Abstract
Background
Prior studies suggested that canonical Activin Receptor II (ActRII) and BMP receptor (BMPR) ligands can have opposing, distinct effects on skeletal muscle depending in part on differential downstream SMAD activation. It was therefore of interest to test ActRII ligands versus BMP ligands in settings of muscle differentiation and in vivo.
Methods and results
In human skeletal muscle cells, both ActRII ligands and BMP ligands inhibited myogenic differentiation: ActRII ligands in a SMAD2/3-dependent manner, and BMP ligands via SMAD1/5. Surprisingly, a neutralizing ActRIIA/B antibody mitigated the negative effects of both classes of ligands, indicating that some BMPs act at least partially through the ActRII receptors in skeletal muscle. Gene expression analysis showed that both ActRII and BMP ligands repress muscle differentiation genes in human myoblasts and myotubes. In mice, hepatic BMP9 over-expression induced liver toxicity, caused multi-organ wasting, and promoted a pro-atrophy gene signature despite elevated SMAD1/5 signaling in skeletal muscle. Local overexpression of BMP7 or BMP9, achieved by intramuscular AAV delivery, induced heterotopic ossification. Elevated SMAD1/5 signaling with increased expression of BMP target genes was also observed in sarcopenic muscles of old rats.
Conclusions
The canonical ActRII ligand-SMAD2/3 and BMP ligand-SMAD1/5 axes can both block human myoblast differentiation. Our observations further demonstrate the osteoinductive function of BMP ligands while pointing to a potential relevancy of blocking the BMP-SMAD1/5 axis in the setting of therapeutic anti-ActRIIA/B inhibition.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13395-025-00373-7.
Keywords: Skeletal muscle, Muscle differentiation, Myostatin, GDF8, GDF11, Activin A, BMP, BMPR2, ActRIIA/B, Heterotopic ossification
Introduction
Adult skeletal muscle mass may fluctuate in size in response to various anabolic or catabolic stimuli. In settings of muscle injury, muscle regeneration occurs—this requires activation of resident satellite cells (myoblasts), which proliferate, differentiate, and either fuse into mature myofibers or, in settings of severe injury, form new myofibers [1, 2]. Myoblast differentiation into myotubes can therefore be studied as a proxy for both developmental and regenerative competence. Proliferating skeletal myoblasts express key myogenic regulatory factors—including MyoD1, myogenin, Myf4, and Myf5—as part of the process of differentiation and fusion [3].
Muscle mass is negatively regulated by myostatin (also known as growth and differentiation factor 8 or GDF8), Activin A (ActA), and GDF11 [4–9], all of which are part of a sub-class of TGFβ family proteins that have a common receptor system–they all bind the Activin type II receptors (ActRIIA or ActRIIB) and the activin receptor-like kinase type I receptors, ALK4, ALK5, and/or ALK7, resulting in phosphorylation and activation of the transcription factors SMAD2 and SMAD3 [4–9]. Myostatin was the first of these ligands to be associated with a skeletal muscle phenotype–namely, animals that are deficient in myostatin have a significant increase in muscle mass, termed ‘double-muscling’ [10–13]. In contrast, global Activin A knockouts are not viable; however, dual pharmacological blockade of Activin A and myostatin in adult animals results in more skeletal muscle mass than blockade of myostatin alone [14, 15]. Moreover, the use of an antibody that blocks both type II receptors ActRIIA and ActRIIB, thereby inhibiting all the ligands that signal through those receptors (e.g., myostatin, GDF11, Activin A, Activin B, among others), also significant increases muscle mass [16, 17].
Bone morphogenic proteins (BMPs) are a distinct sub-class of TGFβ superfamily ligands; the BMPs have their own primary type II receptor, the BMP receptor II (BMPR2/BMPRII), and distinct preferences for type I ALK receptors, including ALK1, ALK2, ALK3, and/or ALK6 [18, 19]. In contrast to canonical ActRII ligands, which cause SMAD2/3 phosphorylation, BMP receptor activation results in SMAD1/5 phosphorylation. Recently, it has been reported that BMP signaling can be beneficial for muscle growth; both BMP7 and BMP14 (or GDF5) were shown to be sufficient for promoting muscle mass, with local over-expression of BMP7 via intramuscular AAV delivery increasing muscle growth in wild-type mice and rescuing a mouse model of denervation-induced muscle atrophy [20, 21]. Mechanistically, it was suggested that the BMP ligand-SMAD1/5 axis can promote the pro-hypertrophy AKT/S6 pathway and dominate over ActRII ligand-SMAD2/3 signaling, in part by competing for the common mediator SMAD4, which hetero-oligomerizes with either SMAD2 and SMAD3 or SMAD1 and SMAD5, a step thought to be necessary for SMAD functionality [19]. Thus, it has been suggested that activation of the BMP ligand-SMAD1/5 axis may be sufficient as a therapeutic modality aimed at ameliorating multiple forms of muscle atrophy [19]. However, others have suggested BMP ligands can have inhibitory effects on muscle. Both BMP2 and BMP7 inhibited primary human myoblast differentiation in a manner comparable to ActRII ligands and BMP9 delayed regeneration in a barium-chloride-induced skeletal muscle injury murine model [13, 22]. Historically, BMPs have been linked to their canonical osteoinductive effects on different cell types, including the induction of heterotopic ossification [23]. In vivo, injection of recombinant BMP9 protein was sufficient to promote heterotopic ossification in the setting of cardiotoxin-induced muscle damage [23], building upon similar results in various models [24–26], which demonstrated that intramuscular adenoviral delivery of multiple human BMPs, including BMP7 and BMP9, was sufficient to promote orthotopic ossification in the muscle of nude rodents.
Given the mixed results across prior published works, the current study aimed to explore differences and similarities between canonical ActRII and BMP/BMPR signaling. We directly compared multiple ligands head-to-head and the functional relevancy of the type II ActRIIA/B receptors, as well as overexpression of the various receptor-activated SMADs (or R-SMADs) on human skeletal myoblasts, by assessing downstream signaling and effects on myoblast differentiation. We then validated these results in human myotubes by assessing key signaling and transcriptional gene targets. We further translated these results in mice with both a hepatic over-expression model as well as an intramuscular AAV-mediated over-expression model to determine the impact of systemic or local BMP elevation on skeletal muscles and other organs in vivo. BMP ligands can inhibit cultured human myoblast differentiation in a SMAD1/5-dependent manner and, notably, at least in part through type II ActRIIA/B signaling, indicating likely physiological relevancy of this type II receptor system in skeletal muscle. BMP ligands failed to directly promote the AKT/S6 signaling pathway in myoblasts or myotubes following acute or longer treatment. SMAD2, SMAD3, SMAD1, and SMAD5 overexpression are each sufficient to recapitulate the inhibitory functional phenotype on myogenic differentiation. In mice, hepatic BMP9 overexpression is associated with liver toxicity and multi-organ wasting/cachexia while local muscle overexpression of BMP7 or BMP9 promoted pathological osteoinductive changes.
Results
ActRII and BMP ligands inhibit human myoblast differentiation
ActRII ligands, including Activin A (ActA), myostatin (or GDF8), and GDF11, have been shown to block myoblast differentiation [13]. We first sought to compare the signaling and functional effects of ActRII ligands versus BMP ligands in human skeletal muscle cell culture.
Primary human muscle cell lines can serve as an in vitro surrogate for how ligands may act in humans [13]. However, primary cultures have several limitations, including restricted proliferation capacity and applicability for stable cell line production. As such, we generated and characterized a human skeletal muscle cell line transduced with human telomerase reverse transcriptase (hTERT) and a temperature-sensitive SV40 large T antigen (tsSV40). In brief, a single cell clone was selected and characterized based on SV40 expression at non-permissive temperatures and molecular profiling was conducted to compare expression of relevant myogenic markers to primary cells at corresponding days of differentiation. These clonal transduced human Skeletal-Muscle Derived Cells (termed iHUSKMDC or iSKMDC in this report) are shown to be myoblasts with an ability to differentiate into myotubes and exhibited similar behavior to primary cells based on molecular profiling (Supplemental Fig. 1A and 1B).
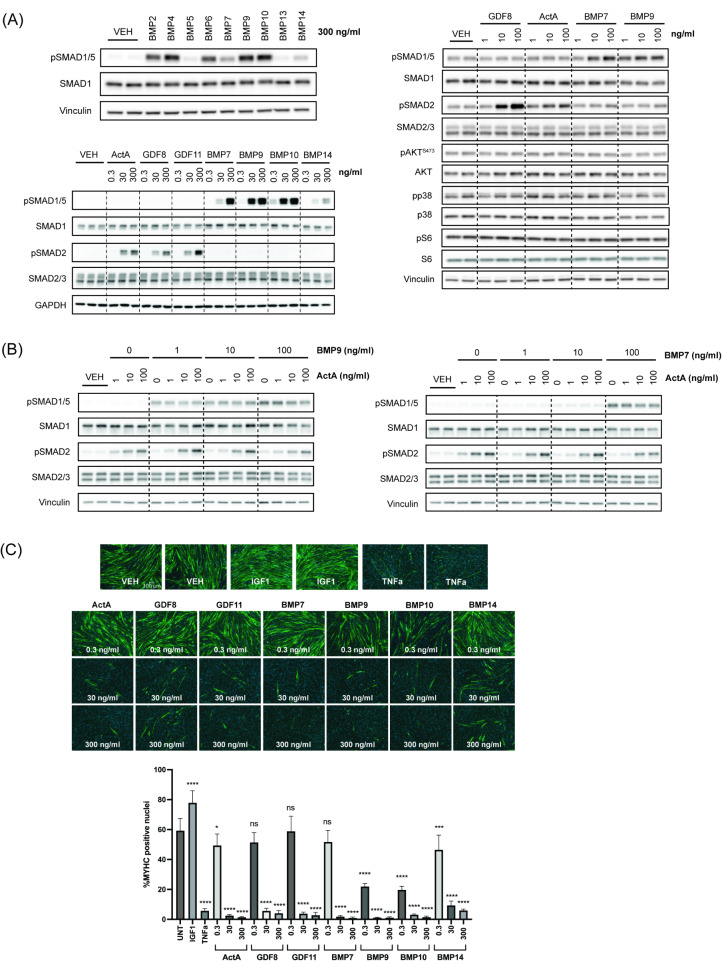
We first compared relative canonical SMAD1/5 signaling across the BMP ligand family and selected ligands representing distinct sub-classes [18] and potencies, including ligands previously linked to muscle growth (Fig. 1A; upper left). Next, we followed up with further experiments to investigate the downstream effects these BMP ligands—BMP7, BMP9, BMP10, or BMP14—compared to ActRII ligands—ActA, GDF8, or GDF11.
