Abstract
Multiple alignments among genomes are becoming increasingly prevalent. This trend motivates the development of tools for efficient homology search between a query sequence and a database of multiple alignments. In this paper, we present an algorithm that uses the information implicit in a multiple alignment to dynamically build an index that is weighted most heavily towards the promising regions of the multiple alignment. We have implemented Typhon, a local alignment tool that incorporates our indexing algorithm, which our test results show to be more sensitive than algorithms that index only a sequence. This suggests that when applied on a whole-genome scale, Typhon should provide improved homology searches in time comparable to existing algorithms.
INTRODUCTION
Sequence alignment is certainly one of the most well-developed and pervasive topics of computational molecular biology. Algorithms in this vein are widely used for tasks varying from the comparative analysis of rodent (1–5) and chicken (6) genomes to the construction of networks of protein interactions (7). With the current sequencing of many genomes (8), fast and sensitive sequence alignment algorithms will likely maintain or increase their role in biological research.
As more and more genomic data has become available, algorithms for locally aligning query sequences to genomic databases have become increasingly important (9–13). Because the exact Smith–Waterman (14) algorithm is impractical for large sequences, database search techniques are almost always based upon the paradigm of seeded alignments. The BLAST algorithm (10) was pivotal in popularizing such a technique, and it has since been incorporated into many tools, a few of which are BLASTZ (4), BLAT (13) and Exonerate (15). In such algorithms, a set of seeds is first generated between the database and the query. Each seed is then extended to determine whether it is a part of high scoring local alignment. Extensions typically consist of two phases: first the seed is extended into an un-gapped alignment, and if this alignment scores above a threshold, the seed is then extended with the allowance of gaps. An enhancement to this simple model is to extend only pairs of seeds close to each other (11). Seeds for the BLAST algorithm are traditionally fixed-length words present in both the database and the query, with the word length referred to as the seed's weight. This leads to an inevitable speed/sensitivity trade-off; heavier seeds prune a larger fraction of the search space but miss more alignments than do seeds with a smaller weight.
In recent years, the introduction of spaced seeds has led to significantly improved local alignment algorithms (12,16–20). Spaced seeds allow non-contiguous patterns of matching nucleotides to initiate a local alignment, and algorithms have been developed (17–22) to compute the probability that a seed will be found within an un-gapped alignment of a given length between two sequences. The optimal seed can then be chosen as the seed that maximizes this probability. It is useful to think of un-gapped alignments of homologous regions as being generated by a probabilistic model that specifies the distribution over matches and mismatches (17,20,22). The model outputs a bit string where each position corresponds to a position in the alignment; the bit is 1 if there is a match in the alignment and 0 if there is a mismatch. While higher-order models are possible (19), in this article we will focus on models that output a 1 independently in each position with a fixed probability, which is called the similarity level (12).
In addition to being provably more sensitive than consecutive seeds in some cases (21), spaced seeds allow an important new speed/sensitivity trade-off. Rather than lowering the weight of a seed to boost sensitivity, one can index multiple seeds per position and obtain a linear, rather than exponential, rise in the size of the search space (18,20). Spaced seed design operates under a resource-constrained paradigm (17), where the weight and number of seeds is specified and the goal is to design an optimal set of seeds that fits these constraints.
In this article, we seek to build on these developments by taking advantage of increasing amounts of available genomic data as well as rapidly improving global multiple sequence alignment algorithms (23–26). We predict that in the near future, these trends will lead to the proliferation of genomic databases consisting of multiple alignments. Information implicit in an alignment has been used to aid in a variety of bioinformatics tasks (27–30), and, similarly, one can hope that a multiple alignment can be utilized to improve database search algorithms. Previous research on searching between multiple alignments has concentrated on position specific scoring schemes (11,31,32). PSI-BLAST (11) is the most popular such program; given a query sequence, it builds a multiple alignment, or profile, from a set of high scoring alignments of the query to the database. It then uses the constructed profile to iterate searches for improved sensitivity. Approaches in this vein have been successful, but in this paper, our focus is orthogonal to such techniques.
The problem we tackle is to align a query sequence to a fixed multiple alignment database. As an example, it may be desirable to augment a multiple alignment of mammalian genomes with a newly sequenced mammalian or vertebrate genome. Our approach uses the multiple alignment database to improve search sensitivity over that obtained using only a sequence database. To do this, we extend the resource-constrained paradigm to apply not only to seed design but also to seed allocation; we allow different positions in the database to index different sets of seeds and determine the best way to do so based on the information implicit in the multiple alignment.
