Highlights
-
•
CRC microenvironment significantly affects tumor development and drug responses.
-
•
Preclinical models are of paramount importance for advances in CRC drug development.
-
•
Preclinical models have different limitations to be considered in drug development.
-
•
Targeting TME in CRC is a promising approach for improving therapeutic outcomes.
Keywords: CRC, Tumor microenvironment, Cancer therapy, Preclinical models
Abstract
Colorectal cancer (CRC) is a common cancer with high mortality rates. Despite progress in treatment, it remains an incurable disease for many patients. In CRC, the tumor microenvironment (TME) plays critical roles in tumor growth, progression, patients’ prognosis, and response to treatments. Understanding TME complexities is important for developing effective therapies. In vitro and in vivo preclinical models are critical in understanding the disease, discovering potential targets, and developing effective therapeutics. In this review, we focus on preclinical research studies associated with modulation of the TME in CRC. These models give insights into understanding the role of stroma and immune cell components of the TME in CRC and improve clinical responses, providing insights in novel treatment options. Various studies have focused on targeting the TME in CRC to improve responses to different therapeutic approaches. These include identifying targets for cancer therapies, targeting molecular signaling, and enhancing the efficacy of immunotherapeutic modalities. Furthermore, targeting stromal and angiogenic factors in the TME may provide new therapeutic options. Overall, understanding and targeting the TME in CRC is a promising approach for improving therapeutic outcomes.
Graphical abstract
Introduction
Colorectal cancer (CRC) is the third most common cancer among adults and the second leading cause of cancer-related death worldwide [1,2]. Many CRC patients are diagnosed with advanced clinical stages, making it challenging to use standard therapeutics, and resulting in poor clinical outcomes [3]. Several therapeutic modalities including chemotherapy, radiotherapy, immunotherapy, and targeted therapy have been used to treat CRC patients. However, despite advances in various therapeutic strategies, metastatic CRC remains challengeable and often incurable. [4]. In lieu of this, understanding the interactions between cancer cells and different components of the tumor microenvironment (TME), that affect tumor cell behavior, could serve as a potential pathway for improving clinical outcomes. The components of the TME play a crucial role in tumor development and progression through reprogramming tumor initiation, progression, and response to treatment. Therefore, it is important to understand the complex interactions of TME in CRC for the development of effective therapeutic targets for better response rates. Since CRC tumors with parental-specific genetic heterogeneity are grafted, the natural interaction of CRC tumor cells with a specific genetic mutational landscape can be observed. This allows a better understanding of the original tumor behavior with respect to its genetic and epigenetic mutational status, histopathological subtypes, metastatic representation, and its biological/genetic conditioning towards various drugs/agents [5]. Therefore, preclinical research models, such as in vitro cell culture and in vivo models, are important for investigating different aspects including the role of TME components and their interaction with CRC cells for identification of potential targets and development of novel and new therapeutic strategies. This review aims to highlight current advances and progress in preclinical models targeting the TME in CRC for the development of novel therapeutic strategies.
Tumor microenvironment in colorectal cancer
The microenvironment in CRC is complex and consists of malignant cells, various immune cells, stromal fibroblasts, intestinal microbiota, extracellular matrix (ECM), blood vessels, and signaling molecules. The majority of these cellular components are well-documented in their ability to contribute to tumor immune escape, leading to the progression and spread of cancer [6]. Specifically, immune cells within the TME can be manipulated by tumor cells to either promote or inhibit tumor growth leading to cancer regression or progression, depending on the type of immune cells involved. For example, intra-tumoral effector cells such as cytotoxic T cells, and M1 macrophages inhibit tumor growth by enhancing antitumor immune responses. On the other hand, tumor-associated immunosuppressive cells, such as M2 tumor-associated macrophages (M2-TAMs), myeloid-derived suppressor cells (MDSCs), some T regulatory cell (Treg) subsets, and other immunosuppressive cells, play critical roles in promoting tumor development by suppressing antitumor immune responses. M2-TAMs play a crucial role in inducing immunosuppression by producing anti-inflammatory cytokines such as CCL20, recruiting CCR6+ Tregs, and contributing to tumor progression [7]. Additionally, M2-TAMs contribute to tumor development by producing vascular endothelial growth factor (VEGF) to promote angiogenesis, releasing fibroblast growth factor-1 and epidermal growth factor to foster cancer cells, and matrix metalloproteinases to enhance tumor invasion [8,9]. MDSCs exert potent immunosuppressive effects via the production of arginase-1, indoleamine 2,3-dioxygenase, and reactive oxygen species, which suppress functions of T and NK cells [10,11]. It has been reported that increased levels of reactive oxygen species in the CRC microenvironment can reduce antigen-specific T cell responses, which are involved in effective anti-tumor immune responses [12]. Furthermore, MDSCs can promote Treg expansion and macrophage polarization towards an M2-phenotype; therefore, reinforcing the immunosuppressive environments and promoting tumor growth through angiogenesis and therapeutic resistance. Many studies investigated the role of MDSCs in CRC [13,14]. It has been reported that mutations in the RAS family (K-RAS, N-RAS, H-RAS), drive releasing of chemokines from cancer cells through the KRAS/IRF2 signaling pathway, which in turn recruits MDSCs into the CRC microenvironment, resulting in inhibiting the anti-tumor immunity, and resistance to immunotherapy [15]. Furthermore, a recent study investigated the effectiveness of chimeric antigen receptor T-cells (CAR-T cells) by targeting immunosuppressive cells which play a crucial role in CRC progression [16]. The authors showed that programmed cell death-1 (PD-1, CD279)-triggering-receptor-expressed on myeloid cells 2 (TREM2)-targeting single-chain variable fragment (scFv) could inhibit the signaling pathway of PD-1/PD-L1 in TAMs and MDSCs by blocking their TREM2 receptors in CRC [16]. Another study explored the feasibility of targeting CD166 with CD6-CAR-T cells in CRC [17]. The study revealed that CD6-CAR-T cells could exert a cytotoxic effect against CRC cells by enhancing interferon-gamma (IFN-γ) production [17].
Stromal cells such as cancer-associated fibroblasts (CAFs) facilitate tumor cell proliferation and progression by supporting tumor angiogenesis for the formation of new blood vessels, modulation of immune cell recruitment, and most importantly initiating ECM modulation to support the structural integrity of the tumor [[18], [19], [20], [21], [22]]. Mainly, ECM within the TME leads to increased stiffness by altering its composition. ECM also promotes tumor cell proliferation and migration through the VEGF, which facilitates angiogenesis by promoting blood vessel formation while proteases facilitate ECM modulation to create pathways for vessel growth [23,24].
The tumor metabolism has essential consequences for the TME. For instance, in CRC, cancer cells utilize aerobic glycolysis to produce more lactate, generating more acidic TME [25]. This, in turn, suppresses the immune cell function including NK cells and CD8 cytotoxic T cells (CTLs), weakening their ability to eradicate cancer cells [26]. Also, excess lactate can polarize TAMs into M2 phenotype, contributing to tumor development via activation of CD47/signal-regulatory protein alpha (SIRPα) [27,28]. Furthermore, amino acid metabolism, such as tryptophan, plays a crucial role in modulating immune responses in CRC. Indoleamine 2,3-dioxygenase plays an important role in mediating tryptophan metabolism. Enhancing tryptophan metabolism can increase the proliferation of immunosuppressive Tregs [29] and promote tumor cell survival [30]. Therefore, understanding and deciphering the role of various TME components can serve as essential tools for developing new and novel therapeutic strategies for improved clinical outcomes in CRC.
Preclinical models in cancer research
Researchers have developed various preclinical models to investigate tumor development and other hallmarks of cancer. Preclinical research models, including cellular and mouse models, help researchers to understand the molecular mechanisms of tumor growth and immune evasion and to evaluate potential therapeutics before moving to clinical trials [31]. Preclinical cancer models encompass a variety of experimental models, starting from in vitro models, which include two-dimensional (2D) monolayer cell culture, coculture, and organoids/three-dimensional (3D) cell culture models, into in vivo models, which include cell-derived xenografts (CDX) and patient-derived xenografts (PDX) models as well as genetically engineered mouse models (GEMMs).
The coculture models of organoids are useful for investigating the interactions between the TME and cancer cells, with 3D cell cocultures providing a more accurate representation of in vivo TME, offering a closer mimicry of in vivo cell settings compared to traditional 2D cell cultures [32], which can help investigations to understand cellular behaviors and interactions. For instance, the tumor organoid models “tumoroid” derived from tumors can be used in coculture systems to study the gene expression involved in cell-cell interaction, the response to immune checkpoint inhibitors (ICIs), and the development of tumor-reactive T cells. A recent study used tumoroid models to investigate carcinoma-TME communication in ex vivo cultures. The authors found that tumoroids could suppress gene expression involved in immune cell migration and inflammation [33]. Thus, these models can serve as critical tools in cancer research for studying cellular interactions within the TME and identifying drug interaction pathways, mechanisms of drug resistance, and drug discovery initiatives against various cancers [[34], [35], [36]].
On the other hand, in vivo models, such as CDX in CRC, depend on the process of implanting or injecting CRC cell lines into immunodeficient mice. These models are utilized to understand CRC development in an advanced stage of the disease [37]. Although CDX has several drawbacks, these preclinical models are still largely used for CRC due to advantages such as the availability of a large number of CRC cell lines, reduced experimental timeline due to rapid tumor growth, ease of genetic manipulation/mutational analysis, and expeditious drug response data available [38]. Although CDX and PDX models can be used for studying a whole tumor in situ [39], PDX closely resembles original tumors and have stromal microenvironments similar to human tumors; therefore, they better recapitulate the complex interactions of in vivo cancer models compared to CDX models [40]. Furthermore, PDX is a high-impact preclinical model especially for studying drug evaluations as it represents similar associated outcomes as expected to be observed in CRC patients [37]. With respect to the TME, it has been observed that upon engraftment of patient stromal cells in the CRC-PDX models, subsequent replacement of these cells with murine equivalents is observed with the recruitment of murine accessory cells to the tumor niche [5]. GEMMs are considered useful preclinical models for studying CRC-TME interactions as these are genetically edited mice models that are created by activating or deactivating certain genes [41]. GEMMs are highly advantageous for studies on CRC development as they have a fully functional natural murine immune system that naturally simulates the development of CRC tumors from adenoma to carcinoma to metastasis. This functionality of GEMMs provides a broader understanding of the role of TME components in CRC development including the development of well-differentiated cancer cells and investigation of the metastatic potential of CRC cells to the lungs and liver. In addition to this, they are useful in studying the function and role of specific genetic/somatic mutations associated with tumor proliferation, invasion, and metastasis such as tumor protein p53 (TP53), kirsten rat sarcoma viral (KRAS), adenomatous polyposis coli (APC), and cyclin-dependent kinase inhibitor 2A (CDKN2A) in CRC and pancreatic ductal adenocarcinoma [42,43].