Fig. 1.
ActRII and BMP ligands promote distinct SMAD signaling yet similar differentiation blockade in human myoblasts. (A) Western blotting analysis for SMAD signaling with BMP ligands at 300 ng/ml (upper left), SMAD signaling with a dose response of ActRII and BMP ligands (bottom left), and non-SMAD signaling with a dose response of ActRII and BMP ligands (right). Human transduced myoblasts (iHUSKMDC) were acutely stimulated with vehicle as a negative control, BMP ligands, or ActRII ligands for 30 min. (B) Western blotting analysis with combined ligands. Human iHUSKMDC myoblasts were stimulated with vehicle as a negative control or with increasing doses of ActA ± BMP9 (left) or ActA ± BMP7 (right) at 1, 10, or 100 ng/ml. (C) Human myoblast differentiation assay with individual ligands. iHUSKMDC myoblasts were differentiated into myotubes with vehicle as negative control, TNFα (30 ng/ml) or IGF1 (10 ng/ml) as positive controls or increasing doses of ActRII or BMP ligands (0.3, 30, or 300 ng/ml). Representative images are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm. Differentiation was quantified by evaluating the percentage of nuclei within myotubes that were positively identified using anti-MyHC antibody staining. Differences between groups were analyzed using one-way ANOVA (compared to UNT group). Data are means ± SD. Asterisks indicate differences with the control. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control
Consistent with previous results in other cell types, in human skeletal muscle cells, ActA, GDF8, and GDF11 acutely induce SMAD2 phosphorylation whereas BMP7, BMP9, BMP10, and BMP14 promote SMAD1/5 phosphorylation in a dose-dependent manner (Fig. 1A; bottom left). Among the BMP ligands tested, BMP9 and BMP10 showed stronger potency compared to BMP7, with BMP14 as the relatively weakest activator of SMAD1/5 signaling.
Given that prior reports have suggested involvement of non-SMAD signaling within the TGFβ superfamily, we next sought to determine the effects of ligands on alternative pathways [27–29]. Of note, it was previously suggested that BMPs may promote positive skeletal muscle regulation via the pro-hypertrophy AKT/S6 pathway [20, 21]. ActRII ligands (GDF8 and ActA) and BMP ligands (BMP7 and BMP9) acutely induced their respective SMAD signaling in fully differentiated myotubes without strong induction of p38 or AKT/S6 signaling (Fig. 1A; right). To determine wither ligands may have longer term effects, we also treated cells for 24-hours and failed to observe consistent BMP-mediated increased pAKT or pS6 in myoblasts or myotubes despite previous studies suggesting activation of this pathway [20, 21] (Supplemental Fig. 1C).
Multiple reports have suggested competition and interplay between the canonical ActRII and BMP axes, both at the receptor and SMAD signaling levels [20, 30, 31]. As such, we selected ActA, BMP7, and BMP9 to further test a dose response of co-treatment in human myoblasts and assess changes in their downstream SMAD response. Combined ActA and either BMP showed minimal, if any, acute crosstalk inhibition of their respective phosphorylated SMAD signaling at the doses tested. (Fig. 1B).
Finally, to determine their functional effects on myogenic differentiation, human myoblasts were allowed to differentiate for 5 days in the presence or absence of ligand (0.3–300 ng/ml) and we assessed differentiation by myosin heavy chain (MYHC) staining. Doses were chosen based on the observed phosphorylated SMAD signaling patterns. As positive controls, the proinflammatory cytokine TNFα blocked differentiation, whereas the pro-anabolic IGF1 promoted differentiation (Fig. 1C). In line with prior reports [4, 13], ActRII ligands significantly inhibited differentiation in a dose-dependent manner. All BMP ligands tested blocked differentiation to varying degrees relative to their phosphorylated SMAD1/5 potencies, confirming a common myogenic functional effect across these TGFβ family members. Taken together, in human muscle cells, ActRII and BMP ligands promote distinct canonical SMAD responses with minimal signaling crosstalk yet show similar blockade of myogenic differentiation.
Receptor-activated SMAD (R-SMAD) overexpression promotes blockade of human myoblast differentiation
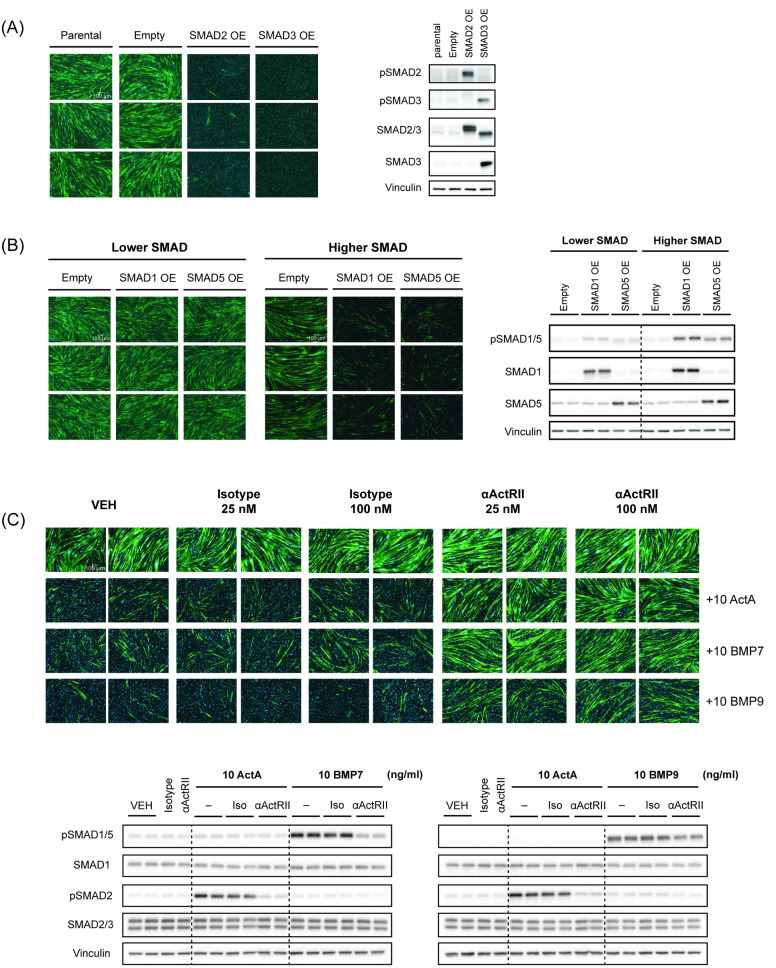
Although non-SMAD pathways have been previously reported for TGFβ family members, 25–27 in our human muscle cells, we failed to see clear, consistent alternative signaling. As such, for the first time, we generated SMAD-overexpressing skeletal muscle cell lines to assess the direct effects of individual SMADs within this cell context. Specifically, we generated stable overexpression cell pools of wild-type receptor activated SMADs or R-SMADs—SMAD1, SMAD5, SMAD2, or SMAD3. The empty vector control line was able to differentiate comparable to the parental iHUSKMDC cells (Supplemental Fig. 2A). Total protein levels of each SMAD were validated by western blotting analysis. Overexpression of each R-SMAD was associated with increased corresponding phosphorylated R-SMAD at baseline, demonstrating induction of SMAD signaling in the absence of exogenous ligand (Fig. 2A and B). Persistent SMAD signaling was observed in each cell line throughout the myogenic differentiation process (Supplemental Fig. 2B.) Expression profiling of this skeletal muscle cell system showed detectable levels of endogenous ActRII and BMP ligands, suggesting an autocrine feedback response (Supplemental Fig. 1B).
Fig. 2.
R-SMAD overexpression blocks human myoblast differentiation and ActRII inhibition mitigates ligand inhibitory effects. (A) Differentiation assay and western blotting analysis validation for SMAD2 and SMAD3 over-expressing lines. Stable skeletal muscle cell lines (iHUSKMDC) with SMAD2 or SMAD3 over-expression (OE) were differentiated into myotubes. Representative images (n = 3 replicates) are shown for each treatment, using anti-MyHC antibody staining for myotubes (green) and nuclei (blue) using DAPI staining. Scale bar, 100 μm. Western blotting analysis at day 0 of differentiation was used to confirm overexpression and signaling with SMAD-selective antibodies. (B) Differentiation assay and western blotting analysis validation for SMAD1 and SMAD5 over-expressing lines. Stable skeletal muscle cell lines (iHUSKMDC) with SMAD1 or SMAD5 over-expression (OE) at lower and higher levels were differentiated into myotubes. Representative images (n = 3 replicates) are shown for each treatment, using anti-MyHC antibody staining for myotubes (green) and nuclei (blue) using DAPI staining. Scale bar, 100 μm. Western blotting analysis at day 0 of differentiation was used to confirm relative overexpression and signaling with SMAD-selective antibodies across the lines. (C) Differentiation assay and western blotting analysis to assess ActRII inhibition effects. Human myoblasts (iHUSKMDC) were pre-treated with a selective anti-ActRIIA/B neutralizing antibody or isotype control (25 or 100 nM) followed by stimulation with 10 ng/ml ActA, BMP7, or BMP9. Scale bar, 100 μm. Representative images (n = 2) are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm
To assess functional implications of R-SMAD overexpression, myoblasts were allowed to differentiate for 5 days without added recombinant protein ligands that activated SMAD signaling (ActRII ligands and BMPs), followed by anti-MYHC staining. Overexpression of SMAD2 or SMAD3 alone blocked myoblast differentiation into myotubes, similar to exogenous ActRII ligand treatment and in line with prior reports [4, 13], demonstrating that SMAD2 or SMAD3 signaling is sufficient to inhibit myogenic differentiation (Fig. 2A). Overexpression of SMAD1 or SMAD5 blocked myoblast differentiation in a dose-dependent manner, with higher SMAD1/5 signaling corresponding to the inhibitory phenotype, suggesting a SMAD signaling threshold effect (Fig. 2B). Together these data indicate that R-SMAD signaling through either the SMAD2/3 or SMAD1/5 axes mediates the inhibitory effects of ActRII and BMP ligands on myogenic differentiation.