We have implemented a local alignment tool, Typhon, which incorporates our indexing algorithm. Tests on real world data shows that Typhon is substantially more sensitive than standard sequence indexing algorithms as well as algorithms that index multiple alignments without using our dynamic indexing methodology. The performance improvement is most dramatic for indexes with a small number of spaced seeds, which is important for large-scale database searches. Source code for Typhon is available under the GNU public license at http://typhon.stanford.edu.
ALGORITHMS
Overview
We are initially given a multiple alignment, a hypothetical query sequence and a phylogenetic tree of all species in the alignment as well as the query. We convert the multiple alignment into a probabilistic profile, where each position in the profile is a tuple of six numbers (ppresent, pA, pC, pT, pG, pid). The first number, ppresent, is the existence probability. It represents the probability that a homologue of the position is present in the query or, equivalently, the probability that the query would align to the position without a gap. The next four values indicate the respective conditional probabilities that the homologous position contains an A, C, G or T, given that there exists a homologous position in the query. The nucleotide with the highest such value is called the consensus character; gaps are ignored when determining this. Note that the consensus character in principle depends on the position of the query in the tree. The final value, pid, is the conditional similarity level (12) of the position given that there exists a homologous position in the query. In other words, it represents the probability that the corresponding position in the query sequence contains the consensus character of the multiple alignment. For the remainder of this paper, we will use the terms profile and probabilistic profile interchangeably.
To begin with, we define several terms. A seed of weight w is a sequence of possibly non-consecutive positions (i1 < ·· < iw); by convention, i1 = 0. The span of a seed is defined as iw − i1 + 1. We define an un-gapped homology h of length l beginning at position p in sequence s and position q in sequence t as two sub-sequences (s[p], …, s[p + l]) and (t[q], …, t[q + l]) that have descended from a common ancestor; one can think of such a homology as a bit string of length l with h[i] = 1 if and only if s[p + i] = t[q + i]. A seed is said to match the homology at offset j if h[j + i1] = 1, …, h[j + iw] = 1, and indexing a seed at position p in a sequence corresponds to recording in the index the presence of (s[p + i1], …, s[p + iw]) at position p. A seed matches a homology if we index the seed at every position in the homology and the seed matches the homology at at least one offset. A set of seeds matches a homology if we index every seed in the set at every position in the homology and at least one seed in the set matches the homology.
We extend these notions to a multiple alignment in a simple manner; homologies are defined to exist between the query and the alignment. An un-gapped homology beginning at position p in the alignment and position q in the query is a set of sub-sequences, one from each species in the alignment as well as the query, that have all descended from a common ancestor. In this case, we define h[i] = 1 if and only if the consensus character at position p + i in the alignment matches the query character at position q + i. Indexing a seed at a position in a multiple alignment corresponds to recording the presence of the string consisting of the consensus characters of the multiple alignment. The notion of a seed matching a homology follows from these definitions. We observe that more complex definitions of homology between a query and an alignment are possible, but we do not address them here.
With these definitions, we can formulate our problem as follows:
Given a probabilistic profile, a set of candidate seeds and a budget B, index a subset of the candidate set at each position in the profile such that the average number of seeds indexed per position of the database does not exceed B. The goal is to maximize the expected number of homologies matched by at least one seed.
Without a budget constraint, this value would obviously be maximized by indexing every seed in the candidate set at every position in the alignment. The value of the budget determines the size of the index and, therefore, the expected number of seed matches; higher values will result in more seed extensions and therefore lead to larger running times. Algorithms that build indexes from sequence databases assign the same set of seeds, with cardinality equal to the budget, to each position in the database. Because we have extra information in the multiple alignment, we can be more flexible. Intuitively, we would like to assign more seeds to positions where this increase is most likely to result in additional detected homologies. We must respect the constraint that the average number of seeds indexed per position does not exceed our budget.
Within a multiple alignment, local rates of conservation vary widely due to both random fluctuations in the number of accumulated mutations as well as differential selective pressures. Both of these effects cause some portions of the multiple alignment to be naturally less likely to contain a match to a homologue in a query sequence. Our algorithm exploits this property to determine how to vary the subset of candidate seeds indexed at each position. Specifically, we use local conservation rates to partition the multiple alignment into regions, which are contiguous blocks of positions whose boundaries reflect changes in the conservation level among species in the alignment (Figure 1).
Figure 1.
Sample region boundaries. Boundaries between the two regions in the multiple alignment reflect the changes in conservation among the species in the alignment. Both (a) and (b) are taken from real data. In the first case, the latter region is more likely to yield alignments to a query sequence, while in the second case, the former region is more likely to yield alignments.
