Summary
Plant m6A modification is highly dynamic, with plant hormones serving as key regulators that induce these dynamics. Here, we present a protocol for detecting phytohormone-induced changes in m6A modification using RNA dot blot and m6A sequencing (m6A-seq) techniques. We describe steps for the auxin treatment, RNA dot blot and m6A-seq library preparation, and data analysis. This protocol holds potential applications for analyzing changes in m6A modification induced by plant hormones.
For complete details on the use and execution of this protocol, please refer to Bin et al.1
Subject areas: Developmental biology, Genetics, Plant sciences
Graphical abstract

Highlights
-
•
Protocol for auxin treatment of Arabidopsis seedlings
-
•
Steps for analyzing m6A abundance by RNA dot blot
-
•
Instructions for m6A-seq library preparation and data analysis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Plant m6A modification is highly dynamic, with plant hormones serving as key regulators that induce these dynamics. Here, we present a protocol for detecting phytohormone-induced changes in m6A modification using RNA dot blot and m6A sequencing (m6A-seq) techniques. We describe steps for the auxin treatment, RNA dot blot and m6A-seq library preparation, and data analysis. This protocol holds potential applications for analyzing changes in m6A modification induced by plant hormones.
Before you begin
N6-methyladenosine (m6A) plays a crucial role in various biological processes in plants, including plant growth and development, disease resistance, flowering and fertility.2 The addition of m6A marks is facilitated by a methyltransferase complex, while demethylases are responsible for their removal.3 The abundance and distribution of plant m6A vary across different developmental stages and environmental conditions. Numerous studies have concentrated on the modification of m6A in response to biotic and abiotic stressors.2 However, the mechanisms by which endogenous signals, such as phytohormones, regulate m6A modification remain inadequately understood, and the established protocol to study this regulation is not fully described.
Recently, we employed the procedures outlined in this protocol, in conjunction with genetic analyses and biochemical experiments, to demonstrate how the activity of the m6A methyltransferase complex is regulated by auxin. This protocol, which encompasses auxin treatment, RNA extraction and mRNA enrichment, RNA dot blotting, and m6A sequencing (m6A-seq), offers a comprehensive workflow for detecting auxin-induced changes in m6A abundance and distribution. Other more accurate quantification methods such as LC-MS/MS should be used to confirm the RNA dot blotting result. Additionally, this protocol can be adapted to explore m6A dynamics in different development stages and in response to other phytohormones in plants.
Prepare 1/2 MS media plates
Timing: 2 h
-
1.
Add 2.21 g MS powder and dissolve it with 1000 mL water.
-
2.
Add 10 g sucrose to dissolve.
-
3.
Adjust pH to 5.8.
-
4.
Add 10 g Agar.
-
5.
Sterilization in 115°C for 15 min.
-
6.
Mix and pour into 90 mm square plates.
Store at 4°C for up to two weeks.
Preparations of stock solutions
-
7.Prepare 1 mL of auxin (1000x) stock solution.
-
a.Dissolve 0.18 g auxin powder in 1 mL 75% alcohol.
-
b.Mix on a magnetic stirrer.
-
c.Store away from light at −20°C.
-
a.
CRITICAL: Auxin is easily decomposed in light, so it should be kept away from light.
-
8.Prepare 100 mL 1 M Tris-HCl (pH7.5).
-
a.Dissolve 12.11 g Tris-base powder in 90 mL ddH20.
-
b.Adjust pH value to 7.5 with HCl.
-
c.Adjust volume to 100 mL.
-
a.
-
9.Prepare 100 mL of 0.5 M EDTA (pH8.0).
-
a.Dissolve 14.55 g EDTA powder in 90 mL ddH2O.
-
b.Adjust pH value to 8.0 with NaOH.
-
c.Adjust volume to 100 mL.
-
a.
-
10.Prepare 100 mL of 1 M LiCl.
-
a.Dissolve 4.24 g LiCl powder in 90 mL ddH2O.