Preclinical studies using therapeutics for modulating the TME in CRC
Recent preclinical studies have focused on modulating the TME of CRC to develop more effective therapies using different models, including human CRC cell lines, CDX, PDX, and GEMMs [37,43,44]. These studies have explored different therapeutic approaches aimed at altering the cellular microenvironment and signaling pathways of the TME, as illustrated in Fig. 1. For instance, some therapeutics act as metabolic modulators, affecting the TME by regulating the interaction between stromal components, immune cells, and tumor cells. This regulation has been evidenced in several studies, which reported that metformin, though an anti-diabetic drug, also exhibits anti-tumor effects against various cancers, including CRC [45]. A preclinical study in an orthotopic CRC model demonstrated that metformin could reprogram the TME [46]. The study reported that metformin enhances chemoimmunotherapy activity, including MIL-100/mitoxantrone/hyaluronic acid nanoparticles (MMH-NPs) chemotherapy, combined with anti-OX40 agonist monoclonal antibody (αOX40) immunotherapy [46] leading to improved delivery of MMH-NPs and increased T-cell infiltration into the TME of CRC while decreasing tumor-associated immunosuppressive cells (e.g. MDSCs and M2-TAMs), which are responsible for inhibiting CTLs in the TME [46]. The mechanism of metformin in enhancing anti-tumor immunity is less well understood. However, a recent study showed that metformin has the potential to regulate the activation of AMP-activated protein kinase (AMPK), which is related to inhibiting immunosuppressive cells [47]. In the same study, it was reported that metformin inhibited 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase of the mevalonate pathway, which is important for immunosuppressive cell differentiation within the TME [47]. These factors may have the potential to suppress tumor cell growth and enhance antitumor immunity [48]. Another study reported that metformin plays a role in reprogramming tryptophan metabolism in colorectal cancer xenograft models [49]. It could reduce cancer cell growth via decreasing solute carrier 7A5 (SLC7A5) through MYC, preventing tryptophan uptake in cancer cells [49]. Therefore, blocking tryptophan metabolism could lead to remodeling the anti-tumor immunity via expansion of CD8+ T cells and enhancing their activation [49]. Another study by Wu et al. investigated the effect of cordycepin (an adenosine analogue) on tumor development through its role in regulating biological function in murine CRC cell lines [50]. The study reported that cordycepin effectively inhibited the proliferation, invasion, and migration of CRC by reducing programmed death-ligand-1 (PD-L1) expression in cancer cells [50]. The study highlights the potential of cordycepin as a valuable tool in cancer therapy by offering a positive effect against CRC development [50]. Furthermore, it has been reported that methionine enkephalin (an opioid peptide) inhibited MC38 colorectal tumor growth in mice through different mechanisms including modulation of the tumor immune microenvironment (TIME) by increasing the infiltration of M1 macrophages and CD4+/CD8+ T effector cells and reducing the infiltration of MDSCs and M2 macrophages [51]. A recent study explored the role of M2-TAMs in inducing chemotherapy resistance in CRC [52]. It was reported that M2-TAMs were responsible for producing CXCL17 and CXCL22, which in turn play key roles in activating the chemokine (C–C motif) receptor 4 (CCR4) receptor in CRC cells [52]. This interaction triggers the intracellular signaling via ATF6 and GRP78, resulting in upregulation of MRP1 [52]. The increased MRP1 in CRC cells leads to the efflux of 5-FU from cancer cells, decreasing its cytotoxic effects [52]. Therefore, targeting this signaling pathway and M2-TAMs within the TME of CRC represents a promising strategy for improving the effectiveness of cancer therapy. Furthermore, it was reported that CD155 plays a crucial role in promoting macrophage polarization towards the M2-TAM phenotype, which contributes to immunosuppressive activities and enhances colorectal tumor progression [53]. A recent study found that TAMs within the CRC TME have high expression levels of CD155 [53]. Role of CD155 in TAMs was explored by using genetically engineered murine models of CRC [53]. It was reported that knockdown of CD155-TAM in CRC ectopic mice models have fewer and smaller tumors with high infiltration of CD8+ T cells and M1-TAMs and increased CD8+/CD4+ T cells ratio, compared to wild-type mice [53]. Collectively, these findings suggest that targeting CD155-TAMs can be a promising strategy for modulating the immune landscape in the TME of CRC and enhancing clinical outcomes.
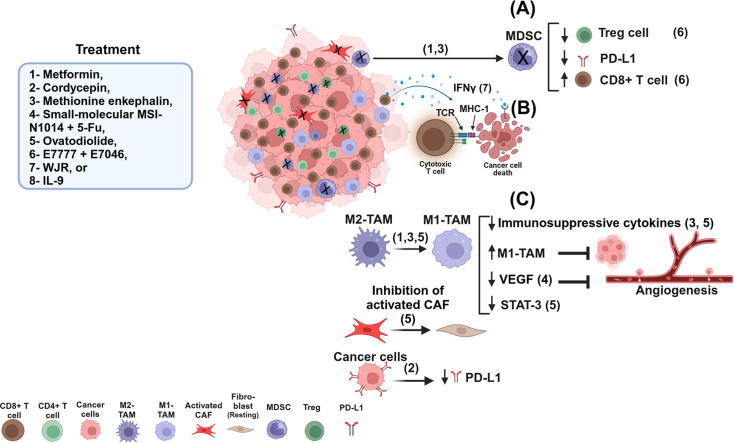
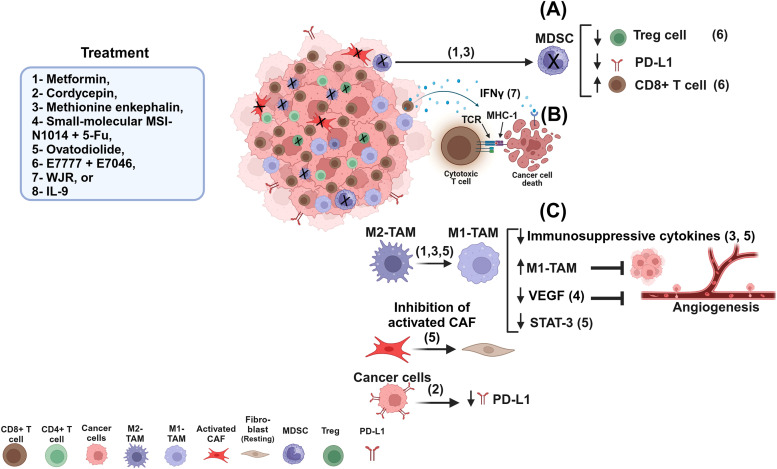
Fig. 1.
A schematic diagram shows different treatments modulating the TME. Treatments (1 and 3) can modulate the TME via decreasing MDSCs and increasing the percentage of CD8+ T cells, while decreasing the Tregs and PD-L1 in cancer cells (A). Treatment (7) modulates the TME by activating CD8+ T cells, leading TCR to bind with MHC-I via peptides derived from neoantigen-reactive CD8+ T cells and inducing IFN-γ production, which can bind to IFN-γ receptor in cancer cells, resulting in destroying cancer cells (B). Treatments (3,5) can modulate the TME via shifting M2-TAM into M1-TAM, decreasing immunosuppressive cytokines and inhibiting tumor growth (C).
Different studies have shown that CAFs are associated with tumor development and progression, and resistance to anticancer therapies. A study by Yadav et al. evaluated the therapeutic efficacy of the small-molecule MSI-N1014 in suppressing the generation of CAFs and oncogenic markers, and inhibiting CRC [54]. It was observed that MSI-N1014 treatment delayed tumor growth, when combined with 5-FU, and showed a strong suppressive effect on tumor development in a drug-resistant colon cancer model [54]. Additionally, this combinational therapy suppressed CAF transformation and decreased tumor-promoting signaling, including the expression of β-catenin, leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5), mammalian target of rapamycin (mTOR), VEGF, CD44 and IL-6 [54].
A traditional Chinese Medicine (TCM), known as Wenzi Jiedu Recipe (WJR), has been reported to exhibit promising potential against different types of cancers [55]. In CRC, WJR treatment has been reported to demonstrate superior therapeutic efficacy in countering CRC development in both in vitro and in vivo models [55]. The study results showed that WJR treatment increased the proportion of CD8+ T cells and enhanced levels of key cytokines including interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), and interleukin-10 (IL-10) indicating its role in modulation of the immune response [55]. Some preclinical studies have also investigated different therapeutic tools such as bacterial minicell therapy, cytokine therapy, immune checkpoint inhibitors, and others in treating CRC through modulation of the TIME. This modulation has the potential to increase infiltration of T cells, reduce tumor-associated immunosuppressive cell subsets, and enhance anti-tumor immune responses. Moreover, current evidence suggests that bacteria-derived minicells may play a key role in enhancing chemotherapeutic drug efficacy, inhibiting chemotherapy-resistant tumor cell growth, and facilitating the delivery of chemotherapeutic agents into hypoxic and necrotic environments within solid tumors [56,57]. For example, VAX014 is an oncolytic bacterial minicell agent that has anti-tumor effects with immunoregulatory properties [58,59]. It has been shown that intralesional injection of VAX014 improved the efficacy of immune checkpoint inhibition in cold tumors of murine melanoma models [58]. A recent study highlighted the potential of VAX014 as an anti-cancer agent against mouse colon adenocarcinoma cell lines [60]. In vivo investigations demonstrated that VAX014 treatment effectively inhibited colon adenoma development by reducing the number and size of colon polyps and inhibiting cell proliferation in a CRC mouse model [60]. Furthermore, in the presence of adenomas, VAX014 exhibited a favorable immune response by activating cell-mediated immunity and increased CTL activity and modifying the CRC microenvironment by promoting activation of pro-inflammatory cytokines such as IL-6 and IL-17 expression levels associated with inflammation [60]. In addition to these cytokines, IL-9 is a multifunctional cytokine that can control the function and activity of different types of cells by displaying both anti-tumor and pro-tumor functions. Notably, IL-9 has been found to have an anti-tumor effect by stimulating innate and adaptive immune responses within the solid TME [61]. Recent studies have suggested that IL-9 may play a beneficial role against colon cancer growth and reshaping the TME in the preclinical allograft animal models [62].