ActRII inhibition rescues activin A and BMP ligand-mediated effects on human skeletal muscle cells
Receptor binding and signaling by ActRII and BMP ligands are highly promiscuous, given that ligands across sub-families have been shown to bind and engage with activin type II receptors (ActRIIA and ActRIIB) and the BMP type II receptor (BMPR2) [32, 33]. Multiple BMP ligands, including BMP7 and BMP9, can reportedly bind and signal through ActRII receptors within some cell types [34, 35]. Previous studies demonstrated that BMP7 can bind with high affinity to ActRIIA relative to BMPR2 whereas BMP9 has a relatively higher affinity to BMPR2 [33, 36].
Prior publications have demonstrated that antibody-mediated ActRIIA/B inhibition (αActRII) is sufficient to reduce ActA and GDF8 downstream SMAD signaling as well as attenuate their blockade of skeletal muscle differentiation [16]. Whether ActRII signaling is functionally relevant to BMP ligands within skeletal muscle cells, however, is unclear. As such, myoblasts were treated with ActA, BMP7, or BMP9 in the presence or absence of an αActRII antibody, either acutely for signaling or for multiple days to assess myogenic differentiation. Consistent with previous results, αActRII blocked ActA-mediated SMAD2 phosphorylation and rescued myogenic differentiation [13, 17] (Fig. 2C). Surprisingly, both BMP7- and BMP9-mediated signaling and differentiation blockade were at least partially rescued with the αActRII antibody. BMP7 was more responsive to antibody treatment, showing similar rescue compared to the canonical ActRII ligand, ActA; this is notable since these cells also express BMPR2, which is not bound by the anti-ActRIIA/B antibody and, thus, amenable for ligand binding (data not shown) (Fig. 2C). Similarly, αActRII-mediated blockade of SMAD1/5 signaling was also observed with other members of the larger BMP ligand family (Supplemental Fig. 2C). Thus, in skeletal muscle cells, BMP ligands can promote downstream effects in a partly ActRII-dependent fashion, demonstrating a putative key role for BMP inhibition with the use of anti-ActRIIA/B neutralizing antibodies.
ActRII and BMP ligands suppress myogenic markers in differentiating human myoblasts and differentiated human myotubes
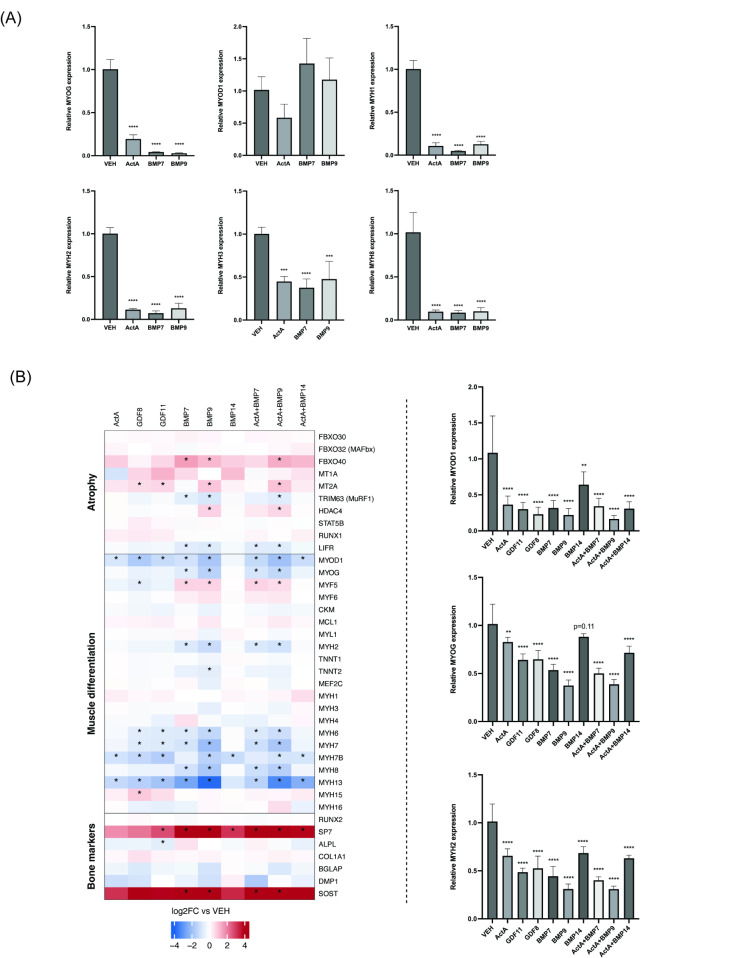
Since we found that both ActRII ligands and BMPs functionally inhibit human myoblast differentiation into myotubes, we next sought to assess the effects of select representative ligands on the expression of key myogenesis markers. Treatment of differentiating (day 0 of differentiation) iHUSKMDC myoblasts with ActA, BMP7, and BMP9 for 24 h suppressed mRNA levels for the myogenic marker myogenin (MYOG), as well as the mRNA levels for myosin heavy chain (MYHC) genes (MYH1, MYH2, MYH3, MYH8) (Fig. 3A). Prior data demonstrated that BMP signaling can have pro-osteoinductive effects on multiple cell types, including resident Sca1+ muscle stromal cells [37]. To determine whether BMPs might have an early osteo-inductive influence on myoblasts, expression of osteogenic differentiation markers were also assessed. Sclerostin (SOST), a negative regulator of bone formation, was significantly increased in response to BMP7 and BMP9; however, expression of the osteoblast marker osteocalcin (BGALP) was significantly repressed and RUNX2, an early osteogenic differentiation marker, was unchanged (Supplemental Fig. 3).
Fig. 3.
ActRII and BMP ligands similarly repress myogenic markers in human myoblasts and differentiated human myotubes. (A) qPCR analysis of myogenic differentiation gene markers in ligand-treated myoblasts. Human myoblasts (iHUSKMDC) were stimulated with vehicle as a negative control or single 300 ng/ml dose of ActA, BMP7, or BMP9 for 24 h with downstream gene expression determined by qPCR (n = 4/treatment). Data are mean ± standard deviation (SD) from the mean. Asterisks indicate differences with the control. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. (B) Heatmap for muscle and bone marker gene expression profiles in ligand-treated human myotubes. ActRII and BMP ligands show similar transcriptional signature on myogenic markers (left). Primary human myotubes (HUSKMDC) stimulated as above were analyzed by qPCR for select genes (n = 5/treatment) (right). Data are mean ± standard deviation (SD) from the mean. Asterisks in the heatmap indicate significant differential expression between the control and the treatment. Asterisks in the bar plot indicate differences with the control group. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control
To further elucidate the transcriptional networks associated with ActRII- and BMP-ligand-mediated responses in differentiated muscle cells (day 8 of differentiation), we performed RNA sequencing using single or combined ligand treatment across multiple ligands in each family. Primary human HUSKMDC myotubes were treated for 24-hours with ActA, GDF8, GDF11, BMP7, BMP9, BMP14 or combined ActA with BMP7, BMP9, or BMP14 to confirm our data in myoblasts. Genes were considered differentially expressed if their expression was perturbed > 1.5-fold compared to vehicle with a Benjamini-Hochberg adjusted p value < 0.05.
In differentiated myotubes, pro-atrophy marker MAFBX expression was notably unperturbed whereas MURF1 expression was significantly reduced with BMP7 or BMP9 and trended toward a decrease with all other ligands (Fig. 3B; left). However, all ligands significantly repressed MYOD1 as compared to baseline, except for BMP14, which showed a downward trend. A similar trend was observed with MYOG across ligand treatment. Subsequent qPCR validated these biomarkers, showing a significant repression of both MYOD1 and MYOG with all ligands, along with decreased expression of MYH2 (Fig. 3B; right)—similar perturbations of myogenic marker in myoblasts and consistent with prior reports on the mechanism underlying the negative regulation of human muscle cells by GDF8 [38].
Hepatic over-expression of BMP9 in mice results in liver toxicity with a cachexia-like phenotype and an induction of a pro-atrophy gene program in skeletal muscles
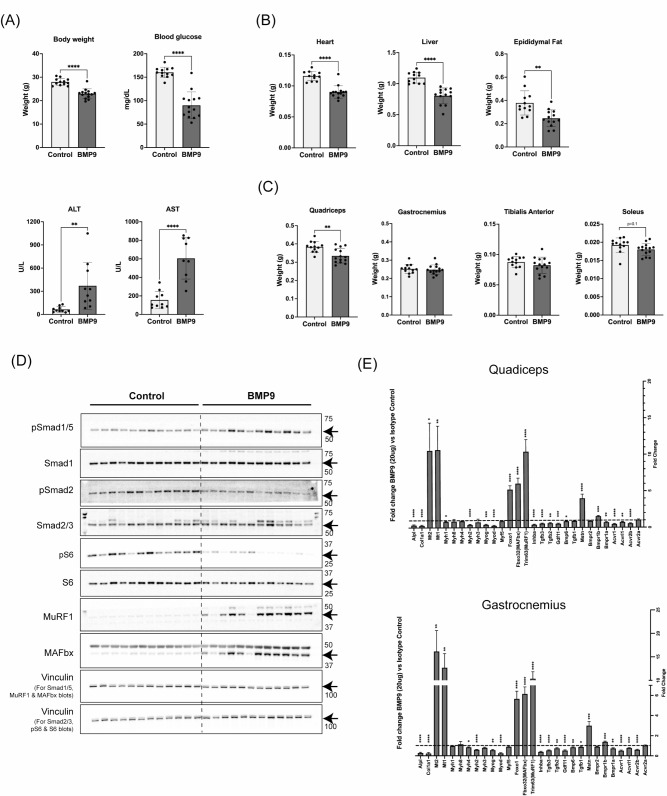
BMP9 promotes heterotopic ossification of isolated mouse muscle resident Sca1+ stromal cells ex vivo [37] and was recently shown to block isolated primary murine skeletal muscle cell differentiation, but its full effects on adult skeletal muscle remain unclear [22]. Given our findings in cultured iHUSKMDCs, we investigated the effects of hepatic over-expression of BMP9 in mice. We delivered 20 ug of murine BMP9 DNA plasmid into C57BL6J male mice by hydrodynamic tail vein injection. The control group received an empty DNA vector. Hydrodynamic delivery (HDD) of BMP9 plasmid induced hepatic overexpression of the BMP9 gene, Gdf2 (Supplemental Fig. 4A). Seven days following BMP9 over-expression, mice lost ∼ 20% body weight and displayed a reduction in blood glucose levels (Fig. 4A), which is consistent with prior reports on BMP9-mediated glucose metabolism [39, 40]. Of note, BMP9 HDD increased liver damage enzymes: alanine aminotransferase (ALT) and aspartate aminotransferase (AST) (Fig. 4A), suggesting that local BMP9 over-expression was associated with hepatotoxicity that could have had systemic effects. Weights of heart, liver, and epididymal fat were lower in BMP9 over-expressing mice compared to controls by ∼ 22%, ∼ 27%, and ∼ 35% respectively (Fig. 4B), demonstrating a global wasting syndrome.