-
b.Adjust volume to 100 mL.
-
a.
Preparations of working solutions
-
11.Prepare 10 mL Binding buffer.
-
a.Add 200 μL 1 M Tris-HCl.
-
b.Add 5 mL 1 M LiCl.
-
c.Add 40 μL 0.5 M EDTA.
-
d.Adjust volume to 10 mL.
-
e.Mix on a magnetic stirrer at 25°C.
-
a.
-
12.Prepare 10 mL Wash buffer.
-
a.Add 100 μL 1 M Tris-HCl.
-
b.Add 300 μL 1 M LiCl.
-
c.Add 20 μL 0.5 M EDTA.
-
d.Mix on a magnetic stirrer at 25°C.
-
a.
-
13.Prepare 1000 mL 1x TBST.
-
a.Add 100 mL 10x TBST.
-
b.Add 900 mL ddH2O.
-
c.Mix on a magnetic stirrer.
-
a.
-
14.Prepare 50 mL 5% milk solution.
-
a.Add 2.5 g skim milk in 50 mL 1x TBST.
-
b.Mix on a magnetic stirrer.
-
c.Store at 4°C.
-
a.
-
15.Prepare 10 mL m6A antibody solution.
-
a.Add 0.3 g BSA in 10 mL 1x TBST.
-
b.Mix on a magnetic stirrer.
-
c.Add 10 μL Anti-N6-methyladenosine antibody, final concentration is 1 μg/mL.
-
d.Mix well and store at 4°C.
-
a.
-
16.Prepare 10 mL secondary antibody solution.
-
a.Add 0.5 g skim milk in 10 mL 1x TBST.
-
b.Mix on a magnetic stirrer.
-
c.Add 2 μL HRP conjugated Goat Anti-Rabbit antibody.
-
d.Mix well and store at 4°C.
-
a.
-
17.Prepare 100 mL TE buffer.
-
a.Add 1 mL 1 M Tris-HCl.
-
b.Add 200 μL 0.5 M EDTA.
-
c.Adjust volume to 100 mL.
-
a.
-
18.Prepare 10 mL 5x RNA binding buffer.
-
a.Add 0.5 mL 1 M Tris-HCl.
-
b.Add 1.5 mL 3 M NaCl.
-
c.Add 0.5 mL Igepal CA-630.
-
d.Adjust volume to 10 mL.
-
a.
-
19.Prepare 10 mL 1x RNA binding buffer.
-
a.Add 2 mL 5x RNA binding buffer.
-
b.Add 8 mL ddH2O.
-
c.Mix on a magnetic stirrer.
-
a.
-
20.Prepare RNA elution buffer.
-
a.Add 90 μL 5x RNA binding buffer.
-
b.Add 150 μL m6A (20 mM stock).
-
c.Add 7 μL RNasin Plus.
-
d.Adjust volume to 450 μL.
-
a.
Note: All buffer preparations for RNA extraction and mRNA enrichment are to be working in RNase free conditions.