Targeting tumor-associated immunosuppressive cells in the TME for reprogramming the TIME by using therapeutic drugs is a crucial strategy, offering promises for innovative cancer immunotherapies. For instance, a recombinant fusion protein of diphtheria toxin and human interleukin- 2 targeting CD25 (denileukin diftitox; E7777) has been shown to improve anti-tumor activity through reprogramming the TIME [63]. The study reported that treatment of animal models with E7777 alone or in combining with E7046 treatment could reduce percentages of intra-tumoral CD4+CD25+FoxP3+ Tregs [63]. Furthermore, therapeutic combination of E7777 with E7046 also increased the frequencies of intra-tumoral CD8+ T cells [63] indicating promising anti-tumor effects and immune regulatory properties in preclinical studies, stressing their role as potential targets for salvage therapies in CRC patients.
Several studies have emphasized the significance of recognizing genetic and nongenetic markers in CRC patients to identify new biomolecular signaling targets for novel therapy protocols [64,65]. For instance, a bioactive compound Ovatodiolide (Ova), derived from the herb Anisomeles, exhibits notable antitumor potentials in several cancer types, including CRC, bladder cancer, and renal cell carcinoma [[66], [67], [68]]. Ova exerts its inhibitory effects on tumor development by targeting oncogenic signaling pathways, such as Yes-associated protein (YAP-1), thereby impeding tumor growth. An in vivo study revealed that Ova, either alone or in combination with fluorouracil (5-FU) treatment, could effectively hinder CRC development by suppressing the oncogenic YAP-1 signaling pathway and inhibiting polarization M2-tumor-associated macrophage (TAM) [68], as illustrated in Fig. 1. Another preclinical investigation showed that Ova can reduce the release of exosomes from colon tumor spheres (Exoph), thereby mitigating M2-TAM polarization and CAF generation in vitro, consequently fostering the efficacy of chemotherapy [69]. Additionally, the investigators observed that Ova treatment inhibited tumor progression by diminishing the expression of STAT-3, β-catenin, and IL-6, alongside reducing the level of miR-1246 in serum exosomes in a mouse xenograft model [69]. A more recent study combined organoids and organ-on-chip models to examine the invasion of CRC within γ-aminobutyric acidergic (GABAergic) TME. It was reported that GABAergic signaling can affect cancer cell behavior, providing insights into potential therapeutic targets [70]. G protein-coupled receptors (GPR56) play a key role in immune regulation by reducing the cytotoxicity of natural killer (NK) cells and the secretion of cytokines [71]. Some studies reported that high levels of GPR56 in tumors were correlated with tumorigenesis, immunoregulation, drug resistance, CRC microsatellite stability (MSS) tumor, and poor survival [[72], [73], [74], [75]]. A recent study revealed that treatment with the GPR56-targeted antibody-drug conjugates (ADCs) showed antitumor efficacy in CRC mouse xenograft models by inhibiting tumor growth and improving survival rates [74]. This suggests that the efficacy of GPR56-targeted ADCs may also contribute to reshaping the TME in xenograft models. Furthermore, a recent study investigated the function of cullin 4B (CUL4B) deletion in the intestinal epithelium of preclinical ApcMin/positive mouse models [76]. The authors found that blocking the CUL4B can enhance adenoma formation and the accumulation of MDSCs through the regulation of granulocyte colony-stimulating factor (G-CSF) transcriptionally [76]. Metastasis is the major cause of related death in patients with CRC. Different genes can contribute to promoting CRC metastasis, including leucine-rich alpha-2 glycoprotein 1 (LRG1)-human epidermal growth factor receptor 3 (HER-3) signaling pathway, which has the potential to promote liver metastasis in CRC patients [77]. A recent study found that LRG-1 released from liver endothelial cells (ECs) can activate CRC-associated HER-3 and promote tumor growth [77]. The preclinical study revealed the blocking of the LRG-1/HER-3 axis could decrease CRC metastasis in the liver, and prolong mouse survival in LRG-1 knockout mouse models [77]. Therefore, targeting LRG-1 may contribute to the development of new therapeutic strategies for improved clinical outcomes in CRC.
Preclinical studies using therapeutics to improve immune checkpoint inhibitors
Immune checkpoint inhibitors (ICIs) are a form of cancer immunotherapy used to treat various malignant tumors by targeting immunologic receptors, such as PD-1 and its ligand (CD274, PD-L1). Though ICIs show impressive clinical outcomes in cancer patients, the response rates are observed in a fraction of the patients indicating that additional strategies must be explored to manipulate the TIME and subsequently increase the response rates [78]. Therefore, combining ICIs with other therapeutics may enhance their efficacy against cancers. Fig. 2 summarizes effects of anti-PD-1/L1, alone or in combination with other therapeutics, on different immune cells and molecules. A recent study investigated the effect of foretinib (receptor tyrosine kinases (RTKs) inhibitor) on anti-tumor immunity in murine colon cancer cells [79], and reported that foretinib can modify the TME and enhance effect of immune checkpoint inhibitors [79]. The study evidenced that combining foretinib with an anti-PD-1 monoclonal antibody ameliorated the TME by enhancing the infiltration and function of effector CD4+ T cells and CTLs (Fig. 2). It also induced overexpression of PD-L1 on cancer cells by activating the JAK2-STAT1 signaling pathway in the animal model [79]. Moreover, inhibition of metastasis of murine colon cancer cells to the mouse lung was observed with a reduction in the proportions of M2-TAMs and Tregs [79] indicating that combinational therapies can potentially enhance the effect of anti-tumor immunotherapies in CRC patients [79]. Another study reported that regorafenib, a kinase inhibitor with anti-angiogenic activity, can play a beneficial role in improving ICI treatment efficacy [80]. Compared to monotherapy, combining regorafenib with anti-PD-1 was shown to enhance the anti-tumor activity of anti-PD-1 immunotherapy by modulating the TME, reducing the percentage of tumor-associated immunosuppressive Tregs and M2-TAMs, and enhancing M1 polarization by activating CD8+ T cells resulting in inhibition of tumor growth and liver metastasis in the orthotopic colon cancer model (Fig. 2) [80]. Similar results were observed with MTL-CEBPA; a novel therapy known as the first small activating RNA (saRNA) therapy, which targets the transcription factor CCAAT enhancer binding protein alpha (C/EBPα) [81] and exhibits beneficial activity against inflammation and cancer [82,83]. A study demonstrated that the combination of MTL-CEBPα with anti-PD-1 can minimize tumor size by regulating the TME leading to increased tumor-infiltrating T cells in pre-clinical models (Fig. 2) [84]. Furthermore, a recent study in colon and liver cancer animal models showed that a combination of E7777 treatment with anti-PD-1 therapy leads to enhanced CD8+ T cell levels (Fig. 2), increased anti-tumor activity, and improved overall survival [85]. The CD1d-restricted invariant natural killer T (iNKT) cells play a crucial role in regulating a wide range of immune responses, including those in cancer [86]. The activation of these cells by alpha-galactosylceramide (alpha-GalCer) leads to the release of various cytokines, including IFN-γ, which is related to killing tumor cells and activating other immune cells, contributing to cancer regression [87]. The presence of these cells in the TME can stimulate the activation of CTLs, preventing cancer's worsening. A recent study in a preclinical model of CRC showed that combined treatment with anti-PD-1 and alpha-GalCer can improve immunotherapy [88]. It was reported that this treatment could prevent the loss of iNKT cells and promote CD4+ TH1 T cells, contributing to the reduction of polyps’ development in ApcMin/+ mice, compared to anti-PD-1 blockade alone [88].
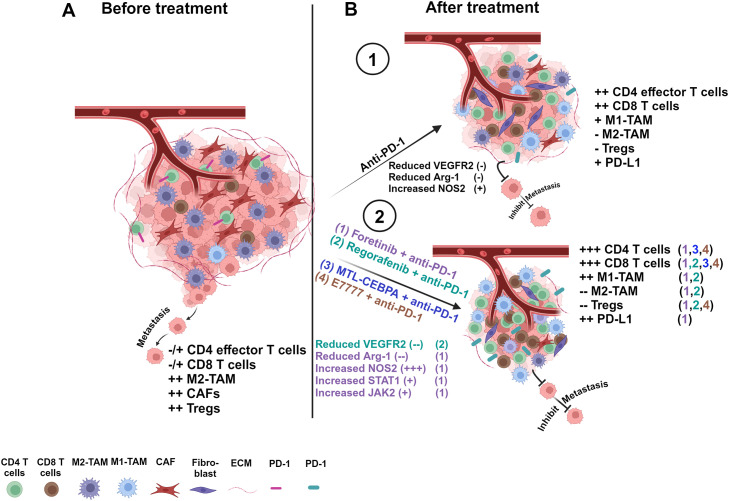
Fig. 2.
A schematic diagram shows different treatments improving anti-PD-1/L1 and modulating the TME. (A) represents the TME before treatment. (B) represents the TME after treatment. Targeting tumor cells and the TME by using immunotherapy alone can induce moderate modulation by increasing effector CD4+ and CD8+ T cells (1). Targeting tumor cells and the TME by combining anti-PD-1 with another therapeutic induces higher modulation in the TME through enhancing effector T cells, activating CD8+ T cells, and decreasing M2-TAM (2).