Fig. 4.
BMP9 overexpression promotes liver toxicity, multi-organ wasting, and elevated muscle atrophy markers despite BMP-SMAD1/5 signaling. (A) Body weight, blood glucose, and levels of liver damage enzymes: alanine aminotransferase (ALT) and aspartate aminotransferase (AST). (B) Heart, liver and epididymal fat pat weights. (C) Weights of lower limb skeletal muscles: quadriceps, gastrocnemius, tibialis anterior, and soleus. In all graphs, light gray bars represent a control group that received an empty vector via a hydrodynamic tail vein injection (n = 11–12) and dark gray bars represent a group that received 20 ug of BMP9 DNA (n = 14). (D) Impact of hepatic BMP9 over-expression for 7 days on signaling in quadriceps muscles. Immunoblots for phosphorylated SMAD1/5 and phosphorylated SMAD2 and total SMAD1 and total SMAD2/3; phosphorylated ribosomal protein 6 S and total S6, MuRF1 and MAFbx in quadriceps muscles of mice that received DNA with an empty vector (control) or BMP9 DNA (BMP9). Immunoblots for vinculin as shown as protein loading controls. Molecular weights are shown on the left-hand side for each blot and arrows point to the protein bands of interest. (E) Impact of hepatic BMP9 over-expression for 7 days on gene expression in quadriceps and gastrocnemius muscles. mRNA amounts for specific genes were quantified by real-time quantitative polymerase chain reaction and expression levels were normalized to a geometric mean of reference genes. Vertical dotted lines in each graph show mRNA levels for each gene in the control group that received DNA with an empty vector (n = 12). Dark gray bars show mRNA levels for specific genes in the group that received BMP9 DNA (n = 14), relative to the control group. Data are mean ± standard deviation (SD) from the mean. Statistical analyses were conducted with an un-paired t-test. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control
Hepatic BMP9 over-expression resulted in skeletal muscle atrophy with quadriceps muscle weights lower in BMP9 over-expressing mice compared to controls by ∼ 13% (Fig. 4C). Weights of the gastrocnemius, the soleus, and tibialis anterior muscles were not significantly affected at the time of takedown (Fig. 4C). To determine the impact of high levels of BMP9 on the regulation of protein synthesis and degradation markers in skeletal muscles, we performed immunoblotting in quadriceps because this muscle showed the weight loss (Fig. 4D). Compared to control animals, in BMP9 over-expressing mice, phosphorylation of SMAD1/5 was higher, while phosphorylation of SMAD2 was lower in quadriceps muscles (Fig. 4D), demonstrating on-target BMP effects in muscle without compensatory SMAD2 pathway activation. Levels of phosphorylated as well as total ribosomal protein S6, a downstream target of the mTORC1/S6K1 protein synthesis pathway, were lower in BMP9 overexpressing mice compared to a control group by ∼ 82% and ∼ 30% respectively (Fig. 4D). We also quantified protein amounts for the E3 ubiquitin ligases MuRF1 and MAFbx, which are transcriptionally upregulated under atrophic conditions, and induce muscle atrophy via proteasomal protein degradation pathway 31. MuRF1 and MAFbx protein levels were higher in quadriceps muscles of BMP9 over-expressing mice compared to controls by 1878% and 374.5% respectively and correlated with SMAD1/5 phosphorylation levels (Fig. 4D; Supplemental Fig. 4B), demonstrating a pro-atrophic condition.
We next performed gene expression analyses in two muscles: quadriceps, which atrophied with BMP9 over-expression, and gastrocnemius, which did not atrophy. Both muscles showed a similar mRNA expression profile with up-regulation of pro-atrophy markers MAFbx/Fbxo32, MuRF1/Trim6331 along with the metallothioneins Mt1 and Mt232,33 (Fig. 4E). Of the canonical SMAD2/3 activating myokines, only Mstn (or Gdf8) levels were increased in BMP9 over-expressing mice. In line with the myogenic biomarkers observed in cell culture, we observed reduced expression of Myod and Myog as well as the myosin Myh2 with BMP9 over-expressing mice, further supporting a direct on-target BMP9 effect in skeletal muscle (Fig. 4E). Thus, despite differences in observed muscle mass effects, both gastrocnemius and quadricep muscles showed a near identical pro-atrophy gene signature with a concomitant suppression of myogenic genes and myosins, suggesting that skeletal muscle is primed for wasting in the setting of elevated systemic BMP9 levels.
Taken together, despite the increased SMAD1/5 phosphorylation observed, there were increased levels of Mt1 and Mt2 along with Foxo1, MAFbx/Fbxo32 and MuRF1/Trim63, as well as reduced levels of phosphorylated S6, indicating decreased mTORC1/S6K1 signaling in a pro-atrophic state in the absence of SMAD2/3 signaling. In addition, SMAD1/5 phosphorylation levels correlated with elevated atrophy signaling and repressed Myh2 expression (Supplemental Fig. 4B). Therefore, our results contrast prior models showing that elevated BMP-SMAD1/5 signaling is dominant and sufficient to promote a pro-hypertrophy response [19].
Intramuscular over-expression of BMP7 and BMP9 in mice results in heterotopic ossification in skeletal muscles
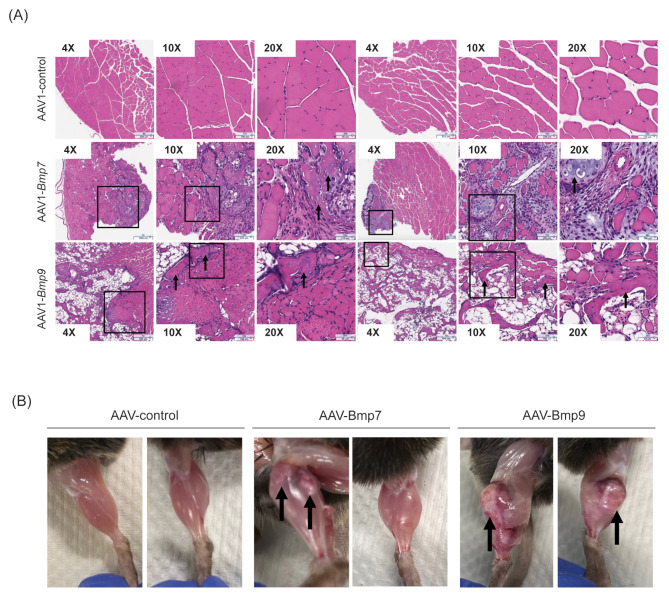
Given the observed liver toxicity with hepatic BMP9 overexpression, we next assessed the intramuscular overexpression of BMP9 and BMP7 to determine local effects and compare functionalities across ligands. Previously, it was shown that local intramuscular AAV: BMP7 delivery was sufficient to promote skeletal muscle hypertrophy 28 days following injection with increased Igf1 expression and elevated AKT/S6 signaling in wild-type mice. In addition, muscle specific over-expression of BMP7 was shown to have a protective effect in the setting of denervation-induced muscle atrophy19 [20]. We designed rAAV1 vectors encoding either murine BMP9 or BMP7, which were delivered to the tibialis anterior (TA) muscles of adult 13-week-old mice at a similar dose to prior studies to assess chronic 28-day changes. Both AAV1:BMP7 and AAV1:BMP9 increased mRNA expression of their respective BMP ligands by day 7 as evident by RNAscope (Supplemental Fig. 5A). Of note, given our prior observations with hepatic BMP9 by HDD, there was no evidence of hepatoxicity by histology following intramuscular AAV gene delivery at 1 month (Supplemental Fig. 5B). After 1 month, muscle-specific over-expression of both AAV1:BMP7 and AAV1:BMP9 had osteoinductive effects, with a more pronounced phenotype observed with BMP9. AAV1:BMP7 injected mice showed partial ossification of the TA, while AAV1:Bmp9 injected mice had widespread TA ossification (Fig. 5A and B). Thus, at an equivalent AAV dose and treatment time to prior reports, we failed to demonstrate a beneficial effect of muscle, instead observing osteogenic changes.
Fig. 5.
Local AAV delivery of BMP7 and BMP9 promotes heterotopic ossification in mouse skeletal muscle. (A) H&E staining of tibialis anterior (TA) cross-sections from mice 1-month post-intramuscular AAV delivery. Representative images of n = 2 mice per group are shown, each at 4X, 10X, and 20X magnification (total numbers of mice were, n = 11 for AAV: BMP7 and n = 12 for AAV1: control or AAV1:BMP9). Boxes indicate select corresponding magnified areas. Arrows indicate areas of ossification. (B) Images of dissected lower limbs from mice1-month post-intramuscular AAV delivery. Representative images of n = 2 mice per group are shown. Arrows indicate areas of ossification
In aged rodent muscles, the TGFβ pathway is upregulated with increased expression of BMP gene markers co-incident with increased pSMAD1/5
Previous studies have shown increased SMAD2/3 activation with age in rat muscles [38]. In addition, increased circulating levels of the activin/TGFβ signaling marker follistatin-like 3 (FSTL3/FLRG) have been shown in old human plasma [41]. These observations suggest a regulatory role of activin/TGFβ signaling with age [38, 41]. Of note, BMP7 was among the conserved age-related plasma protein that increased in both humans and mice [41]. Skeletal muscle SMAD1/5 activation was shown to decrease in mice from 1-week to 6-months of age [19], but changes to the pathway in the setting of age-related muscle loss (or sarcopenia) are unclear.