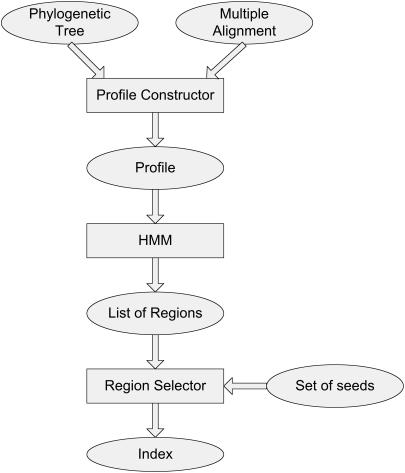
Our approach, then, is to change the set of seeds assigned on a region-by-region basis. By assigning fewer seeds to unpromising regions, we can pay more attention to promising regions and increase sensitivity while still respecting our budget. A high-level outline of our algorithm is shown in Figure 2. First, we convert the multiple alignment into a probabilistic profile. We then use a hidden Markov model to partition the profile into a set of regions where each region gives us the necessary information to evaluate the probability that a homology will exist in that region as well as the probability that a seed will match the homology. Finally, we choose the set of seeds to assign to each position based upon the region to which it belongs. We aim to assign enough seeds to ensure a high probability of finding a match but not too many to waste our budget when it can be used more effectively elsewhere.
Figure 2.
High-level diagram of the Typhon algorithm for indexing a multiple alignment. The overall flow of Typhon consists of three main algorithmic components; above, data is shown in ovals and methods are shown in rectangles. Given a tree and query, the multiple alignment is first converted into a probabilistic profile. Then, the profile is decoded recursively using a simple Hidden Markov Model. Finally, the regions are assigned a set of seeds to index.
We note that in theory one could use a different candidate set of seeds for each position. Such an approach would be particularly useful in cases where highly variable similarity levels strongly influence the optimal shape of the candidate seeds. A typical case is coding exons, where seeds with significant 3-periodicity such as the (0,1,2,8,9,11,12,14,15,17,18) seed (17) have been shown to perform well because they accommodate 3rd-base wobble positions. We do not pursue these options here; rather, we fix the candidate set for all regions and vary only the number of seeds assigned to each position.
Generation of the profile
Our first goal is to convert the alignment into a probabilistic profile. We assume that we are given a phylogenetic tree and the position in the tree at which we expect the query to lie; we root this tree at the query. For each position, we work our way up from the leaves, obtaining ppresent and pN for each nucleotide N ∈ {A, C, T, G} at each node in the tree; we obtain pid only at the root. A leaf has ppresent = 1 if the corresponding sequence is un-gapped at the position in the multiple alignment and 0 otherwise. Furthermore, it has probability pN = 1 for the nucleotide present in the sequence; if ppresent is 0, then we set pN = 0 for all N.
As we work up the tree, we obtain ppresent and pN independently. For each internal node, we obtain pN by applying Felsenstein's algorithm with a Kimura rate matrix (34–36). Since some children can have pN = 0 for each nucleotide, we consider only children for which at least one pN is positive. The task of obtaining ppresent is somewhat more problematic because there is not as well-developed an evolutionary theory for insertions and deletions as there is for nucleotide substitutions. For a node n, our method sets ppresent(n) to be a weighted average ∑i wi × ppresent(ni), where the sum is taken over all children of n. We choose the weight assigned to a child to be proportional to the inverse of the branch length between the parent and the child and normalize the weights sum to one.
When we have reached the root, we choose pid as maxN∈{A, C, T, G}(pN). This is because the maximum pN corresponds to the conditional probability that the consensus character of the multiple alignment is present at the homologous position in the query, given that there is a homologous position in the query. Because at a given position in the multiple alignment we choose the consensus character as the character to record in the index, the average value of pid obtained in such a way over a region will correspond to the conditional similarity level of the alignment of that region to a homologue in the query.
It turns out that reconstructing the profile in this manner results in empirically slightly inaccurate values. In particular, it tends to overestimate actual values of pid, which presents difficulties because small changes in the value of pid can have large effects on the estimated hit probability of a seed (12,18). Furthermore, such an estimate for ppresent is heuristic and is not guaranteed to yield accurate predictions.
A complete treatment of profile reconstruction is beyond the scope of this paper. For our purposes, we plotted the predicted versus experimental values of ppresent and pid for several species and assessed accuracy. To do this, we began with a multiple alignment, removed one species as the test species, and converted the remaining alignment to a probabilistic profile using the method described above. We then grouped the values of ppresent and pid as predicted by the profile into a finite set of buckets, each representing a discrete value.
