. 1989;177:318–322. doi: 10.1016/0003-2697(89)90059-6. [DOI] [PubMed] [Google Scholar]
- 37.Annan R. S., Carr S. A. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 38.Finley E. L., Dillon J., Crouch R. K., Schey K. L. Identification of tryptophan oxidation products in bovine α-crystallin. Protein Sci. 1998;7:2391–2397. doi: 10.1002/pro.5560071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sørensen E. S., Petersen T. E. Phosphorylation, glycosylation and amino acid sequence of component PP3 from the proteose peptone fraction of bovine milk. J. Dairy Res. 1993;60:535–542. doi: 10.1017/s0022029900027886. [DOI] [PubMed] [Google Scholar]
- 40.Brunati A. M., Marin O., Bisinella A., Salviati A., Pinna L. A. Novel consensus sequence for the Golgi apparatus casein kinase, revealed using proline-rich protein-1 (PRP1)-derived peptide substrates. Biochem. J. 2000;351:765–768. [PMC free article] [PubMed] [Google Scholar]
- 41.Kemp B. E., Pearson R. B. Protein kinase recognition sequence motifs. Trends Biochem. Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 42.Salih E., Fluckiger R. Complete topographical distribution of both the in vivo and in vitro phosphorylation sites of bone sialoprotein and their biological implications. J. Biol. Chem. 2004;279:19808–19815. doi: 10.1074/jbc.M310299200. [DOI] [PubMed] [Google Scholar]
- 43.Qin C., Cook R. G., Orkiszewski R. S., Butler W. T. Identification and characterization of the carboxyl-terminal region of rat dentin sialoprotein. J. Biol. Chem. 2001;276:904–909. doi: 10.1074/jbc.M006271200. [DOI] [PubMed] [Google Scholar]
- 44.Andersson G., Ek-Rylander B., Hollberg K., Ljusberg-Sjölander J., Lång P., Norgård M., Wang Y., Zhang S. J. TRACP as an osteopontin phosphatase. J. Bone Miner. Res. 2003;18:1912–1915. doi: 10.1359/jbmr.2003.18.10.1912. [DOI] [PubMed] [Google Scholar]
- 45.Salih E., Wang J., Mah J., Fluckiger R. Natural variation in the extent of phosphorylation of bone phosphoproteins as a function of in vivo new bone formation induced by demineralized bone matrix in soft tissue and bony environments. Biochem. J. 2002;364:465–474. doi: 10.1042/BJ20011272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda K., Takahashi N., Tsukamoto Y., Honma H., Kohri K. N-glycan structures of an osteopontin from human bone. Biochem. Biophys. Res. Commun. 2000;268:814–817. doi: 10.1006/bbrc.2000.2224. [DOI] [PubMed] [Google Scholar]