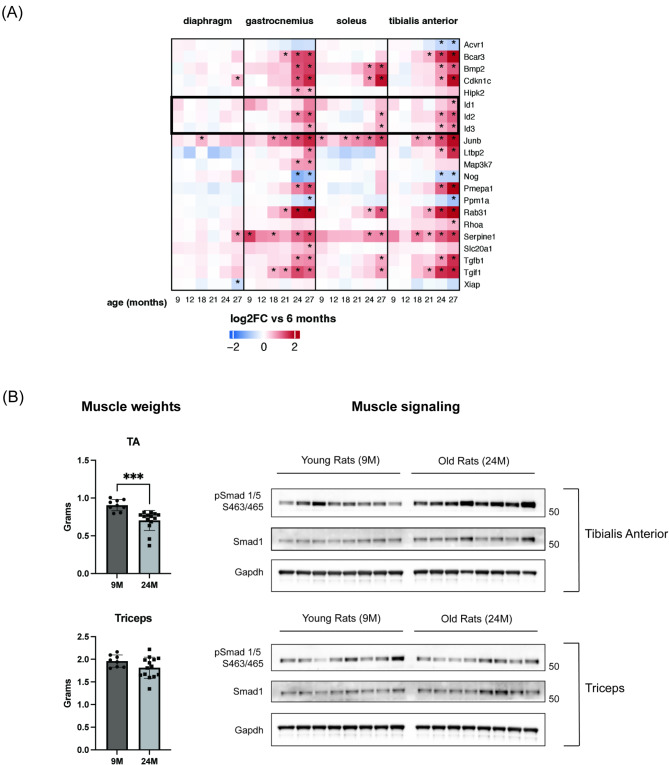
We previously performed comprehensive transcriptional profiling of multiple muscle groups form old rats and identified up-regulated markers of TGFβ signaling co-incident with sarcopenia, suggesting a putative causal role for the pathway in the setting of aging [42, 43]. In these profiled sarcopenic skeletal muscles, we observed consistently elevated BMP markers Id2 and Id3, indicative of an increased BMP signaling axis (Fig. 6A). To confirm changes in BMP signaling with sarcopenia, we assessed SMAD1/5 phosphorylation by immunoblotting in a sarcopenic muscle (tibialis anterior) and a non-sarcopenic muscle (triceps) from rats (Fig. 6B; left). SMAD1/5 phosphorylation was increased in aged rat tibialis anterior, but unperturbed in triceps, demonstrating that the BMP-SMAD1/5 axis is in fact elevated with sarcopenia in rats (Fig. 6B; right).
Fig. 6.
Elevated SMAD1/5 signaling and BMP gene targets are observed in aged rat sarcopenic muscle. (A) Heatmap for TGFB pathway gene expression profiles in aged rat diaphragm, gastrocnemius, soleus, and tibialis anterior muscles. Tissues were profiled from multiple time points across the lifespan of rats (from 6 to 27 months). For age-related genes (see the original paper for details), the time points where the genes show significant differential expression vs. 6 months are marked by asterisks. Id1-3, BMP target genes, are highlighted in a black box. (B) SMAD1/5 immunoblotting analysis in select rat sarcopenic and non-sarcopenic muscles. Muscle weights for sarcopenic tibialis anterior (or TA) and non-sarcopenic triceps are shown on the left. Phosphorylated SMAD1/5 and total SMAD1 levels for each muscle group are shown on the right. Data are mean ± standard deviation (SD) from the mean. Statistical analyses were conducted with an un-paired t-test. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control
Discussion
Our group has a longstanding interest in mechanisms controlling skeletal muscle mass, particularly through the TGFβ superfamily, and have previously demonstrated that ActRII-SMAD2/3 signaling ligands, such as myostatin (GDF8) and Activin A, act predominantly by inhibiting genes that are normally induced during myogenesis and upon myoblast differentiation28. However, ActRII ligands can induce muscle atrophy in certain settings, as was shown with liver-induced over-expression of GDF11, which was sufficient to induce full-body cachexia 39–42. Importantly, a neutralizing antibody that blocks signaling through ActRIIA/B was shown to promote significant skeletal muscle growth in rodents as well as increase lean mass and reduce adiposity in humans [44]. Recently, however, studies have suggested an alternative means of drugging the TGFβ family for similar purposes—namely that BMP-SMAD1/5 signaling opposes ActRII-SMAD2/3 activation and promotes skeletal muscle hypertrophy, including via intramuscular AAV delivery of BMP ligands, thereby emphasizing a potential role as a clinical intervention [20]. As such, it was of interest to better understand the relevant roles of the ActRII-SMAD2/3 and BMP-SMAD1/5 axes within skeletal muscle, especially given prior data on the anti-myogenic differentiation and pro-osteoinductive effects of BMP signaling [20, 24–26].
In line with other past studies and similar to canonical ActRII-SMAD2/3 ligands (ActA, GDF8, and GDF11), we found that multiple BMP ligands inhibit human myoblast differentiation at doses sufficient to activate SMAD1/5 signaling. Other groups have suggested that BMP ligands may stimulate mTORC1 signaling through AKT activation, thereby promoting anabolic signaling in skeletal muscle [20, 21]. To test this, we examined the activation of alternative pathways in response to acute and prolonged BMP stimulation. ActRII ligands (GDF8 and Activin A) and BMP ligands (BMP7 and BMP9) acutely induced their respective canonical SMAD signaling in both differentiating myoblasts and fully differentiated myotubes. However, in contradiction to these prior reports, BMP ligands did not directly promote AKT/S6 phosphorylation. This discrepancy challenges the premise that BMP signaling acts as a functional antagonist to ActRII-SMAD2/3 signaling in skeletal muscle cells. Instead, BMP ligands appear to share a similar inhibitory effect on differentiation and myogenic gene expression. This was confirmed in our qPCR and RNAseq analyses, which showed that both BMP and ActRII ligands can downregulate key myogenic regulators, including MyoD1 and myogenin, as well as critical myosin heavy chain isoforms required for sarcomere formation.
Given the likely essential role of canonical downstream SMAD signaling, we also wanted to understand if SMAD1/5 over-expression itself was able to mediate the inhibition of myoblast differentiation, as was the case with SMAD2 and SMAD3 over-expression. We found that SMAD1 or SMAD5 were able to block myoblast differentiation when sufficiently over-expressed. We chose SMAD1 and SMAD5 as representatives of their R-SMAD family—we did not include SMAD8 in our analysis given recent data suggesting it may act in a dominant negative fashion in some cell contexts.
Prior studies demonstrated that BMP ligands can promote downstream SMAD1/5 phosphorylation through both BMPR2 as well as ActRII receptor complexes in non-muscle cell types [34, 35]. It was previously shown that a selective anti-ActRIIA/B monoclonal antibody was sufficient to block downstream signaling and effects of ActA and GDF8 in skeletal muscle [16]. As such, we assessed the consequences of a neutralizing ActRIIA/B (αActRII) antibody on different ligands. We confirmed published reports showing prevention of ActA-mediated signaling and differentiation blockade. Importantly, however, we also saw that ActRII inhibition could mitigate the effects of multiple BMPs, including both BMP9 and BMP7, albeit with a stronger rescue for BMP7, in line with its known relative binding affinities for type II receptors [33, 36]. While many BMP ligands have been shown to bind and signal through ActRIIA and/or ActRIIB [32, 34, 35], it is surprising that neutralization of just the ActRIIA/B receptors, leaving the BMPR2 unblocked, would have such a significant effect on SMAD1/5 signaling and myoblast differentiation. The partial rescue of the BMP9 effect with selective ActRII inhibition is in line with its higher affinity for BMPR2 relative to either ActRII receptor [33, 36]. However, although BMP7 preferentially binds ActRIIA over BMPR2 [33, 36], it is notable that BMPR2 could not compensate for downstream function—the anti-ActRII antibody significantly reduced BMP7-mediated signaling and alleviated the inhibitory differentiation phenotype on par with the canonical ActRII ligand ActA. These unexpected results suggest that the clinical therapeutic benefit observed with ActRII inhibition may in fact also involve blockade of relevant BMP signaling [16].
Our in vitro data in isolated skeletal muscle cells emphasized the need of transability in vivo to determine the physiological effects of the BMP-SMAD1/5 axis on adult tissue within the skeletal muscle micro-environment. As such, we first asked what the effects were of a BMP ligand when over-expressed systemically. Hepatic overexpression of BMP9 resulted in liver toxicity, body weight loss, and a pro-atrophy gene signature with suppressed mTORC1 signaling in skeletal muscle despite robust SMAD1/5 activation. Given the observed hepatotoxicity with BMP9 hydrodynamic gene delivery, we could not rule out the possibility of indirect effects on tissues secondary to the liver injury. However, our initial findings demonstrated that activated BMP9-SMAD1/5 signaling in skeletal muscle is, at minimum, insufficient to mitigate the pro-wasting phenotype promoted under the observed conditions.
We next sought to confirm effects of local intramuscular BMP administration. This approach allowed us to achieve sustained, localized BMP activity without hepatoxic effects. Prior published work utilized AAV6:BMP7 at 109-2.5 × 1010 vg per IM injection into young juvenile mice (2–8 weeks) [19, 20]—we injected AAV1:BMP7 or AAV1:BMP9 into young fully mature adult mice (13 week) at 1 × 1010 vg. Despite utilizing a similar dose, we showed that intramuscular BMP gene delivery leading to chronic over-expression caused heterotopic ossification with both ligands after 1 month. These results are in line with prior findings [24, 25]and in contrast to the beneficial pro-hypertrophy results observed by others [20].
Taken together, while our study cannot rule out the possibility that BMP signaling may have therapeutic benefits on skeletal muscle when used at very specific, tightly controlled doses, our findings confirm the inhibitory effects of BMPs on myogenic differentiation alongside the osteoinductive and systemic toxicity associated with BMP ligand over-expression. These effects emphasize the difficulty of achieving a therapeutic effect without incurring significant risks even if there is a narrow therapeutic index, as previously claimed [20]. This concern is particularly relevant given SMAD1/5 signaling is often already elevated in muscle disease settings [19], including in sarcopenia from our data, amplifying the relative risk of adverse effects when further activating this pathway therapeutically. In contrast, blockade of GDF8, GDF8 in combination with Activin A, and multiple ligands via the ActRIIA/B pathways, have been shown to be effective in increasing muscle mass across species. The anti-ActRIIA/B antibody demonstrated efficacy while blocking particular BMPs in addition to Myostatin, GDF8 and GDF11, as evidenced by this manuscript.
Methods
Cell culture and treatment
Primary human skeletal muscle derived cells (HUSKMDC or SKDMC; Cook MyoSite) were transduced for stable expression of hTERT (human telomerase) and a temperature-sensitive SV40 large T antigen (SV40). Transduced HUSKMDCs (iHUSKMDCs or iSKMDCs) were cultured in growth medium consisting of MyoTonic Basal Medium with Growth Supplement (Cook MyoSite) and Pen-Strep (Gibco) at a permissive temperature of 33ºC. For experiments, cells were grown in T-75 flasks and switched to a non-permissive 37ºC temperature for five days (Day − 5; D-5) in total. Cells were seeded at D-2 to 6-well or 12-well collagen-coated plates at 1 × 106 or 4.3 × 105 cells/well, respectively, and fed with fresh growth medium at D-1.