Tregs increase within the TME can weaken the effectiveness of anti-cancer immune responses in some cancers. Therefore, by reducing the suppressive influence of Tregs, the immune system may have better chances of effectively targeting and eliminating cancer cells. CCR8 expressed on Tregs, can act as a driver of immunosuppression [89,90]. It was reported that high levels of CCR8+ Tregs were associated with tumor progression in different cancers [[91], [92], [93]]. A recent study showed that a novel Fc-optimized anti-CCR8 antibody (BAY 3,375,968) can potentially deplete tumor-infiltrating Tregs in colon cancer xenograft mouse models [94]. This anti-mouse CCR8 antibody could recognize and bind to Tregs, inhibiting their suppressive function and enhancing anti-tumor immune responses [94]. Furthermore, combining anti-mouse CCR8 antibody with anti-PD-L1 led to minimizing the tumor mass, decreased intra-tumoral CD4+CD25+FoxP3+ Tregs, increased the ratio of CD8+ T cells to Tregs, and enhanced IFN-γ production within the TME, as well as improved the survival rate in colon cancer mouse model [94]. Another study reported that CCR8+ Tregs can induce the exhaustion of anti-tumor immune cells and enhance the immune suppressive functions of Tregs in the CRC microenvironment [95]. Overexpression of CCR8 on tumor-infiltrating Tregs was capable of enhancing TIGIT and CTLA4 immune checkpoints and inducing strong immune suppression of conventional CD4+ T cells and CTLs in the TME of CRC animal models [95]. Depletion of CCR8+ cells enhanced the anti-tumor activation of conventional CD4+ and CD8+ T cells by reducing expression of PD-1, T-cell immunoglobulin and mucin domain 3 (TIM-3), and lymphocyte activation gene 3 protein (LAG-3) [95]. Collectively, these findings suggest that an anti-CCR8 antibody has the potential to enhance the efficacy of cancer therapy.
Cytokines modulate immune responses and their utility as combinational tools with immunotherapy are well studied. For example, IL-15 has been evidenced to enhance proliferation and differentiation of CD8+ T cells and NK cells, thereby exerting significant anti-tumor effects [96]. A recent study generated a recombinant IL-15 fusion albumin binding domain (hIL15-ABD) and explored the therapeutic efficacy and immune regulation of combination fusion protein with anti-PD-L1 immunotherapy in murine colon cancer and melanoma models [97]. It was observed that the combination of hIL15-ABD with anti-PD-L1 enhanced anti-tumor effector cell activity, increased the production of granzyme-B and levels of CD8+IFN-γ+ T cells and CD8+IL-2+ T cells, and reduced populations of tumor-associated immunosuppressive cells [97]. The mechanism of inhibiting tumor growth may be due to secreting granzyme-B by CD8+ T cells and NKT cells, inducing apoptosis in cancer cells via regulating the BAX/BAK pro-apoptotic pathway [98]. Similarly, another study explored the role of IL-6 in TME modulation in CRC. The study reported that IL-6 can trigger significant immunosuppression within the TME by recruiting immunosuppressive cells and hindering T-cell infiltration [99]. Consequently, blocking IL-6 improved the effectiveness of anti-PD-L1 therapy in CRC, offering a new strategy to combat anti-PD-L1 resistance in CRC patients [99] and promising anti-tumor effects, highlighting their potential in CRC treatment. On the other hand, a preclinical study revealed that exosomal-PD-L1 can reduce the efficacy of anti-PD-1 therapy, contributing to immune evasion in tumors [100]. Therefore, targeting exosome-PD-L1 may inhibit tumor dissemination by improving the efficacy of anti-PD-1 blockade.
Stromal and angiogenic targets
Several studies have demonstrated the utility of therapeutic approaches targeting stromal structures and angiogenic markers within the TME in preclinical studies on CRC. These approaches target components distinct from cancer or immune cells, offering promising avenues for intervention in the early stages of CRC. Analysis of the TME components has revealed that certain stromal-related genes correlate with poor patient outcomes [101]. Moreover, recent advances have led to the development of sophisticated 3D models for investigating CRC cells, monocytic cells, and stromal mesenchymal cells (MSCs) [102,103]. These models serve the purpose of understanding the TME dynamics and designing innovative therapies. Of particular interest is the identification of deficiencies in DNA mismatch repair (dMMR)/microsatellite instability-high (MSI-H) is most common in CRC patients; and can be detected in other cancer types. These cases often exhibit a TGF-β-rich tumor-promoting TME, associated with dismal survival outcomes [104]. In TGF-β-rich TME, M1-macrophages shift into M2-TAMs, which can express high levels of VEGF and TGF- β, thus contributing to CRC progression via suppressing the infiltration of CD8+ T cells and NK cells [105]. Additionally, TGF-β can enhance CAFs to produce ECM proteins such as collagen and fibronectin, resulting in tumor development and immune evasion [106].
Some preclinical studies have investigated the role of CAFs in CRC growth. CAFs are known to promote tumor growth and metastasis through the production of growth factors and extracellular matrix remodeling [20]. Therefore, targeting CAFs or their signaling pathways could potentially inhibit tumor progression. On the other hand, ECM plays a crucial role in tumor-stromal interactions and tumor progression. In a preclinical study, investigators elucidated the intricate interplay between CAFs and squamous epithelial cells (SECs) within the TME [107]. This study underscores the pivotal role of these cellular components in driving progression of inflammation-induced CRC [107]. Using a mouse model, the researchers showed distinct cellular alterations occurring within the TME, shedding light on potential mechanisms underlying tumor initiation and progression [107]. Such preclinical studies are invaluable for unraveling the complex biology of cancer and identifying novel therapeutic targets. The findings from this work contribute significantly to understanding CRC pathogenesis and hold promise for the development of targeted interventions aimed at disrupting the tumor-promoting interactions between CAFs and SECs. The calponin 1 (CNN1) and tropomyosin beta chain isoform 2 (TPM2) are specific stromal markers in tumor-associated stromal cells that hold promise for identifying high-risk CRC patients and enabling more effective targeted therapies [108]. Targeting these specific stromal markers may help to understand cancer pathogenesis. Additionally, research has identified alterations in angiogenesis-related genes (ARGs) that correlate with CRC characteristics and prognosis [109]. Surgical stress has been reported to induce immunosuppression and promote pro-tumorigenic cytokine production such as CCL18, leading to the recruitment of Tregs and the depletion of NK cells, and contributing to tumor recurrence or metastasis [110]. Surgery-induced stress can alter key components and cellular metabolisms of adaptive and innate immune systems, one of which requires a ubiquitin-like protein named neural precursor cell expressed, developmentally downregulated 8 (NEDD8), involved in the neddylation pathway. The NEDD8 protein is important in regulating various cellular processes, such as cell cycle progression, transcriptional factor activation, and promotion of tumor angiogenesis [111]. A recent study found that NEDD8 protein was increased in the Tregs due to surgery-induced stress [112]. Depletion of NEDD8 can reduce the postoperative lung metastasis of CRC cells in mouse models and recover anti-tumor immunity, including NK cells and CD8+ T cells [112]. Therefore, alteration of key components caused by surgical stress can influence cancer cells' behavior and their ability to metastasize. These findings underscore the potential of targeting angiogenic, metastasis, and stromal factors to develop innovative therapies addressing CRC progression-related factors at the molecular level within the TME.
Computational methodologies for identifying therapeutic targets in CRC-TME
The application of bioinformatics in the study of colorectal tumors involves the use of computational tools to analyze genetic and molecular data for a deeper understanding of the tumor's underlying mechanisms. Several bioinformatics approaches exist to identify potential biomarkers for the early detection of CRC. Detection of potential targets is important for drug discovery. Bioinformatics approaches and integrated statistics have been applied to explore molecular signatures of CRC and their receptors as drug targets [113]. Lin et al., found through bioinformatics analysis that NDUFA4L2, an important mitochondrial respiratory chain subunit, is a novel biomarker for colorectal cancer. The study detected 20 hub genes from the protein-protein interaction, using the maximum clique centrality algorithm [114]. Another study by Yue et al., used bioinformatics tools and Venn diagrams to detect upregulated and downregulated intersection genes of differentially expressed genes. They investigated the correlation between the TME-related genes and CRC by using the ESTIMATE data algorithm based on The Cancer Genome Atlas (TCGA) database. Interestingly, the study reported that CX3CR1 can act as a protective biomarker in the TME of CRC. These bioinformatics results were further validated by analyzing the expression of CX3CR1 in CRC tissues and cell lines [115].
Yu et al., combined bioinformatics tools and surface-enhanced laser desorption/ionization mass spectrometry to discover new biomarkers and patterns with high sensitivity and specificity for the detection of colorectal cancer. A multi-layer perceptron artificial neural network with a scaled conjugate gradient optimized backpropagation algorithm was applied to differentiate colorectal cancer from healthy individuals. Additionally, a linear support vector machine was used to differentiate CRC from colorectal adenoma [116]. The identification of genetic variations in hereditary CRC screening can also be carried out using bioinformatics tools [117]. A bioinformatics algorithm known as the multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) effectively categorizes MLH1/MSH2 deleterious and neutral missense variants. This algorithm is helpful in the evaluation of the pathogenicity of hereditary CRC gene variants [118]. Furthermore, Thompson et al., calibrated several in silico methods to predict the effects of mismatch repair gene missense substitutions. They found that a bioinformatics tool is adequately predictive to be applied as a continuous variable in the quantitative Bayesian integrated evaluation for the clinical categorization of missense substitutions in mismatch repair gene [119].
It is reported that there is a high number of hyper-methylated and silenced genes in colon cancer, the majority of which are tumor-suppressor genes [120]. Progress in sequencing techniques and the evolution of bioinformatics tools have facilitated the creation of single-base resolution maps of human methylomes [121]. Using bioinformatics analysis and next-generation bisulfite sequencing technology, researchers were able to create single-base pair resolution methylation maps representing various stages of cellular differentiation. A recent pan-omics study on commercially available CRC organoids highlighted the role of SMAD4 inactivation in enhancing cell migration, proliferation, and tumorigenesis. CRC patients with SMAD4 mutations and elevated DKK4 (Dickkopf family) expression show poor prognosis, suggesting new therapeutic approaches for advanced colorectal cancers [122]. The data generated by these pan-omics techniques are filtered, aligned, and analyzed using various bioinformatics tools and compared with parent tumor tissue. This information can be used to identify new therapeutic targets for the cancer diagnostics and therapeutic monitoring. Additionally, the data can support clinical decision-making regarding the administration of chemotherapeutic agents, thus contributing to precision medicine [123].