Primer for RT-qPCR
| Primer (5′-3′) | Sequence |
|---|---|
| GH3-RT-F | CAATTTAAGTTATTTAGCCG |
| GH3-RT-R | TGTTCGTCATTAACATCGAC |
| ARF1-RT-F | CCAATCATTCATCTGGTAAACCT |
| ARF1-RT-R | CATCTGTTGCTCCAAACCTTGG |
RNA-to-cDNA mix
| Reagent | Amount |
|---|---|
| 10X ezDNase Buffer | 1 μL |
| ezDNase enzyme | 1 μL |
| 500 μg/μL | 1 μL |
| Nuclease-free Water | to 10 μL |
Cycling conditions
| Steps | Temperature | Time |
|---|---|---|
| Anneal primers | 25°C | 10 min |
| Reverse transcribe RNA | 50°C | 10 min |
| Inactivate enzyme | 85°C | 5 min |
| Hold | 4°C | One week |
qPCR reaction master mix
| Reagent | Amount |
|---|---|
| 2x SYBR | 10 μL |
| Primer 1 | 2 μL |
| Primer 2 | 2 μL |
| cDNA | 1 μL |
| ddH2O | 4 μL |
| Total | 20 μL |
qPCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 98°C | 30 s | 1 |
| Denaturation | 98°C | 10 s | 40 cycles |
| Annealing | 60°C | 20 s | |
| Extension | 72°C | 20 s | |
| Final extension | 72°C | 5 min | 1 |
| Hold | 4°C | forever | |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-N6-methyladenosine | Abcam | Ab151230 (1:1000 dilution) |
| Goat anti-rabbit IgG H&L (HRP) | Abcam | Ab6721 (1:5000 dilution) |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | Invitrogen | Cat# 15596026CN |
| Murashige & Skoog medium | Solarbio | Cat# M8521 |
| Oligo (dT) magnetic beads | NEB | Cat# S1419S |
| Trichloromethane | SCRC | Cat# 10006818 |
| Western ECL substrate | Bio-Rad | Cat# 170-5061 |
| Tris base | Sigma | Cat# 77-86-1 |
| LiCl | Merck | Cat# 7447-41-8 |
| EDTA | Merck | Cat# 60-00-4 |
| NAA | Merck | Cat# 86-87-3 |
| Skim milk | BD Difco | Cat# 232100 |
| 10X TBST | Biosharp | Cat# BL315B |
| DNase I | Thermo Fisher Scientific | Cat# EN0525 |
| Nitrocellulose membrane | Pall Corporation | Cat# 69095847 |
| BSA | Sigma | Cat# B667 |
| RNA fragment module | NEB | E6150S |
| Protein-G beads | Thermo Fisher Scientific | Cat# 88848 |
| IGEPAL CA-630 | Sigma | Cat# I8896 |
| m6A | Sigma | Cat# M2780 |
| RNasin Plus | Promega | Cat# N2611 |
| Software and algorithms | ||
| ImageJ | Schneider et al.4 | https://imagej.nih.gov/ij/ |
| AdapterRemoval | Schubert et al.5 | https://adapterremoval.readthedocs.io/en/stable/ |
| STAR | Dobin et al.6 | https://github.com/alexdobin/STAR |
| DEGseq | Wang et al.7 | https://www.bioconductor.org/packages/release/bioc/html/DEGseq.html |
| MeRIPtools | Zhang et al.8 | https://github.com/scottzijiezhang/MeRIPtools |
Materials and equipment
Binding buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) pH 7.5 | 20 mM | 200 μL |
| LiCl (1 M) | 0.5 M | 5 mL |
| EDTA (0.5 M) pH 8.0 | 2 mM | 40 μL |
| ddH2O | N/A | 4.76 mL |
| Total | N/A | 10 mL |
Storage conditions: Store at 4°C for six months.
Wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) pH 7.5 | 10 mM | 100 μL |
| LiCl (1 M) | 30 mM | 300 μL |
| EDTA (0.5 M) pH 8.0 | 1 mM | 20 μL |
| ddH2O | N/A | 9.58 mL |
| Total | N/A | 10 mL |
Storage conditions: Store at 4°C for six months.
5 x RNA binding buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) pH 7.5 | 50 mM | 0.5 mL |
| NaCl (3 M) | 0.45 M | 1.5 mL |
| Igepal CA-630 | 5% (v/v) | 0.5 mL |
| ddH2O | N/A | 7.5 mL |
| Total | N/A | 10 mL |
Storage conditions: Store at 4°C for six months.
RNA elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) pH 7.5 | 10 mM | 4.5 μL |
| NaCl (3 M) | 0.15 M | 13.5 μL |
| Igepal CA-630 | 1% (v/v) | 4.5 μL |
| m6A (20 mM) | 6.67 mM | 150 μL |
| RNasin Plus | 0.62 U/μL | 7 μL |
| ddH2O | N/A | 270.5 μL |
| Total | N/A | 450 μL |
Storage conditions: Store at 4°C for six months. RNasin Plus and m6A should be added to the buffer immediately before use, after which the buffer can be stored at 4°C for up to 2 h.