To initiate differentiation, confluent D0 myoblasts were washed twice with PBS (Ca+ Mg+) and switched to differentiation medium consisting of serum-free MyoTonic medium (Cook MyoSite) supplemented with 2% horse serum (Gibco). To assess effects on acute signaling, D0 myoblasts were serum starved in MB-2222 alone for 4 h prior to ligand treatment for 30 min. To assess effects on differentiation, ligands were added at the onset of differentiation at D0 in MD-9999 + 2% horse serum and cells were allowed to differentiate for a total of 5 days with a media and fresh ligand change at D2. For molecular profiling, primary human HUSKMDCs were allowed to differentiate for 8 days in MD-9999 + 2% horse serum prior to a 24-hour ligand treatment. For the comparison of transcriptomic signatures of the iHUSKMDC and the primary HUSKMDC myoblasts and myotubes, we differentiated primary HUSKMDC cultures and iHUSKMDC cultures to 4 different time points following a switch to differentiation media: day 0, 4, 7, and 11.
Human SMAD (SMAD1, SMAD5, SMAD2, SMAD3, or SMAD4) expressing stable cells were generated in the transduced iHUSKMDCs using lentiviral expression constructs followed by standard lentiviral vector production and infection protocol [32]. In brief, SMAD1, SMAD5, SMAD2, or SMAD3 cDNA (OriGene) were cloned into the EcoRI-BamHI blunt sites of PLVX-EF1a-IRES-Puro vector. All constructs were verified by Sanger sequencing. Stable cell pools were selected in Cook MyoSite growth media supplemented with 1.0 µg/mL puromycin.
Animal experiments
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of Regeneron and New York Medical College (NY, USA). Male C57BL/6 J mice were purchased from The Jackson Laboratory (USA) and housed in groups of 5 at Regeneron animal holding facilities under specific pathogen free (SPF) conditions, with controlled temperature and light (22℃, 12-h light/12-h dark cycle: lights on at 0600 h/lights off at 1800 h) and with ad libitum access to food (PicoLab Rodent Diet 20, Lab Supply) and water.
For hydrodynamic gene delivery (HDD), mice at 14 weeks of age were randomized into experimental groups by body weight criterion and were subjected to HDD. Control and mouse BMP9 (Gdf2) with an internal myc-myc-His tag constructs were generated in a mammalian expression vector pRG977 and confirmed by DNA sequencing (Regeneron Pharmaceuticals, Tarrytown, NY). On study day 0, mice were injected via tail vein with twenty micrograms (20 µg)/28g mouse of control empty vector or mouse BMP9 plasmids in sterile saline. Seven days following DNA delivery, mice were sacrificed by CO2 asphyxiation and tissues were weighted and collected (flash frozen or fixed for paraffin-embedding).
For intramuscular AAV delivery, we designed rAAV1 vectors encoding either murine BMP9 (Gdf2) or BMP7 (Bmp7) under the CMV promoter, followed by an optimized post-transcriptional regulatory element (oPRE).These vectors were delivered to the tibialis anterior (TA) muscles of 13-week-old adult male mice at a dose similar to prior studies [19]: 1e10 vg in 50 µL of saline buffer. The control vector contained the promoter, an ORF stuffer, and oPRE. The AAV and plasmids were produced by VectorBuilder with the following IDs: pAAVExp]-CMV > ORF_Stuffer: oPRE (Vector ID: VB900151-4351qxj), AAV[Exp]-CMV > mBmp7[NM_007557.3*:oPRE (Vector ID: VB240923-1665xpt), and pAAV[Exp]-CMV> [Bmp9-mmh]: oPRE (Vector ID: VB240920-1558nge). Seven days or twenty-eight days following AAV delivery, mice were sacrificed by CO2 asphyxiation and tissues were weighted and collected (flash frozen or fixed for paraffin-embedding).
For rat studies, male Sprague Dawley (SD) rats were purchased at 3–4 weeks of age from Envigo (Indianapolis, USA) and aged at Envigo under specific pathogen free (SPF) conditions before being imported to New York Medical College (NYMC) animal holding facilities 8–12 weeks prior to tissue collection. At NYMC, rats were housed two per cage at the SPF facility with controlled temperature and light (22℃, 12-h light/12-h dark cycle: lights on at 0600 h/lights off at 1800 h) and with ad libitum access to food (2014 Teklad Global 14% Protein diet, Envigo) and water. Skeletal muscle tissues were collected on the same day between 9am and 1 pm. Prior to tissue collection, rats were anesthetized with 3.5% isoflurane and sacrificed by exsanguination and thoracotomy [43].
Histology and RNAscope
Tissues (livers and muscles) were fixed in 10% normal buffered formalin (NBF) for 24 h followed by 70% ethanol for 24 h. The tissues were then sent to Histoserv, Inc. for paraffin-embedding, sectioning and staining with Hematoxylin and Eosin (H&E). Slides were scanned using the AperioScan at 20X magnification.
RNAscope was performed according to the manufacturer’s protocol using the RNAscope Multiplex Fluorescent reagent kit V2 and the ACD HybEZ II hybridization system. Probes Mm-Bmp7-O1-C1 (#1326831-C1) and Mm-Gdf2-O1-C1 (#1326821-C1) were used. Slides were scanned using the Zeiss AxioScan at 40X magnification.
Biochemicals and antibodies
Information on all reagents and antibodies are listed the Supplementary Table 3. Recombinant human Activin A, GDF8/myostatin, GDF11, BMP2, BMP4, BMP5, BMP6, BMP7, BMP9, BMP10, BMP13, BMP14, long-R3-insulin-like growth factor I (IGF1) and tumor necrosis factor alpha (TNFa) were were sourced from R&D Systems. The anti-ActRIIA/B monoclonal antibody (αActRII) was generated in-house at Regeneron. Stock solutions were prepared in either 4 mM HCl supplemented with bovine serum albumin (BSA) and 5% glycerol, PBS with 0.1% BSA (TNFa), or 100 mM acetic acid (R3-IGF1) as per vendor protocol. The stock solution for the αActRII antibody was prepared in 10 mM histidine, pH 5.8. For Western blotting analysis, primary antibodies against the following were used: phospho-SMAD2 (Ser465/467), phospho-SMAD3 (Ser423/425), phospho-SMAD1/5 (S463/465), total SMAD2/3, total SMAD3, total SMAD1, total SMAD5, phospho-S6 (S240/244), total S6, MuRF1, MAFbx, GAPDH, and vinculin were from Cell Signaling. For secondary antibodies, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Jackson ImmunoResearch were used with either Pierce™ ECL Western Blotting Substrate (Thermo Scientific) or Immobilon Western Chemiluminescent HRP Substrate (Millipore). For immunostaining, primary antibody against myosin heavy chain (anti-MYHC, clone A4.1025, Abcam) and secondary antibody (Alexa Fluor 488 F (AB’)) from Invitrogen were used.
Western blotting
For iHUSKMDCs, cells were washed in ice-cold PBS and lysed in RIPA buffer (Thermo) supplemented with Halt™ protease and phosphatase inhibitor cocktail (Thermo). Lysates were sonicated and centrifuged at 15,000 rpm for 10–15 min at 4 °C. For mouse tissues, frozen samples were pulverized with a mortar and pestle in liquid nitrogen, then homogenized in a Precellys® Cryolys Evolution. Homogenates were rotated for 30 min then centrifuged at 15,000 rpm for 10–15 min at 4 °C, supernatants collected and sonicated. Total protein concentrations were determined using bicinchoninic acid (BCA) method as per manufacturer’s protocol (Thermo). Protein samples were prepared in sample buffer and denatured as per standard protocol. Equal amounts of protein were loaded per lane of 4–12% polyacrylamide Bis-Tris NuPAGE gels, separated by electrophoresis, and transferred onto PVDF membranes. Membranes were blocked in 5% w/v non-fat milk powder in TBST (TBS with 0.1% Tween-20) or Immobilon® Block Chemiluminescent Blocker (Millipore) for 30–60 min. Membranes were incubated in primary antibodies in 5% milk TBST or 1.5% FBS with 0.05% sodium azide in TBS followed by secondary antibodies. Protein bands were visualized using enhanced chemiluminescent detection (Thermo or Millipore) and membranes were imaged by Azure Biosystem. Densitometry analysis was conducted as per standard protocol with Fiji [45].
Immunostaining and imaging for myogenic differentiation assay
Cells were fixed with 4% paraformaldehyde for 15 minutes at room temperature prior to permeabilization with 0.3% Triton-X100 in PBS for 15 minutes. Nonspecific binding was blocked with 10% normal goat serum (Invitrogen) for 1 hour followed by incubation with anti-MyHC (Abcam) diluted in PBS with 1% goat serum and 0.1% Triton-X100 overnight at 4ºC. Cells were subsequently incubated with secondary antibody Alexa Fluor® 488 F (AB’) for 1 h at room temperature. DAPI was added for 15 min at room temperature and cells were stored in PBS prior to imaging. Images were acquired using fluorescence microscopy on an Axio Observer 7 to assess effects of molecules on muscle differentiation.
Percentage of nuclei of each image that were MYHC-positive was quantified in the HALO Image Analysis Platform (v3.6.4134.309, Indica Labs, Albuquerque, New Mexico). Using the CytoNuclear FL v2.0.12 algorithm, nuclei were identified from the DAPI channel. Each nucleus was classified as positive or negative via visually tuned thresholding of the average nuclear MYHC fluorescent signal intensity. Identical analysis settings were applied for all images of each experiment. The resulting markup images were inspected for accuracy in a blinded fashion.
RNA extraction and RT-qPCR
RNA was extracted from cultured transduced iHUSKMDCs or mouse tissues preserved in RNA later using MagMax-96 total RNA isolation kit (Thermo Fisher). The RNA concentration was quantified using NanoDrop Spectrophotometer (NanoDrop Technologies), and the integrity was assessed using Fragment Analyzer (Agilent). RNA samples were reverse transcribed to cDNA using SuperScript IV VILO kit (Thermofisher Scientific). The cDNA was then used for RT-qPCR to determine the expression profile of genes of interest using an ABI Prism 7900 sequence detection system in combination with Taqman® fast PCR Master Mix (Applied Biosystems). Transcript levels were normalized to a geometric mean of 3–10 reference genes that showed STD ≤ 0.5 across samples within an experiment. Fold changes were relative to controls and calculated as 2−ΔΔCT. Supplemental Tables S1 and S2 list specific probes and primers used in this study.