Recently, Wills et al., utilized a series of tools to conduct bioinformatics analysis on a whole genome-wide association study involving a very large cohort of patients. They identified a link between overall survival and rs79612564 in the receptor tyrosine kinase ERBB4. They found that patients with high ERBB4 expression in colon tumors had poorer survival rates. Both the rs79612564 variant and ERBB4 were proposed as predictive biomarkers for survival [124]. In a study, Xi et al., reported the development of a competing endogenous RNA network and identified novel molecular biomarkers in colon cancer. They conducted a bioinformatics analysis to construct a competing endogenous RNA network, utilizing differentially expressed long non-coding RNAs and RNAs from two colon cancer gene expression datasets [125]. They identified new regulatory pathways, such as LINC00114/miR-107/PCSK5, UCA1/miR-107/PCSK5, and UCA1/miR-129–5p/SEMA6A. Furthermore, bioinformatics analysis revealed two novel lncRNAs, LINC00114 and UCA1, and found that LINC00114 might be linked to the overall survival of colon cancer patients. Liang et al., investigated gene signatures related to immunity in colon adenocarcinoma to predict the immunotherapy effectiveness using non-negative matrix factorization and weighted gene co-expression network analysis algorithms [126]. They developed a prognostic model employing various bioinformatics tools and validated its accuracy with data from the Gene Expression Omnibus (GEO) database. The model's ability to predict treatment outcomes for cancer patients was assessed, and distinct immunological profiles among subgroups were characterized. Bioinformatics methods were also used to study the development of liver metastases in colon adenocarcinoma patients, exploring possible mechanisms and identifying key therapeutic genes. A basic flowchart for the identification of hub genes and targets of colorectal cancers, as well as the screening of inhibitors, is presented in Fig. 3.
Fig. 3.
The figure illustrates the identification of hub genes for colorectal cancer, with subsequent stages including High-Throughput Virtual Screening, discovery of inhibitors, identification of therapeutic targets, assembly of drug-target complexes, and molecular dynamic simulations for comprehensive analysis.
Conclusions
Major preclinical CRC models such as organoids, CDX, PDX, and GEMMs aim to incur various advantages in the exploration of various molecular and immune targets for personalized CRC therapy. The drawbacks and advantages discussed above of these models highlight the importance of transitioning from one model to another as per requirement. Although an important preclinical model, there are several limitations associated with CDX. Firstly, in vitro passage is a mandatory step when using cell lines. This brings an inherent drawback of recapitulating the parental or initial tumor genetic heterogeneity. Passaging of CRC cell lines before implantation, can lead to loss or absence of the original TME as subclones develop genetic and epigenetic changes that may not be representative of the original tumor [37,127]. On the other hand, species mismatches in human CRC cell lines and mouse stromal cells may limit crosstalk. In such cases, drug development and precise targeting may be compromised due to clonal selection and differences in inter-specific cell-to-cell communication [38]. In addition to this, due to the use of immunodeficient mice, studying the role of vascular, lymphatic, and immune environments is limited [128]. In the context of remodeling of the CRC-TME, PDX suffices an unparcelled advantage in terms of studying patient-specific mutational status, investigating the metastatic potential of CRC, and studying patient-specific TME components, using novel mechanistic approaches. Several limitations have to be taken into consideration while working with PDX CRC models. Firstly, it is not a fast drug screening method for patients as it has a lengthy engraftment phase of 4–8 months and requires high technical expertise to avoid losing precious patient samples [37]. Secondly, stromal and immune interactions are under-represented due to distinct differences in model species compatibility as well as inherent immune/cellular component deficiencies that are associated with using an immunodeficient animal model [5]. Alternatively, GEMMs provide an advantage in understanding the natural history of CRC from stage I to advanced stages, providing a broad understanding of CRC cells and their interaction at cellular and molecular levels. In terms of testing novel therapeutics in CRC, preclinical models provide a platform that could be used extensively for studying treatment responses and the development of novel therapeutic targets in CRC. Therefore, preclinical models are of paramount importance for advances in CRC drug development and improved patient outcomes. Furthermore, precision medicine strategies targeting the TME in CRC show promises in improving therapeutic outcomes.
Authorship contribution statement
All authors reviewed the whole work and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Funding
This paper was not funded.
Data availability
Not applicable.
CRediT authorship contribution statement
Abdo Meyiah: Writing – review & editing, Writing – original draft, Visualization, Investigation. Faez Iqbal Khan: Writing – review & editing, Writing – original draft, Visualization. Dia Aldeen Alfaki: Writing – review & editing. Khaled Murshed: Writing – review & editing. Afsheen Raza: Writing – review & editing. Eyad Elkord: Writing – review & editing, Writing – original draft, Visualization, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that there are no financial or nonfinancial conflicts of interest related to this work.
Acknowledgements
Not applicable.
References
- 1.Maida M., Macaluso F.S., Ianiro G., Mangiola F., Sinagra E., Hold G., Maida C., Cammarota G., Gasbarrini A., Scarpulla G. Screening of colorectal cancer: present and future. Expert. Rev. AntiCancer Ther. 2017;17:1131–1146. doi: 10.1080/14737140.2017.1392243. [DOI] [PubMed] [Google Scholar]
- 2.Shaukat A., Levin T.R. Current and future colorectal cancer screening strategies. Nature Rev. Gastroenterol. Hepatol. 2022;19:521–531. doi: 10.1038/s41575-022-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrew A.S., Parker S., Anderson J.C., Rees J.R., Robinson C., Riddle B., Butterly L.F. Risk factors for diagnosis of colorectal cancer at a late stage: a population-based study. J. Gen. Intern. Med. 2018;33:2100–2105. doi: 10.1007/s11606-018-4648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A., Gautam V., Sandhu A., Rawat K., Sharma A., Saha L. Current and emerging therapeutic approaches for colorectal cancer: a comprehensive review. World J. Gastrointest. Surg. 2023;15:495–519. doi: 10.4240/wjgs.v15.i4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassidy J.W., Caldas C., Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75:2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nallasamy P., Nimmakayala R.K., Parte S., Are A.C., Batra S.K., Ponnusamy M.P. Tumor microenvironment enriches the stemness features: the architectural event of therapy resistance and metastasis. Mol. Cancer. 2022;21:225. doi: 10.1186/s12943-022-01682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Zhang N., Li Q., Zhang W., Ke F., Leng Q., Wang H., Chen J., Wang H. Tumor-associated macrophages recruit CCR6+ regulatory T cells and promote the development of colorectal cancer via enhancing CCL20 production in mice. PLoS. One. 2011;6:e19495. doi: 10.1371/journal.pone.0019495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basak U., Sarkar T., Mukherjee S., Chakraborty S., Dutta A., Dutta S., Nayak D., Kaushik S., Das T., Sa G. Tumor-associated macrophages: an effective player of the tumor microenvironment. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1295257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Min A.K.T., Mimura K., Nakajima S., Okayama H., Saito K., Sakamoto W., Fujita S., Endo H., Saito M., Saze Z., et al. Therapeutic potential of anti-VEGF receptor 2 therapy targeting for M2-tumor-associated macrophages in colorectal cancer. Cancer Immunol. Immunother.: CII. 2021;70:289–298. doi: 10.1007/s00262-020-02676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., Luo Y., Rao D., Wang T., Lei Z., Chen X., Zhang B., Li Y., Liu B., Xia L., et al. Myeloid-derived suppressor cells in cancer: therapeutic targets to overcome tumor immune evasion. Exp. Hematol. Oncol. 2024;13:39. doi: 10.1186/s40164-024-00505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OuYang L.Y., Wu X.J., Ye S.B., Zhang R.X., Li Z.L., Liao W., Pan Z.Z., Zheng L.M., Zhang X.S., Wang Z., et al. Tumor-induced myeloid-derived suppressor cells promote tumor progression through oxidative metabolism in human colorectal cancer. J. Transl. Med. 2015;13:47. doi: 10.1186/s12967-015-0410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui C., Lan P., Fu L. The role of myeloid-derived suppressor cells in gastrointestinal cancer. Cancer Commun. (Lond) 2021;41:442–471. doi: 10.1002/cac2.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieminska I., Baran J. Myeloid-derived suppressor cells in colorectal cancer. Front. Immunol. 2020;11:1526. doi: 10.3389/fimmu.2020.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zwing N., Failmezger H., Ooi C.H., Hibar D.P., Cañamero M., Gomes B., Gaire F., Korski K. Analysis of spatial organization of suppressive myeloid cells and effector T cells in colorectal cancer-a potential tool for discovering prognostic biomarkers in clinical research. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.550250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao W., Overman M.J., Boutin A.T., Shang X., Zhao D., Dey P., Li J., Wang G., Lan Z., Li J., et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35 doi: 10.1016/j.ccell.2019.02.008. 559-572.e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Zhu T., Jiang G., Zeng Q., Li Z., Huang X. Target delivery of a PD-1-TREM2 scFv by CAR-T cells enhances anti-tumor efficacy in colorectal cancer. Mol. Cancer. 2023;22:131. doi: 10.1186/s12943-023-01830-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S., Li S., Guo J., Zeng X., Liang D., Zhu Y., Li Y., Yang D., Zhao X. CD166-specific CAR-T cells potently target colorectal cancer cells. Transl. Oncol. 2023;27 doi: 10.1016/j.tranon.2022.101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casey T., Bond J., Tighe S., Hunter T., Lintault L., Patel O., Eneman J., Crocker A., White J., Tessitore J., et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast. Cancer Res. Treat. 2009;114:47–62. doi: 10.1007/s10549-008-9982-8. [DOI] [PubMed] [Google Scholar]