Step-by-step method details
We provide a detailed step-by-step protocol for auxin treatment, followed by RNA extraction and mRNA purification. The m6A dynamics are analyzed by RNA dot blot and m6A-seq.
Arabidopsis seedlings preparation
Timing: 7 days
This step describes how to prepare Arabidopsis seedlings for auxin treatment.
-
1.
Sterilize 0.1 g Arabidopsis seeds in 1 mL 75% alcohol for 10 min.
-
2.
Remove the supernatant.
-
3.
Add 1 mL sterile water to wash seeds for 10 min.
-
4.
Remove the supernatant.
-
5.
Repeat step c and d, plate sterilized seeds on the 1/2 MS solid medium.
-
6.
Put the plates in a fridge at 4°C in the dark for 3 days.
-
7.
Place in incubator at 22°C for 7 days under long day condition (16 h light and 8 h dark).
Auxin treatment and material collection
Timing: 24 h
This step describes how to treat Arabidopsis seedlings with NAA.
-
8.
Submerge 30 7-day-old Arabidopsis seedlings in 10 mL H2O with 50 μM NAA in the dark at 21°C without stirring for indicated time (Figure 1).
-
9.
Harvest and freeze control and NAA treated seedlings in liquid nitrogen (Figure 1).
Note: As short time treatment of Arabidopsis seedlings in water will not affect their status, for convenience, we used water for NAA treatment. However, for long time treatment, liquid 1/2 MS medium should be used instead.
Figure 1.
Scheme of auxin treatment
Seven-day-old Arabidopsis seedlings are submerged in H2O with or without 50 μM NAA for 0, 4, 8, 16 and 24 h. Seedlings are collected by RNA extraction.
RNA extraction
Timing: 3 h
This step describes how to extract total RNA from NAA treated Arabidopsis seedlings.
-
10.
Homogenize the collected seedlings into powder in liquid nitrogen.
-
11.
For each 100 μL powder, add 1000 μL TRIzol reagent and incubate for 10 min on ice.
-
12.
Add 200 μL chloroform and mix well.
-
13.
Centrifuge at 12,000 x g for 5 min.
-
14.
Transfer the supernatant to a new 1.5 mL EP tube.
-
15.
Add 500 μL of isopropyl alcohol and incubate at −80°C for 30 min.
-
16.
Centrifuge at 12,000 x g for 20 min at 4°C, remove the supernatant.
-
17.
Wash the RNA pellet twice with 75% ethanol.
-
18.
Remove the remaining ethanol and dry the pellet in the air.
-
19.
Dissolve the RNA pellet in 20 μL RNase-free water.
CRITICAL: TRIzol and chloroform are highly toxic, avoid direct contact, wear double gloves for skin protection and remove them immediately after handling TRIzol or chloroform. Always wear safety glasses, a lab coat, and closed-toe shoes and do it in hood. The RNA extraction must be conducted at 4°C, and the tubes need to be ice-chilled before use.
Deplete DNA from RNA
Timing: 2 h
This step describes how to remove DNA from total RNA.
-
20.
Aliquot 50 μg RNA and adjust the RNA solution to 38 μL with H2O.
-
21.
Add 10 μL 10x DNase buffer and 2 μL DNase I (1 U/μL) and incubate at 37°C for 5 min.
-
22.
Stop the reaction by adding 1000 μL TRIzol.
-
23.
Add 200 μL of chloroform and mix.
-
24.
Centrifuge at 12,000 x g for 5 min.
-
25.
Transfer the supernatant to a new 1.5 mL EP tube.
-
26.
Add 500 μL of isopropyl alcohol and incubate at −80°C for 20 min.
-
27.
Centrifuge at 12,000 x g for 20 min, remove the supernatant.
-
28.
Wash the RNA pellet twice with 75% ethanol.
-
29.