For each discrete value of ppresent and pid as predicted by the profile, we calculated the experimental values of ppresent and pid for the test species as follows. To obtain the experimental ppresent for a given discrete predicted value, we first counted the total number of positions in the profile at which ppresent was that value. Of those positions, we counted the number of positions that were un-gapped in the test species. The experimental value was then determined as the latter number divided by the former. The experimental values of pid were obtained similarly.
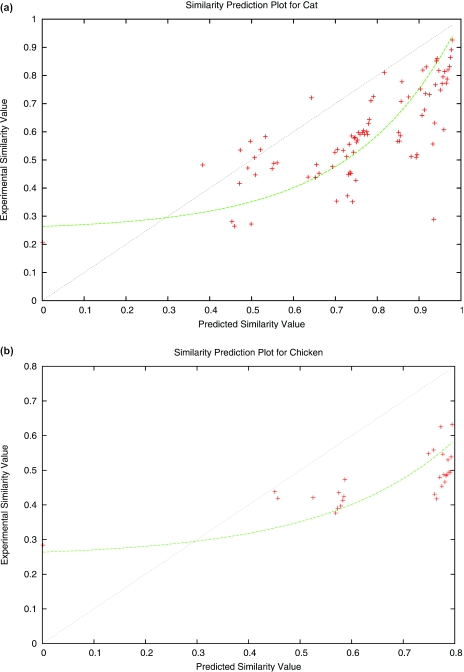
If our predictions were perfectly accurate, resulting plots of experimental versus predicted values would show a linear relationship with slope 1. Plots for ppresent had an extremely high variance and did not appear to follow an obvious pattern, and, based upon these results, we kept our predictions for ppresent unchanged. Improved predictions are likely possible and can only improve the performance of our algorithm; however, they do not appear immediately available. The plots for pid, on the other hand, did appear to follow a pattern; Figure 3 shows sample plots for pid for two different test species, cat and chicken, obtained from an alignment consisting of human, chimp, baboon, dog, cat, pig, cow and chicken. These plots are similar to plots obtained using other species as test cases.
Figure 3.
Plots of predicted versus experimental profile values. For use in correcting predictions of profile values, we plotted predicted versus experimental values of (a) pid for cat and (b) pid for chicken. Although not shown, we examined plots for other species, which are similar. Plots for ppresent did not obey an immediate pattern and thus did not lead us to change our predictions. Each cross represents a plotted data point; shown also is the function we used for converting our initial predictions of pid to our final predictions, as well as the linear fit that would be suitable if our predictions matched the experimental values.
We do not attempt to address any possible theoretical foundations for the above plots here, as that would take us far from our current focus. Currently, our chief concern is only to convert our initial predictions into values that will work well when given as input to our indexing algorithm, and we found that fitting an exponential curve of the form g(x) = αeβ(x) + δ to our data was more than adequate for this purpose in practice. We chose α and δ to fix g(0) = 0.25 and g(1) = 1; this leaves β as an adjustable parameter. Based upon our observations, a value of β = 4 seems to work fairly well for a variety of species, and we fixed this parameter for all of our tests.
Region decomposition
As mentioned above, one advantage of a multiple alignment is that it delineates different regions, each of which can be characterized by a conservation level among the species in the alignment. Therefore, before building an index, our algorithm partitions the probabilistic profile into a set of such regions. For simplicity, we group regions into a finite number of region classes; a region class is a pair of characteristics (, ), which represent typical values of ppresent and pid, respectively, for each region in the region class. All regions in a region class index the same set of seeds.
Partitioning a profile into regions can be done with the aid of a simple Hidden Markov Model, where the states are region classes that emit values of ppresent and pid. Similar ideas have been explored before (19,36); for our purposes here, it is enough that all regions in a region class possess roughly the same properties. It is important that the cardinalities of the region classes be roughly equal so that we have maximum flexibility when assigning seeds; if one region class is enormous, then in order to free enough budget to assign extra seeds to it we must choose to assign fewer seeds to a large number of smaller region classes, which may be undesirable.
We begin this section by considering a conceptually straightforward approach for decomposing a profile in order to introduce the basic ideas of our method. We then describe how our particular approach extends this idea.
Suppose that we build an HMM consisting of states for each region class (, ). Each state emits a position of the profile with values (ppresent, pid) with probability proportional to and transitions to every state other than itself with equal probability. This probability can be chosen to ensure that the optimal Viterbi parse (33) gives no region shorter than a minimum length; this length should be large enough to ensure that every region is at least larger than the span of our seeds, and we found that a minimum region length of 64 works reasonably well for seeds of span ∼20.