- 19.Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cellul. Proteomic.: MCP. 2012;11 doi: 10.1074/mcp.M111.014647. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi H., Gieniec K.A., Lannagan T.R.M., Wang T., Asai N., Mizutani Y., Iida T., Ando R., Thomas E.M., Sakai A., et al. The origin and contribution of cancer-associated fibroblasts in colorectal carcinogenesis. Gastroenterology. 2022;162:890–906. doi: 10.1053/j.gastro.2021.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang B., Huang M., Li Q. Cancer-associated fibroblasts promote angiogenesis of hepatocellular carcinoma by VEGF-mediated EZH2/VASH1 pathway. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819879905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holter J.C., Chang C.W., Avendano A., Garg A.A., Verma A.K., Charan M., Ahirwar D.K., Ganju R.K., Song J.W. Fibroblast-derived CXCL12 increases vascular permeability in a 3-D microfluidic model independent of extracellular matrix contractility. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.888431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Socovich A.M., Naba A. The cancer matrisome: from comprehensive characterization to biomarker discovery. Semin. Cell Dev. Biol. 2019;89:157–166. doi: 10.1016/j.semcdb.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z., Li Y., Zhang S., Wang X., Dou H., Yu X., Zhang Z., Yang S., Xiao M. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol. Cancer. 2023;22:48. doi: 10.1186/s12943-023-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng F., Zhou R., Lin C., Yang S., Wang H., Li W., Zheng K., Lin W., Li X., Yao X., et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics. 2019;9:1001–1014. doi: 10.7150/thno.30056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q., Wu J., Zhu G., Li T., Zhu X., Ni B., Xu B., Ma X., Li J. Lactate-related metabolic reprogramming and immune regulation in colorectal cancer. Front. Endocrinol. (Lausanne) 2022;13 doi: 10.3389/fendo.2022.1089918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Sime W., Juhas M., Sjölander A. Crosstalk between colon cancer cells and macrophages via inflammatory mediators and CD47 promotes tumour cell migration. Eur. J. Cancer (1965) 2013;49:3320–3334. doi: 10.1016/j.ejca.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Wei Y., Huang Y., Qin G., Zhao C., Ren J., Qu X. Lactate-responsive gene editing to synergistically enhance macrophage-mediated cancer immunotherapy. Small. 2023;19 doi: 10.1002/smll.202301519. [DOI] [PubMed] [Google Scholar]
- 29.Stone T.W., Williams R.O. Modulation of T cells by tryptophan metabolites in the kynurenine pathway. Trends. Pharmacol. Sci. 2023;44:442–456. doi: 10.1016/j.tips.2023.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Shi D., Wu X., Jian Y., Wang J., Huang C., Mo S., Li Y., Li F., Zhang C., Zhang D., et al. USP14 promotes tryptophan metabolism and immune suppression by stabilizing IDO1 in colorectal cancer. Nat. Commun. 2022;13:5644. doi: 10.1038/s41467-022-33285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katt M.E., Placone A.L., Wong A.D., Xu Z.S., Searson P.C.In. Vitro Tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen C., Teng Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020;7:33. doi: 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N., Zhu Q., Tian Y., Ahn K.J., Wang X., Cramer Z., Jou J., Folkert I.W., Yu P., Adams-Tzivelekidis S., et al. Mapping and modeling human colorectal carcinoma interactions with the tumor microenvironment. Nat. Commun. 2023;14:7915. doi: 10.1038/s41467-023-43746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jubelin C., Muñoz-Garcia J., Griscom L., Cochonneau D., Ollivier E., Heymann M.-F., Vallette F.M., Oliver L., Heymann D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022;12:155. doi: 10.1186/s13578-022-00887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poornima K., Francis A.P., Hoda M., Eladl M.A., Subramanian S., Veeraraghavan V.P., El-Sherbiny M., Asseri S.M., Hussamuldin A.B.A., Surapaneni K.M., et al. Implications of three-dimensional cell culture in cancer therapeutic research. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.891673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoetemelk M., Rausch M., Colin D.J., Dormond O., Nowak-Sliwinska P. Short-term 3D culture systems of various complexity for treatment optimization of colorectal carcinoma. Sci. Rep. 2019;9:7103. doi: 10.1038/s41598-019-42836-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Xin Z., Wang K. Patient-derived xenograft model in colorectal cancer basic and translational research. Animal. Model. Exp. Med. 2023;6:26–40. doi: 10.1002/ame2.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIntyre R.E., Buczacki S.J., Arends M.J., Adams D.J. Mouse models of colorectal cancer as preclinical models. Bioessays. 2015;37:909–920. doi: 10.1002/bies.201500032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma G., Goyal Y., Bhatia S. In: Handbook of Animal Models and Its Uses in Cancer Research. Pathak S., Banerjee A., Bisgin A., editors. Springer Nature Singapore; Singapore: 2023. Preclinical animal models of cancer: applications and limitations; pp. 1051–1071. [Google Scholar]
- 40.Abdolahi S., Ghazvinian Z., Muhammadnejad S., Saleh M., Asadzadeh Aghdaei H., Baghaei K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022;20:206. doi: 10.1186/s12967-022-03405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung K.E., Maricevich M.A., Richard L.G., Chen W.Y., Richardson M.P., Kunin A., Bronson R.T., Mahmood U., Kucherlapati R. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gopinathan, A.; Morton, J.P.; Jodrell, D.I.; Sansom, O.J. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis. Models. Mech. 2015, 8, 1185–1200, doi:10.1242/dmm.021055. [DOI] [PMC free article] [PubMed]
- 43.Neto Í., Rocha J., Gaspar M.M., Reis C.P. Experimental murine models for colorectal cancer research. Cancers. (Basel) 2023;15 doi: 10.3390/cancers15092570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helmer R.A., Martinez-Zaguilan R., Kaur G., Smith L.A., Dufour J.M., Chilton B.S. Helicase-like transcription factor-deletion from the tumor microenvironment in a cell line-derived xenograft model of colorectal cancer reprogrammed the human transcriptome-S-nitroso-proteome to promote inflammation and redirect metastasis. PLoS. One. 2021;16 doi: 10.1371/journal.pone.0251132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higurashi T., Nakajima A. Metformin and colorectal cancer. Front. Endocrinol. (Lausanne) 2018;9:622. doi: 10.3389/fendo.2018.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni W., Wu J., Feng Y., Hu Y., Liu H., Chen J., Chen F., Tian H. Metformin reprograms tumor microenvironment and boosts chemoimmunotherapy in colorectal cancer. Biomater. Sci. 2022;10:5596–5607. doi: 10.1039/D2BM00988A. [DOI] [PubMed] [Google Scholar]
- 47.Kang J., Lee D., Lee K.J., Yoon J.E., Kwon J.H., Seo Y., Kim J., Chang S.Y., Park J., Kang E.A., et al. Tumor-suppressive effect of metformin via the regulation of M2 macrophages and myeloid-derived suppressor cells in the Tumor microenvironment of colorectal cancer. Cancers. (Basel) 2022;14 doi: 10.3390/cancers14122881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu C., Chen H., Hu B., Shi J., Chen Y., Huang K. New insights into the therapeutic potentials of statins in cancer. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1188926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X., Sun T., Wang J., Hong X., Chen H., Yan T., Zhou C., Sun D., Yang C., Yu T., et al. Metformin reprograms tryptophan metabolism to stimulate CD8+ T-cell function in colorectal cancer. Cancer Res. 2023;83:2358–2371. doi: 10.1158/0008-5472.Can-22-3042. [DOI] [PubMed] [Google Scholar]
- 50.Wu S., Fang W., Chen L., Feng C., Chen R., Ying H., Zheng X., Jiang J. Cordycepin remodels the tumor microenvironment of colorectal cancer by down-regulating the expression of PD-L1. J. Cancer Res. Clin. Oncol. 2023;149:17567–17579. doi: 10.1007/s00432-023-05460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Li S., Yan S., Shan Y., Wang X., Jingbo Z., Wang Y., Shan F., Griffin N., Sun X. Methionine enkephalin inhibits colorectal cancer by remodeling the immune status of the tumor microenvironment. Int. Immunopharmacol. 2022;111 doi: 10.1016/j.intimp.2022.109125. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L., Lu X., Xu Y., La X., Tian J., Li A., Li H., Wu C., Xi Y., Song G., et al. Tumor-associated macrophages confer colorectal cancer 5-fluorouracil resistance by promoting MRP1 membrane translocation via an intercellular CXCL17/CXCL22-CCR4-ATF6-GRP78 axis. Cell Death. Dis. 2023;14:582. doi: 10.1038/s41419-023-06108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X., Liang R., Lan T., Ding D., Huang S., Shao J., Zheng Z., Chen T., Huang Y., Liu J., et al. Tumor-associated macrophage-specific CD155 contributes to M2-phenotype transition, immunosuppression, and tumor progression in colorectal cancer. J. ImmunOther Cancer. 2022;10 doi: 10.1136/jitc-2021-004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yadav V.K., Huang Y.J., George T.A., Wei P.L., Sumitra M.R., Ho C.L., Chang T.H., Wu A.T.H., Huang H.S. Preclinical evaluation of the novel small-molecule MSI-N1014 for treating drug-resistant colon cancer via the LGR5/β-catenin/miR-142-3p network and reducing cancer-associated fibroblast transformation. Cancers. (Basel) 2020;12 doi: 10.3390/cancers12061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu W., Sang T., Chen H., Zhou H., Wang Z., Zhou H. Wenzi Jiedu Recipe ameliorates colorectal cancer by remodeling the gut microbiota and tumor microenvironment. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.915498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali M.K., Liu Q., Liang K., Li P., Kong Q. Bacteria-derived minicells for cancer therapy. Cancer Lett. 2020;491:11–21. doi: 10.1016/j.canlet.2020.07.024. [DOI] [PubMed] [Google Scholar]