Remove the remaining ethanol and dry the pellet in the air.
-
30.
Dissolve the RNA pellet in 50 μL RNase-free water.
-
31.
Measure the RNA concentration with Nanodrop.
Note: RNA concentration should be higher than 2 μg/μL: firstly, the obtained material can propose RNA greater than 100 μg, secondly, the subsequent experiment needs RNA of not less than 2 μg/μL.
mRNA enrichment
Timing: 3 h
This step describes how to purify mRNA from total RNA.
-
32.
Aliquot 75 μg DNA free RNA, adjust the volume to 100 μL with TE buffer.
-
33.
Incubate at 65°C for 5 min and immediately place on ice.
-
34.
Wash 200 μL oligo-(dT) magnetic beads once with 1 mL binding buffer.
-
35.
Resuspend the beads in 100 μL of binding buffer.
-
36.
Add the beads into 100 μL RNA solution and incubate at 25°C for 10 min.
-
37.
Separate the beads on a magnetic rack and remove the supernatant.
-
38.
Wash beads twice with 200 μL of wash buffer, remove the wash buffer on a magnetic rack.
-
39.
Add 10 μL TE buffer and incubate at 80°C for 2 min.
-
40.
Separate the beads on a magnetic rack and collect the supernatant.
Note: Supernatant is mRNA, which needs to be stored at −80°C. The expected yield mRNA from 75 μg total RNA is 0.75 μg.
RNA dot blot
Timing: 8 h
This step describes how to perform RNA dot blot.
-
41.
Adjust the concentration of the mRNA solution to 500 ng/μL.
-
42.
Spot 1 μL mRNA onto the nitrocellulose membrane using a pipette.
-
43.
Air-dry the membrane at 25°C for 10 min and then then incubate at 85°C for 1 h in a drying oven.
-
44.
Cool the membrane to 25°C and block it with 5% milk solution for 30 min at 4°C.
-
45.
Remove milk solution and incubate the membrane with m6A antibody solution and leave at 4°C standing for 4 h.
-
46.
Wash the membrane three times with 1x TBST.
-
47.
Incubate the membrane with 10 mL secondary antibody solution for 1 h at 25°C.
-
48.
Wash the membrane three times with 1x TBST.
-
49.
The membrane is now ready for detection.
-
50.
Mix 2 mL of liquid A and B of western ECL Substrate at a ratio of 1:1.
-
51.
Drop the mixed solution onto the membrane.
-
52.
Detect the signal in a ChemiDoc imaging system (Bio-Rad).
RNA dot blot data analysis
Timing: 1 h
This step describes how to quantify RNA dot blot signal. ImageJ can be used according to the published protocol.4
-
53.
Indicate the area of the selected area.
-
54.
Average gray value of the selected area.
-
55.
Derive gray value.
-
56.
Analyze data.
Construct m6A-seq library
Timing: 2 days
This step describes how to construct m6A-seq library.
-
57.
Isolate poly(A) mRNA using oligo-(dT) magnetic beads from 200 μg total RNA.
-
58.
Fragment mRNA into ∼200 nt using RNA fragment module according to the manufacturer’s instructions.
-
59.
Adjust the volume of RNA fragment to 400 μL.
-
60.
Add 400 μL isopropyl alcohol, 40 μL 3M sodium acetate salt (pH 5.2) and incubate at −80°C for 20 min.
-
61.
Centrifuge at 12,000 x g for 20 min, remove the supernatant.
-
62.
Wash the RNA pellet twice with 75% ethanol.
-
63.
Remove the remaining ethanol and dry the pellet in the air.
-
64.
Dissolve the RNA pellet in 50 μL RNase-free water.
-
65.
Dilute the RNA in 400 μL RNA binding buffer with 2 μg m6A antibody and incubate at 4°C for 4 h.
-
66.
Aliquot 20 μL Protein G dynabeads and wash twice with RNA binding buffer.
-
67.
Add Protein G dynabeads into RNA-antibody mixture and incubate at 4°C for 1 h.