Bulk RNA-seq
RNA was extracted from primary HUSKDMC cells using MagMax-96 total RNA isolation kit (Thermo Fisher). The RNA concentration was quantified using NanoDrop Spectrophotometer (NanoDrop Technologies), and the integrity was assessed using Fragment Analyzer (Agilent). Strand-specific RNA-seq libraries were prepared from 500 ng RNA using KAPA mRNA HyperPrep Kit for Illumina Platforms (Roche). Twelve-cycle PCR was performed to amplify libraries. Sequencing was performed on NovaSeq 6000 (Illumina) using a 2 × 76, paired-end sequencing recipe. Sequencing reads were processed with ArrayStudio RNA-seq pipeline to quantify gene expression levels. For reference, we used hg19 genome reference.
Differential expression analysis
We used DESeq2 to process the raw count without filtering and calculate a gene’s fold change and p value between the vehicle condition and a ligand treated condition. We then filtered out low-expression genes from DESeq2’s output. For each gene, we used DESeq2’s output to determine the condition where the gene had a low mean expression level. If the gene had less than 10 reads in more than 20% of the samples in the higher expressing condition, the gene was excluded. The p values of the remaining genes were adjusted by the Benjamini-Hochberg procedure. Genes were considered differentially expressed if their fold changes are greater than 1.5 and adjusted p value were smaller than 0.05.
Rat gene expression data
We took the age-related genes and fold changes of genes from the original paper [43] without modification. Note that the fold changes had been calculated by edgeR [43] and limma [43], and significant differential expression had been determined by a fold change greater than 1.5 and a Benjamini-Hochberg adjusted p value smaller than 0.05.
Statistical analysis
Data are expressed as mean ± SD. Statistical analyses were performed using GraphPad Prism (v. 9). Unpaired two-tailed Student t-tests were performed for statistical comparisons between two groups with p <.05 being considered statistically significant. One-way ANOVA tests were performed for statistical comparisons between more than two groups with p <.05 being considered statistically significant. Asterisks indicate the following: *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Fig. 1. Ligand differentiation blockade in primary HUSKMDCs, SKDMC gene expression profiles, and signaling in transduced iHUSKMDCs. (A) Primary human myoblast (HUSKMDC) differentiation assay with individual ligands. Myoblasts were differentiated into myotubes with vehicle as negative control versus ActRII or BMP ligands (300 ng/ml). Representative images (n = 3) are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm. (B) Relative expression of activin/GDF/TGFB ligands, BMP ligands, and type I and type II receptors in primary HUSKMDCs and clonal transduced iHUSKMDCs. TPM (transcripts per million) values from RNA sequencing at 4 time points across differentiation (day 0, 4, 7, and 11) for primary and clonal transduced cells. (C) Western blotting analysis for non-SMAD signaling with ActRII or BMP ligands. Human myoblasts oy myotubes (iHUSKMDC) were stimulated with vehicle as a negative control, BMP ligands, or ActRII ligands, for 24 h. Fig. 2. Characterization of stable iHUSKMDC cells, αActRII-mediated BMP blockade, differentiation assay quantification. (A) Differentiation assay in parental iHUSKMDC cells and empty vector control stable line. Parental and stable skeletal muscle (iHUSKMDC) cell lines were differentiated into myotubes. Representative images (n = 3 replicates) are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm. (B) Western blotting analysis for SMAD signaling time course in SMAD over-expression (OE) lines. Western blotting analysis at day 0–5 of differentiation was used to confirm overexpression and signaling with SMAD-selective antibodies. (C) Western blotting analysis of anti-ActRII (αActRII) mediated blockade of BMP signaling in myoblasts. Human myoblasts (iHUSKMDC) were stimulated with vehicle as a negative control, BMP2, or BMP6 for 30 min in the presence of an isotype control antibody or (αActRII). Ligands were used at 10 ng/ml and antibodies at 100 nM. Densitometry analysis by Fiji (Schindelin et al. 2012) are presented. (D and E) Quantification of differentiation assay from SMAD over-expression (OE) cells or ligands plus an isotype control antibody versus αActRII. Differentiation was quantified in the bar graph by evaluating the percentage of nuclei within myotubes that were positively identified using anti-MyHC antibody staining. Asterisks indicate differences between control and treatment groups. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 3. Effects of ActRII and BMP ligands on early induction of osteogenic markers in human myoblasts. qPCR analysis of osteogenic differentiation gene markers in ligand treated myoblasts. Human myoblasts (iHUSKMDCs) were stimulated with vehicle as a negative control or single 300 ng/ml dose of ActA, BMP7, or BMP9 for 24 h with downstream gene expression determined by qPCR (n = 4/treatment). Asterisks indicate differences between control and BMP9 groups. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 4. Validation of hepatic BMP9 following HDD and quadricep pSMAD1/5 signaling correlations. (A) qPCR analysis of BMP9 gene Gdf2 in liver from HDD mice. mRNA amounts for Gdf2 were quantified by real-time quantitative polymerase chain reaction and expression levels were normalized to a geometric mean of reference genes. Control group received DNA with an empty vector (n = 12) and dark gray bar show mRNA levels for Gdf2 genes in the group that received BMP9 DNA (n = 14). Data are mean ± standard deviation (SD) from the mean. Statistical analyses were conducted with an un-paired t-test. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. (B) Correlation analyses for quadricep phosphor-SMAD1/5 signaling. Correlation plots are shown between pSMAD1/5 and MURF1 expression, MAFbx expression, or phospho-S6 expression in western blot by densitometry (relative density) as well as with Myh2 expression by qPCR (relative fold-change). Fig. 5. RNAscope validation at 1 week and liver histology at 1 month following IM AAV delivery. (A) BMP7 and BMP9 RNA over-expression visualized in the muscle by RNAscope. RNAscope of BMP7 (Bmp7) or BMP9 (Bmp9) in tibialis anterior (TA) cross-sections from mice 1-week post-intramuscular AAV delivery. Representative images of n = 4 mice per group are shown, at 4X and/or 20X magnification (total numbers of mice were, n = 11 for AAV: BMP7 and n = 12 for AAV1: control or AAV1:BMP9). DAPI staining for nuclei is in blue and BMP-specific mRNA staining is in pink. (B) H&E staining of liver cross-sections from mice 1-month post-intramuscular AAV delivery. Representative images of n = 3 mice per group are shown, each at 10X magnification. Fig. 6. Densitometry analyses for western blots in Figs. 1 and 2. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012). Fig. 7. Densitometry analyses for western blots in Figs. 4 and 6. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012). Statistical analyses were conducted with a One-way ANOVA (S7A) or unpaired T test (S7B). Data are means ± SD. Asterisks indicate differences with the control. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 8. Densitometry analyses for western blots in Supplemental Fig. 1. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012).
Acknowledgements
We thank Drs. L.S. Schleifer, GD. Yancopoulos and A. Murphy, along with the rest of the Regeneron Community for their support. We would also like thank the Molecular Profiling Core, including Michelle Ascencio, Shuo Li, Nicole Negron, and Min Ni, for their work on the RNA sequencing study. Thanks as well to the entirety of the Aging/Age-Related Disorders Therapeutic Functional Area.
Author contributions
Conceptualization: MAE and DJGContent Refinement: MAE and TS and DJGManuscript Writing: all authors, but primarily MAE and DJGManuscript Editing: all authors, but primarily MAE and DJGIn Vitro work: MAE, YZ, JXIn Vivo work: TS, AG, CMcE, RD, HS, KB, YJ, PMImaging analysis: MGMolecular Profiling: KX, YB.
Funding
This study was funded by Regeneron.
Data Availability
Raw data files and datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Animal studies were approved by the Institutional Animal Care and Use Committee of Regeneron and New York Medical College (NY, USA).