- 57.MacDiarmid J.A., Langova V., Bailey D., Pattison S.T., Pattison S.L., Christensen N., Armstrong L.R., Brahmbhatt V.N., Smolarczyk K., Harrison M.T., et al. Targeted doxorubicin delivery to brain tumors via minicells: proof of principle using dogs with spontaneously occurring tumors as a model. PLoS. One. 2016;11 doi: 10.1371/journal.pone.0151832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reil K.A., Tsuji S., Molina E., Nelson K.L., McGuire K.L., Giacalone M.J. Intralesional administration of VAX014 facilitates in situ immunization and potentiates immune checkpoint blockade in immunologically cold tumors. J. ImmunOther Cancer. 2023;11 doi: 10.1136/jitc-2023-006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hancock B.M., McGuire K.L., Tsuji S., Reil K., Hernandez V., Giacalone M.J., Godbey W.T. A single intravesical instillation of VAX014 inhibits orthotopic superficial bladder tumor implantation to increase survival. Anticancer Res. 2016;36:6243–6248. doi: 10.21873/anticanres.11218. [DOI] [PubMed] [Google Scholar]
- 60.Grenier S.F., Khan M.W., Reil K.A., Sawaged S., Tsuji S., Giacalone M.J., Tian M., McGuire K.L. VAX014, an oncolytic therapy, reduces adenomas and modifies colon microenvironment in mouse model of CRC. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24129993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim I.K., Kim B.S., Koh C.H., Seok J.W., Park J.S., Shin K.S., Bae E.A., Lee G.E., Jeon H., Cho J., et al. Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells. Nat. Med. 2015;21:1010–1017. doi: 10.1038/nm.3922. [DOI] [PubMed] [Google Scholar]
- 62.Wang J., Sun M., Zhao H., Huang Y., Li D., Mao D., Zhang Z., Zhu X., Dong X., Zhao X. IL-9 exerts antitumor effects in colon cancer and transforms the tumor microenvironment In vivo. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819857737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albu D.I., Wang Z., Huang K.C., Wu J., Twine N., Leacu S., Ingersoll C., Parent L., Lee W., Liu D., et al. EP4 Antagonism by E7046 diminishes Myeloid immunosuppression and synergizes with treg-reducing IL-2-diphtheria toxin fusion protein in restoring anti-tumor immunity. Oncoimmunology. 2017;6 doi: 10.1080/2162402x.2017.1338239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plattner C., Lamberti G., Blattmann P., Kirchmair A., Rieder D., Loncova Z., Sturm G., Scheidl S., Ijsselsteijn M., Fotakis G., et al. Functional and spatial proteomics profiling reveals intra- and intercellular signaling crosstalk in colorectal cancer. iScience. 2023;26 doi: 10.1016/j.isci.2023.108399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nur Ay A. Therapeutic targeting of molecular pathways in colorectal cancer. Exp. Oncol. 2022;44:2–6. doi: 10.32471/exp-oncology.2312-8852.vol-44-no-1.17455. [DOI] [PubMed] [Google Scholar]
- 66.Ho J.Y., Hsu R.J., Wu C.L., Chang W.L., Cha T.L., Yu D.S., Yu C.P. Ovatodiolide targets β -catenin signaling in suppressing tumorigenesis and overcoming drug resistance in renal cell carcinoma. Evid. Based. Complement. Alternat. Med. 2013;2013 doi: 10.1155/2013/161628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu A.T.H., Srivastava P., Yadav V.K., Tzeng D.T.W., Iamsaard S., Su E.C., Hsiao M., Liu M.C. Ovatodiolide, isolated from Anisomeles indica, suppresses bladder carcinogenesis through suppression of mTOR/β-catenin/CDK6 and exosomal miR-21 derived from M2 tumor-associated macrophages. Toxicol. Appl. Pharmacol. 2020;401 doi: 10.1016/j.taap.2020.115109. [DOI] [PubMed] [Google Scholar]
- 68.Huang Y.J., Yang C.K., Wei P.L., Huynh T.T., Whang-Peng J., Meng T.C., Hsiao M., Tzeng Y.M., Wu A.T., Yen Y. Ovatodiolide suppresses colon tumorigenesis and prevents polarization of M2 tumor-associated macrophages through YAP oncogenic pathways. J. Hematol. Oncol. 2017;10:60. doi: 10.1186/s13045-017-0421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang Y.J., Huang T.H., Yadav V.K., Sumitra M.R., Tzeng D.T., Wei P.L., Shih J.W., Wu A.T. Preclinical investigation of ovatodiolide as a potential inhibitor of colon cancer stem cells via downregulating sphere-derived exosomal β-catenin/STAT3/miR-1246 cargoes. Am. J. Cancer Res. 2020;10:2337–2354. [PMC free article] [PubMed] [Google Scholar]
- 70.Strelez C., Perez R., Chlystek J.S., Cherry C., Yoon A.Y., Haliday B., Shah C., Ghaffarian K., Sun R.X., Jiang H., et al. Integration of patient-derived organoids and organ-on-chip systems: investigating colorectal cancer invasion within the mechanical and GABAergic tumor microenvironment. bioRxiv: Preprint Server Biol. 2023 doi: 10.1101/2023.09.14.557797. [DOI] [Google Scholar]
- 71.Chang G.W., Hsiao C.C., Peng Y.M., Vieira Braga F.A., Kragten N.A., Remmerswaal E.B., van de Garde M.D., Straussberg R., König G.M., Kostenis E., et al. The adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on Human NK cells. Cell Rep. 2016;15:1757–1770. doi: 10.1016/j.celrep.2016.04.053. [DOI] [PubMed] [Google Scholar]
- 72.Zhang S., Chatterjee T., Godoy C., Wu L., Liu Q.J., Carmon K.S. GPR56 Drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol. Cancer Res.: MCR. 2019;17:2196–2207. doi: 10.1158/1541-7786.Mcr-19-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng, K.F.; Chen, T.C.; Stacey, M.; Lin, H.H. Role of ADGRG1/GPR56 in tumor progression. Cells 2021, 10, doi:10.3390/cells10123352. [DOI] [PMC free article] [PubMed]
- 74.Jacob J., Francisco L.E., Chatterjee T., Liang Z., Subramanian S., Liu Q.J., Rowe J.H., Carmon K.S. An antibody-drug conjugate targeting GPR56 demonstrates efficacy in preclinical models of colorectal cancer. Br. J. Cancer. 2023;128:1592–1602. doi: 10.1038/s41416-023-02192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francisco L.E., Chatterjee T., Liang Z., Zhang S., Carmon K. Abstract 6328: GPR56 as a therapeutic target for the development of antibody-drug conjugates for the treatment of colorectal cancer. Cancer Res. 2022;82:6328. -6328. [Google Scholar]
- 76.Guo B., Zheng Y., Fan Y., Yang Y., Wang Y., Qin L., An Y., Xu X., Zhang X., Sun G., et al. Enhanced Apc(Min/+) adenoma formation after epithelial CUL4B deletion by recruitment of myeloid-derived suppressor cells. Neoplasia. 2024;53 doi: 10.1016/j.neo.2024.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rathore M., Curry K., Huang W., Wright M., Martin D., Baek J., Taylor D., Miyagi M., Tang W., Feng H., et al. Leucine-rich alpha-2-glycoprotein 1 promotes metastatic colorectal cancer growth through Human epidermal growth factor receptor 3 signaling. Gastroenterology. 2025;168 doi: 10.1053/j.gastro.2024.10.004. 300-315.e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darvin P., Toor S.M., Sasidharan Nair V., Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 2018;50:1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y., Peng Y., Zhao S., Mou J., Zeng L., Jiang X., Yang C., Huang C., Li Y., Lu Y., et al. Combination Foretinib and Anti-PD-1 antibody immunotherapy for colorectal carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.689727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doleschel D., Hoff S., Koletnik S., Rix A., Zopf D., Kiessling F., Lederle W. Regorafenib enhances anti-PD1 immunotherapy efficacy in murine colorectal cancers and their combination prevents tumor regrowth. J. Exp. Clin. Cancer Res.: CR. 2021;40:288. doi: 10.1186/s13046-021-02043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarker D., Plummer R., Meyer T., Sodergren M.H., Basu B., Chee C.E., Huang K.-W., Palmer D.H., Ma Y.T., Evans T.R.J., et al. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-α, in patients with advanced liver cancer: A first-in-Human, multicenter, open-label, phase I trial. Clin. Cancer Res. 2020;26:3936–3946. doi: 10.1158/1078-0432.ccr-20-0414. [DOI] [PubMed] [Google Scholar]
- 82.Zhou J., Li H., Xia X., Herrera A., Pollock N., Reebye V., Sodergren M.H., Dorman S., Littman B.H., Doogan D., et al. Anti-inflammatory activity of MTL-CEBPA, a small activating RNA drug, in LPS-stimulated monocytes and humanized mice. Mol. Therapy: J. Am. Soc. Gene Therapy. 2019;27:999–1016. doi: 10.1016/j.ymthe.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reebye V., Sætrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D., et al. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang K.W., Tan C.P., Reebye V., Chee C.E., Zacharoulis D., Habib R., Blakey D.C., Rossi J.J., Habib N., Sodergren M.H. MTL-CEBPA combined with immunotherapy or RFA enhances immunological anti-tumor response in preclinical models. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22179168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahdi H.S., Woodall-Jappe M., Singh P., Czuczman M.S. Targeting regulatory T cells by E7777 enhances CD8 T-cell-mediated anti-tumor activity and extends survival benefit of anti-PD-1 in solid tumor models. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1268979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McEwen-Smith R.M., Salio M., Cerundolo V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol. Res. 2015;3:425–435. doi: 10.1158/2326-6066.Cir-15-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Look A., Burns D., Tews I., Roghanian A., Mansour S. Towards a better understanding of human iNKT cell subpopulations for improved clinical outcomes. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1176724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y., Bhave M.S., Yagita H., Cardell S.L. Natural killer T-cell agonist α-galactosylceramide and PD-1 blockade synergize to reduce tumor development in a preclinical model of colon cancer. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.581301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campbell J.R., McDonald B.R., Mesko P.B., Siemers N.O., Singh P.B., Selby M., Sproul T.W., Korman A.J., Vlach L.M., Houser J., et al. Fc-optimized anti-CCR8 antibody depletes regulatory T cells in Human tumor models. Cancer Res. 2021;81:2983–2994. doi: 10.1158/0008-5472.Can-20-3585. [DOI] [PubMed] [Google Scholar]