-
68.
Separate Protein G dynabeads in a magnetic rack, remove supernatant.
-
69.
Wash beads three times with RNA binding buffer.
-
70.
Elute RNA with 50 μL elution buffer at 50°C for 1 h.
-
71.
Purify eluted RNA by TRIzol reagent.
-
72.
Build m6A-seq library using NEBNext RNA Library Prep Kit and sequence in 2 × 150 bp paired-end mode. At least six gigabases (Gb) data are acquired (Nova6000).
Analyze m6A-seq data
Timing: 6 h
This step describes how to analyze m6A-seq data.
-
73.
Remove the adapters using the AdapterRemoval tool5 following the parameter settings “--mm 0.5 --minlength 18”.
-
74.
Map the reads against the Arabidopsis genome (TAIR10) by STAR6 following the parameter settings “--alignIntronMax 10000 --outSAMtype BAM Unsorted --outFilterMultimapNmax 1”.
-
75.
Assign reads to gene models using the R package DEGseq7 using default parameter settings.
-
76.
Call m6A peaks using the R package MeRIPtools8 using default parameter settings.
Expected outcomes
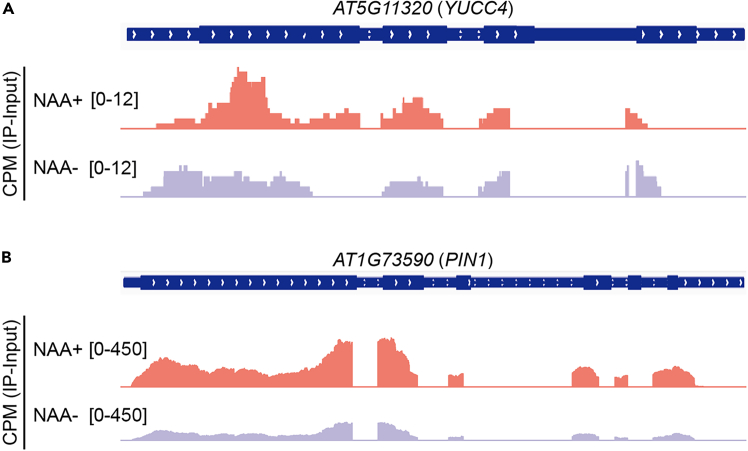
This protocol can be used to analyze auxin-induced changes of abundance and distribution of m6A modification. By utilizing qRT-PCR, this protocol enables the detection of relative expression of auxin responsive genes (Figure 2) to ensure the success of auxin treatment. m6A dot blot is used to measure changes of m6A abundance on different kinds of RNAs upon auxin treatment (Figure 3). Additionally, the m6A dot blot signals are quantified using ImageJ to analyze the relative abundance of m6A modification (Figure 4). This protocol also analyze the changes of m6A distribution by m6A-seq, the transcriptome-wide changes of m6A or changes of m6A at specific genomic loci can be analyzed as shown in the research of Li et al., 2024 (Figure 5).
Figure 2.
Relative expression of auxin response genes
Relative expression of IAA2 (A) and GH3.3 (B) after auxin treatment for indicated time. Relative expression level is determined by quantitative real-time PCR and normalized to the expression of ACTIN. Bars show mean ± SEM of three independent replicates.
Figure 3.
Dot blot analysis of m6A modification abundance
(A). Relative abundance of m6A modification of total RNA. Arabidopsis seedlings are treated by 50 μM NAA for 0, 4, 8,16 and 24 h. RNA is extracted by TRIzol reagent; (B-C). Relative abundance of m6A modification of RNAs with (B) and without (C) poly(A) tail. Arabidopsis seedlings are treated by 50 μM NAA for 0, 4, 8,16 and 24 h. RNA is extracted by TRIzol reagent, poly(A) RNA is purified using oligo(dT) magnetic beads, supernatant is collected and precipitated by ethanol and used as RNAs without poly(A) tail.
Figure 4.