Competing interests
All authors are employees of Regeneron. Some own stock in the company.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Le Grand F, Rudnicki M. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19(6):628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumont N, Bentzinger C, Sincennes M-C, Rudnicki M. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5(3):1027–59. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Hernandez JM, Garcia-Gonzalez EG, Brun CE, Rudnicki MA. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev Biol. 2017;72:10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49(1):59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, McPherron A. Regulation of myostatin activity and muscle growth. PNAS. 2001;98(16):9306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor {beta}-Like Signaling Pathway to Block adipogenesis. Mol Cell Biol. 2003;23(20):7230–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchida K, Nakatani M, Uezumi A, Murakami T, Cui X. Signal Transduction Pathway through activin receptors as a therapeutic target of Musculoskeletal diseases and Cancer. Endocr J. 2008;55(1):11–21. [DOI] [PubMed] [Google Scholar]
- 8.Hinck AP, Mueller TD, Springer TA. Structural Biology and Evolution of the TGF-beta family. Cold Spring Harb Perspect Biol. 2016;8(12). [DOI] [PMC free article] [PubMed]
- 9.Lee SJ, Lehar A, Liu Y, Ly CH, Pham QM, Michaud M, et al. Functional redundancy of type I and type II receptors in the regulation of skeletal muscle growth by myostatin and activin A. Proc Natl Acad Sci U S A. 2020;117(49):30907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83–90. [DOI] [PubMed] [Google Scholar]
- 11.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A. 1997;94(23):12457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R et al. SMAD2 AND 3 TRANSCRIPTION FACTORS CONTROL MUSCLE MASS IN ADULTHOOD. Am J Physiol Cell Physiol. 2009:00104.2009. [DOI] [PubMed]
- 13.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiology-Cell Physiol. 2009;296(6):C1258–70. [DOI] [PubMed] [Google Scholar]
- 14.Brown CW, Li L, Houston-Hawkins DE, Matzuk MM. Activins are critical modulators of growth and survival. Mol Endocrinol. 2003;17(12):2404–17. [DOI] [PubMed] [Google Scholar]
- 15.Latres E, Mastaitis J, Fury W, Miloscio L, Trejos J, Pangilinan J, et al. Activin A more prominently regulates muscle mass in primates than does GDF8. Nat Commun. 2017;8:15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lach-Trifilieff E, Minetti GC, Sheppard K, Ibebunjo C, Feige JN, Hartmann S, et al. An antibody blocking activin type II receptors induces strong skeletal muscle hypertrophy and protects from Atrophy. Mol Cell Biol. 2014;34(4):606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morvan F, Rondeau JM, Zou C, Minetti G, Scheufler C, Scharenberg M, et al. Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy. Proc Natl Acad Sci U S A. 2017;114(47):12448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldin CH, Moustakas A. Signaling receptors for TGF-beta family members. Cold Spring Harb Perspect Biol. 2016;8(8). [DOI] [PMC free article] [PubMed]
- 19.Sartori R, Gregorevic P, Sandri M. TGFbeta and BMP signaling in skeletal muscle: potential significance for muscle-related disease. Trends Endocrinol Metab. 2014;25(9):464–71. [DOI] [PubMed] [Google Scholar]
- 20.Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J Cell Biol. 2013;203(2):345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, et al. BMP signaling controls muscle mass. Nat Genet. 2013;45(11):1309–18. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, Li L, Wu N, Chen W, Hong S, Xu M, et al. BMP9 functions as a negative regulator in the myogenic differentiation of primary mouse myoblasts. Biosci Biotechnol Biochem. 2023;87(11):1255–64. [DOI] [PubMed] [Google Scholar]
- 23.Shore EM. Osteoinductive signals and heterotopic ossification. J Bone Min Res. 2011;26(6):1163–5. [DOI] [PubMed] [Google Scholar]
- 24.Jane JA Jr., Dunford BA, Kron A, Pittman DD, Sasaki T, Li JZ, et al. Ectopic osteogenesis using adenoviral bone morphogenetic protein (BMP)-4 and BMP-6 gene transfer. Mol Ther. 2002;6(4):464–70. [DOI] [PubMed] [Google Scholar]
- 25.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11(17):1312–20. [DOI] [PubMed] [Google Scholar]
- 26.Li JZ, Li H, Dunford B, Holman D, Beres B, Pittman DD, et al. Rat strain differences in the ectopic osteogenic potential of recombinant human BMP adenoviruses. Mol Ther. 2003;8(5):822–9. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z, Chen D, Zhang K, Yu B, Chen X, Meng J. Regulation of myostatin signaling by c-Jun N-terminal kinase in C2C12 cells. Cell Signal. 2007;19(11):2286–95. [DOI] [PubMed] [Google Scholar]
- 28.Tzavlaki K, Moustakas A. TGF-beta signaling. Biomolecules. 2020;10(3). [DOI] [PMC free article] [PubMed]
- 29.Philip B, Lu Z, Gao Y. Regulation of GDF-8 signaling by the p38 MAPK. Cell Signal. 2005;17(3):365–75. [DOI] [PubMed] [Google Scholar]
- 30.Aykul S, Martinez-Hackert E. Transforming growth factor-beta family ligands can function as antagonists by competing for type II receptor binding. J Biol Chem. 2016;291(20):10792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen OE, Wader KF, Hella H, Mylin AK, Turesson I, Nesthus I, et al. Activin A inhibits BMP-signaling by binding ACVR2A and ACVR2B. Cell Commun Signal. 2015;13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbonaro M, Wang K, Huang H, Frleta D, Patel A, Pennington A, et al. IL-6-GP130 signaling protects human hepatocytes against lipid droplet accumulation in humanized liver models. Sci Adv. 2023;9(15):eadf4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012;586(14):1846–59. [DOI] [PubMed] [Google Scholar]
- 34.Lavery K, Swain P, Falb D, Alaoui-Ismaili MH. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem. 2008;283(30):20948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upton PD, Davies RJ, Trembath RC, Morrell NW. Bone morphogenetic protein (BMP) and activin type II receptors balance BMP9 signals mediated by activin receptor-like kinase-1 in human pulmonary artery endothelial cells. J Biol Chem. 2009;284(23):15794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodr V, Machillot P, Migliorini E, Reiser JB, Picart C. High-throughput measurements of bone morphogenetic protein/bone morphogenetic protein receptor interactions using biolayer interferometry. Biointerphases. 2021;16(3):031001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leblanc E, Trensz F, Haroun S, Drouin G, Bergeron E, Penton CM, et al. BMP-9-induced muscle heterotopic ossification requires changes to the skeletal muscle microenvironment. J Bone Min Res. 2011;26(6):1166–77. [DOI] [PubMed] [Google Scholar]
- 38.Trendelenburg A-U, Meyer A, Jacobi C, Feige J, Glass D. TAK-1/p38/NFkappaB signaling inhibits myoblast differentiation by increasing levels of activin A. Skelet Muscle. 2012;2(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C, Grzegorzewski KJ, Barash S, Zhao Q, Schneider H, Wang Q, et al. An integrated functional genomics screening program reveals a role for BMP-9 in glucose homeostasis. Nat Biotechnol. 2003;21(3):294–301. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Ma C, Sun T, Ren L. Potential roles of bone morphogenetic protein-9 in glucose and lipid homeostasis. J Physiol Biochem. 2020;76(4):503–12. [DOI] [PubMed] [Google Scholar]
- 41.Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25(12):1843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shavlakadze T, Morris M, Fang J, Wang SX, Zhu J, Zhou W, et al. Age-related gene expression signature in rats demonstrate early, late, and Linear Transcriptional changes from multiple tissues. Cell Rep. 2019;28(12):3263–e733. [DOI] [PubMed] [Google Scholar]
- 43.Shavlakadze T, Xiong K, Mishra S, McEwen C, Gadi A, Wakai M, et al. Age-related gene expression signatures from limb skeletal muscles and the diaphragm in mice and rats reveal common and species-specific changes. Skelet Muscle. 2023;13(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heymsfield SB, Coleman LA, Miller R, Rooks DS, Laurent D, Petricoul O, et al. Effect of Bimagrumab vs Placebo on Body Fat Mass among adults with type 2 diabetes and obesity: a phase 2 Randomized Clinical Trial. JAMA Netw Open. 2021;4(1):e2033457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Fig. 1. Ligand differentiation blockade in primary HUSKMDCs, SKDMC gene expression profiles, and signaling in transduced iHUSKMDCs. (A) Primary human myoblast (HUSKMDC) differentiation assay with individual ligands. Myoblasts were differentiated into myotubes with vehicle as negative control versus ActRII or BMP ligands (300 ng/ml). Representative images (n = 3) are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm. (B) Relative expression of activin/GDF/TGFB ligands, BMP ligands, and type I and type II receptors in primary HUSKMDCs and clonal transduced iHUSKMDCs. TPM (transcripts per million) values from RNA sequencing at 4 time points across differentiation (day 0, 4, 7, and 11) for primary and clonal transduced cells. (C) Western blotting analysis for non-SMAD signaling with ActRII or BMP ligands. Human myoblasts oy myotubes (iHUSKMDC) were stimulated with vehicle as a negative control, BMP ligands, or ActRII ligands, for 24 h. Fig. 2. Characterization of stable iHUSKMDC cells, αActRII-mediated BMP blockade, differentiation assay quantification. (A) Differentiation assay in parental iHUSKMDC cells and empty vector control stable line. Parental and stable skeletal muscle (iHUSKMDC) cell lines were differentiated into myotubes. Representative images (n = 3 replicates) are shown per treatment, with myotubes (green) identified using anti-MyHC antibody staining and nuclei (blue) using DAPI staining. Scale bar, 100 μm. (B) Western blotting analysis for SMAD signaling time course in SMAD over-expression (OE) lines. Western blotting analysis at day 0–5 of differentiation was used to confirm overexpression and signaling with SMAD-selective antibodies. (C) Western blotting analysis of anti-ActRII (αActRII) mediated blockade of BMP signaling in myoblasts. Human myoblasts (iHUSKMDC) were stimulated with vehicle as a negative control, BMP2, or BMP6 for 30 min in the presence of an isotype control antibody or (αActRII). Ligands were used at 10 ng/ml and antibodies at 100 nM. Densitometry analysis by Fiji (Schindelin et al. 2012) are presented. (D and E) Quantification of differentiation assay from SMAD over-expression (OE) cells or ligands plus an isotype control antibody versus αActRII. Differentiation was quantified in the bar graph by evaluating the percentage of nuclei within myotubes that were positively identified using anti-MyHC antibody staining. Asterisks indicate differences between control and treatment groups. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 3. Effects of ActRII and BMP ligands on early induction of osteogenic markers in human myoblasts. qPCR analysis of osteogenic differentiation gene markers in ligand treated myoblasts. Human myoblasts (iHUSKMDCs) were stimulated with vehicle as a negative control or single 300 ng/ml dose of ActA, BMP7, or BMP9 for 24 h with downstream gene expression determined by qPCR (n = 4/treatment). Asterisks indicate differences between control and BMP9 groups. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 4. Validation of hepatic BMP9 following HDD and quadricep pSMAD1/5 signaling correlations. (A) qPCR analysis of BMP9 gene Gdf2 in liver from HDD mice. mRNA amounts for Gdf2 were quantified by real-time quantitative polymerase chain reaction and expression levels were normalized to a geometric mean of reference genes. Control group received DNA with an empty vector (n = 12) and dark gray bar show mRNA levels for Gdf2 genes in the group that received BMP9 DNA (n = 14). Data are mean ± standard deviation (SD) from the mean. Statistical analyses were conducted with an un-paired t-test. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. (B) Correlation analyses for quadricep phosphor-SMAD1/5 signaling. Correlation plots are shown between pSMAD1/5 and MURF1 expression, MAFbx expression, or phospho-S6 expression in western blot by densitometry (relative density) as well as with Myh2 expression by qPCR (relative fold-change). Fig. 5. RNAscope validation at 1 week and liver histology at 1 month following IM AAV delivery. (A) BMP7 and BMP9 RNA over-expression visualized in the muscle by RNAscope. RNAscope of BMP7 (Bmp7) or BMP9 (Bmp9) in tibialis anterior (TA) cross-sections from mice 1-week post-intramuscular AAV delivery. Representative images of n = 4 mice per group are shown, at 4X and/or 20X magnification (total numbers of mice were, n = 11 for AAV: BMP7 and n = 12 for AAV1: control or AAV1:BMP9). DAPI staining for nuclei is in blue and BMP-specific mRNA staining is in pink. (B) H&E staining of liver cross-sections from mice 1-month post-intramuscular AAV delivery. Representative images of n = 3 mice per group are shown, each at 10X magnification. Fig. 6. Densitometry analyses for western blots in Figs. 1 and 2. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012). Fig. 7. Densitometry analyses for western blots in Figs. 4 and 6. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012). Statistical analyses were conducted with a One-way ANOVA (S7A) or unpaired T test (S7B). Data are means ± SD. Asterisks indicate differences with the control. Statistical analyses were conducted with a One-way ANOVA. *p ≤.05; **p ≤.01; ***p ≤.001; ****p ≤.0001 compared to control. Fig. 8. Densitometry analyses for western blots in Supplemental Fig. 1. Densitometry analysis were conducted by Fiji software (Schindelin et al. 2012).
Data Availability Statement
Raw data files and datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.