- 90.Whiteside S.K., Grant F.M., Gyori D.S., Conti A.G., Imianowski C.J., Kuo P., Nasrallah R., Sadiyah F., Lira S.A., Tacke F., et al. CCR8 marks highly suppressive treg cells within tumours but is dispensable for their accumulation and suppressive function. Immunology. 2021;163:512–520. doi: 10.1111/imm.13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu S., Tao Z., Lou J., Li R., Fu X., Xu J., Wang T., Zhang L., Shang W., Mao Y., et al. CD4(+)CCR8(+) Tregs in ovarian cancer: a potential effector Tregs for immune regulation. J. Transl. Med. 2023;21:803. doi: 10.1186/s12967-023-04686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plitas G., Konopacki C., Wu K., Bos P.D., Morrow M., Putintseva E.V., Chudakov D.M., Rudensky A.Y. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haruna M., Ueyama A., Yamamoto Y., Hirata M., Goto K., Yoshida H., Higuchi N., Yoshida T., Kidani Y., Nakamura Y., et al. The impact of CCR8+ regulatory T cells on cytotoxic T cell function in human lung cancer. Sci. Rep. 2022;12:5377. doi: 10.1038/s41598-022-09458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roider H.G., Hoff S., Tseng S.Y., Berndt S., Trautwein M., Filarsky K., Gritzan U., Camps J., Nadler W.M., Grudzinska-Goebel J., et al. Selective depletion of tumor-infiltrating regulatory T cells with BAY 3375968, a novel Fc-optimized anti-CCR8 antibody. Clin. Exp. Med. 2024;24:122. doi: 10.1007/s10238-024-01362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen Q., Shen M., Yan M., Han X., Mu S., Li Y., Li L., Wang Y., Li S., Li T., et al. Targeting tumor-infiltrating CCR8(+) regulatory T cells induces antitumor immunity through functional restoration of CD4(+) T(convs) and CD8(+) T cells in colorectal cancer. J. Transl. Med. 2024;22:709. doi: 10.1186/s12967-024-05518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai M., Huang X., Huang X., Ju D., Zhu Y.Z., Ye L. Research progress of interleukin-15 in cancer immunotherapy. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1184703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsu F.T., Liu Y.C., Tsai C.L., Yueh P.F., Chang C.H., Lan K.L. Preclinical evaluation of recombinant human IL15 protein fused with albumin binding domain on anti-PD-L1 immunotherapy efficiency and anti-tumor immunity in colon cancer and melanoma. Cancers. (Basel) 2021;13 doi: 10.3390/cancers13081789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vince J.E., De Nardo D., Gao W., Vince A.J., Hall C., McArthur K., Simpson D., Vijayaraj S., Lindqvist L.M., Bouillet P., et al. The mitochondrial apoptotic effectors BAX/BAK activate Caspase-3 and -7 to trigger NLRP3 inflammasome and caspase-8 driven IL-1β activation. Cell Rep. 2018;25 doi: 10.1016/j.celrep.2018.10.103. 2339-2353.e2334. [DOI] [PubMed] [Google Scholar]
- 99.Li J., Xu J., Yan X., Jin K., Li W., Zhang R. Targeting interleukin-6 (IL-6) sensitizes anti-PD-L1 treatment in a colorectal cancer preclinical model. Med. Sci. Monitor: Int. Med. J. Exp. Clin. Res. 2018;24:5501–5508. doi: 10.12659/msm.907439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leonard N.A., Reidy E., Thompson K., McDermott E., Peerani E., Tomas Bort E., Balkwill F.R., Loessner D., Ryan A.E. Stromal cells promote matrix deposition, remodelling and an immunosuppressive tumour microenvironment in a 3D model of colon cancer. Cancers. (Basel) 2021;13 doi: 10.3390/cancers13235998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marques-Magalhães Â., Cruz T., Costa Â.M., Estêvão D., Rios E., Canão P.A., Velho S., Carneiro F., Oliveira M.J., Cardoso A.P. Decellularized colorectal cancer matrices as bioactive scaffolds for studying tumor-stroma interactions. Cancers. (Basel) 2022;14:359. doi: 10.3390/cancers14020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu J.W., Yu F., Tan Y.F., Huo J.P., Liu Z., Wang X.J., Li J.M. Profiling of tumor microenvironment components identifies five stroma-related genes with prognostic implications in colorectal cancer. Cancer BiOther Radiopharm. 2022;37:882–892. doi: 10.1089/cbr.2020.4118. [DOI] [PubMed] [Google Scholar]
- 104.Waldner M.J., Neurath M.F. TGFβ and the tumor microenvironment in colorectal cancer. Cells. 2023:12. doi: 10.3390/cells12081139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Teicher B.A. Transforming growth factor-β and the immune response to malignant disease. Clin. Cancer Res. 2007;13:6247–6251. doi: 10.1158/1078-0432.CCR-07-1654. [DOI] [PubMed] [Google Scholar]
- 106.Batlle E., Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vega P.N., Nilsson A., Kumar M.P., Niitsu H., Simmons A.J., Ro J., Wang J., Chen Z., Joughin B.A., Li W., et al. Cancer-associated fibroblasts and squamous epithelial cells constitute a unique microenvironment in a mouse model of inflammation-induced colon cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.878920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crutcher M.M., Baybutt T.R., Kopenhaver J.S., Snook A.E., Waldman S.A. Emerging drug targets for colon cancer: a preclinical assessment. Expert. Opin. Ther. Targets. 2022;26:207–216. doi: 10.1080/14728222.2022.2039119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang C., Liu T., Yun Z., Liang B., Li X., Zhang J. Identification of angiogenesis-related subtypes, the development of prognostic models, and the landscape of tumor microenvironment infiltration in colorectal cancer. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1103547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Onuma A.E., Zhang H., Gil L., Huang H., Tsung A. Surgical stress promotes tumor progression: a focus on the impact of the immune response. J. Clin. Med. 2020;9 doi: 10.3390/jcm9124096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang T., Li X., Ma R., Sun J., Huang S., Sun Z., Wang M. Advancements in colorectal cancer research: unveiling the cellular and molecular mechanisms of neddylation (Review) Int. J. Oncol. 2024;64 doi: 10.3892/ijo.2024.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang Y., Gao S., Sun H., Wu X., Gu J., Wu H., Liao Y., Ben-Ami R., Miao C., Shen R., et al. Targeting NEDD8 suppresses surgical stress-facilitated metastasis of colon cancer via restraining regulatory T cells. Cell Death. Dis. 2024;15:8. doi: 10.1038/s41419-023-06396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Horaira M.A., Islam M.A., Kibria M.K., Alam M.J., Kabir S.R., Mollah M.N.H. Bioinformatics screening of colorectal-cancer causing molecular signatures through gene expression profiles to discover therapeutic targets and candidate agents. BMC. Med. Genomics. 2023;16:64. doi: 10.1186/s12920-023-01488-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin Y., Xie H., Zhao W., Li Y., Zhang Z. NDUFA4L2 is a novel biomarker for colorectal cancer through bioinformatics analysis. Medicine (Baltimore) 2023;102:e35893. doi: 10.1097/md.0000000000035893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yue Y., Zhang Q., Sun Z. CX3CR1 Acts as a protective biomarker in the tumor microenvironment of colorectal cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.758040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu J.K., Chen Y.D., Zheng S. An integrated approach to the detection of colorectal cancer utilizing proteomics and bioinformatics. World J. Gastroenterol. 2004;10:3127–3131. doi: 10.3748/wjg.v10.i21.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pineda M., González S., Lázaro C., Blanco I., Capellá G. Detection of genetic alterations in hereditary colorectal cancer screening. Mutat. Res. 2010;693:19–31. doi: 10.1016/j.mrfmmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 118.Chao E.C., Velasquez J.L., Witherspoon M.S.L., Rozek L.S., Peel D., Ng P., Gruber S.B., Watson P., Rennert G., Anton-Culver H., et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms–mismatch repair (MAPP-MMR) Hum. Mutat. 2008;29:852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- 119.Thompson B.A., Greenblatt M.S., Vallee M.P., Herkert J.C., Tessereau C., Young E.L., Adzhubey I.A., Li B., Bell R., Feng B., et al. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum. Mutat. 2013;34:255–265. doi: 10.1002/humu.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 121.Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J., et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dijkstra J.J., Neikes H.K., Rezaeifard S., Ma X., Voest E.E., Tauriello D.V.F., Vermeulen M. Multiomics of colorectal cancer organoids reveals putative mediators of cancer progression resulting from SMAD4 inactivation. J. Proteome Res. 2023;22:138–151. doi: 10.1021/acs.jproteome.2c00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jose A., Kulkarni P., Thilakan J., Munisamy M., Malhotra A.G., Singh J., Kumar A., Rangnekar V.M., Arya N., Rao M. Integration of pan-omics technologies and three-dimensional in vitro tumor models: an approach toward drug discovery and precision medicine. Mol. Cancer. 2024;23:50. doi: 10.1186/s12943-023-01916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wills C., He Y., Summers M.G., Lin Y., Phipps A.I., Watts K., Law P.J., Al-Tassan N.A., Maughan T.S., Kaplan R., et al. A genome-wide search for determinants of survival in 1926 patients with advanced colorectal cancer with follow-up in over 22,000 patients. Eur. J. Cancer. 2021;159:247–258. doi: 10.1016/j.ejca.2021.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xi G., Ziyu X., Yiting L., Zonghang L., Lifeng Z. Construction of competing endogenous RNA network and identification of novel molecular biomarkers in colon cancer: A bioinformatic analysis. Medicine (Baltimore) 2021;100:e25369. doi: 10.1097/md.0000000000025369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang W., Yang X., Li X., Wang P., Zhu Z., Liu S., Xu D., Zhi X., Xue J. Investigating gene signatures associated with immunity in colon adenocarcinoma to predict the immunotherapy effectiveness using NFM and WGCNA algorithms. Aging. 2024;16:7596–7621. doi: 10.18632/aging.205763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R., et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pan B., Wei X., Xu X. Patient-derived xenograft models in hepatopancreatobiliary cancer. Cancer Cell Int. 2022;22:41. doi: 10.1186/s12935-022-02454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.