Quantification of m6A signal
(A) Dot blot analysis of the m6A levels. The relative abundance of m6A signal is quantified by image J, and the m6A signals of 4, 6, 16 and 24 h are normalized to that of 0 h
(B) Plot of relative abundance of m6A signal in (A).
Figure 5.
Genomic browser snapshot of the m6A-seq signal
Genomic browser snapshot for input-subtracted m6A-seq signal at the YUCC4 (A) and PIN1 (B) loci in the seedlings with or without NAA treatment. The y axis represents the mean counts per million (CPMs) from replicates of immunoprecipitated samples subtracted from that from input samples.
Limitations
This protocol is designed to analyze the relative abundance of m6A modification rather than provide absolute quantification. Additionally, this protocol employs m6A sequencing to examine changes in the distribution of m6A modification. However, the resolution is limited, and it cannot provide information about the specific nucleotides where m6A modification occur. Given the distinct characteristics of various plant hormones, the protocol should also be applicable to other phytohormones.
Troubleshooting
Problem 1
Auxin responsive genes do not up-regulate after auxin treatment. Related to step 2.
Potential solution
Prepare a new auxin storage solution, aliquot it after preparation, and store it in a −20°C freezer away from light. Ensure that seedlings are fully submerged in H2O with NAA during auxin treatment, and change the solution every 4 h if necessary.
Problem 2
The concentration of mRNA is low, or its purity is poor. Related to step 5.
Potential solution
Increase the amount of total RNA and oligo-dT magnetic beads to enrich a sufficient quantity of mRNA. Repeat the oligo-dT enrichment step to enhance the purity of mRNA.
Problem 3
Noisy background of dot blotting result. Related to step 6.
Potential solution
Replace milk with BSA for membrane blocking and extend the blocking time.
Adjust the dilution ratio of the antibody and reduce the incubation time of membrane in the m6A antibody solution. Before incubating with the secondary antibody solution, repeat the washing step with TBST.
Use a high-sensitivity ECL solution to achieve improved signal detection.
Problem 4
Broad peak of m6A-seq result. Related to step 8.
Potential solution
Fragment the RNA in a PCR machine. Do not place the RNA in the machine until it reaches 95°C, and increase the fragmentation time.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chongsheng He (chongshenghe@outlook.com).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Bin Li (binli369@hnu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any datasets/code.
Acknowledgments
The study has been supported in part by the National Natural Science Foundation of China (32270623), the Natural Science Foundation of Hunan Province (2024JJ2016), and the Hunan agricultural science and technology innovation fund project (2024CX119).
Author contributions
B.L. and C.H. developed the protocol. B.L. performed the experiments with the help of Y.L., Lan Li, Lei Li, and L.C. B.L. and C.H. wrote the manuscript. Li Li and C.H. revised the manuscript. All the authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Lan Li, Email: lilan7@hnu.edu.cn.
Li Li, Email: lili@hhrrc.ac.cn.
Chongsheng He, Email: chongshenghe@outlook.com.
References
- 1.Li B., Zhou Q., Cai L., Li L., Xie C., Li D., Zhu F., Li X., Zhao X., Liu X., et al. TMK4-mediated FIP37 phosphorylation regulates auxin-triggered N6-methyladenosine modification of auxin biosynthetic genes in Arabidopsis. Cell Rep. 2024;43 doi: 10.1016/j.celrep.2024.114597. [DOI] [PubMed] [Google Scholar]
- 2.Shen L., Liang Z., Wong C.E., Yu H. Messenger RNA Modifications in Plants. Trends Plant Sci. 2019;24:328–341. doi: 10.1016/j.tplants.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z., Luo K., Zou Z., Qiu M., Tian J., Sieh L., Shi H., Zou Y., Wang G., Morrison J., et al. Genetic analyses support the contribution of mRNA N6-methyladenosine (m6A) modification to human disease heritability. Nat. Genet. 2020;52:939–949. doi: 10.1038/s41588-020-0644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any datasets/code.