Once this HMM has been constructed, each position can be assigned to the region class corresponding to the state that emits it in the optimal parse obtained via the Viterbi algorithm. Region boundaries then occurs between two positions that belong to different region classes.
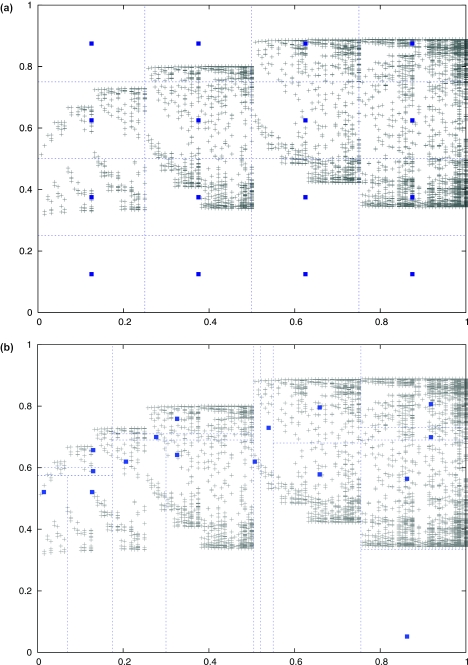
This basic algorithm suffers from the problem that we must determine the set of region classes at the beginning of the algorithm in order to be able to construct the HMM. The weakness of this approach is shown in Figure 4a. If we choose to represent each position in the profile as a point (ppresent, pid), then choosing a set of region classes is conceptually related to partitioning the plane in which the positions lie. All points contained in a rectangle in the partition are closest to the center of a specific region class. By fixing the region classes a priori, we will likely make choices that do not fit the structure of the profile. The chief problem occurs when several region classes are empty and one region class contains many more regions than the others; in this case partitioning achieves little.
Figure 4.
Alternative decomposition of the profile into region classes; region classes can be determined from a profile in several ways. Each cross represents a position in the profile plotted as a point (ppresent, pid). The squares represent the region class values , , and the lines roughly delineate portions of the plane that are closest to a particular region class. (a) When the set of region classes is fixed, the resulting decomposition does not always capture the structure of the profile. In this case, five out of sixteen region classes contain almost no positions and the region class values do not necessarily represent the average profile values of all positions in the region class. (b) An adaptive decomposition can adjust based on the structure of the profile. Here, only one out of sixteen classes contains few positions; the remaining can be distributed to help refine the region of space where most points lie. Furthermore, the region class values more accurately represent the positions in the region class. Note that the goal of this partitioning algorithm is not to cluster points, but to generate a set of region classes that each contain similar number of positions.
An alternative method is to adaptively choose region classes to match the manner in which the positions are distributed, as shown in Figure 4b. One way of doing this is to use k-means clustering (37) and choose region classes corresponding to the resulting clusters. This does not translate directly to our problem, however, as choosing region classes in this manner cannot predict how the profile will actually be decoded by the HMM. Instead, we use a related algorithm that somewhat corrects this problem. This is related to the fundamental idea behind wavelets (38), which can analyze data dynamically by decomposing a signal into pieces that can be represented at different scales of resolution.
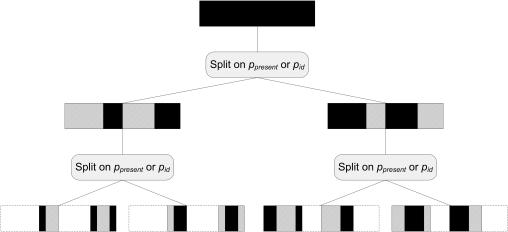
Our algorithm is shown graphically in Figure 5. At a high-level, we progressively split the profile into regions belonging to one of two region classes. We perform the decoding at each stage using an HMM exactly as described previously but consisting of only two states. We then decompose each region class further as long as the number of total region classes is less than the maximum number of region classes.
Figure 5.
High-level outline of our algorithm for decoding the profile. The algorithm we use for decoding the probabilistic profile into a set of regions consists of a series of recursive stages. At each level, we choose to partition the portion of the profile shown in black into two different region classes that differ either in or . We then recursively split each of the two classes until we have partitioned the profile into enough different region classes.
In detail, at each stage we are faced with decomposing a set of regions, all of which belong to the same region class; in the first stage, there is one region consisting of the entire profile. We construct a simple HMM consisting of two states, which correspond to two new region classes; the transition probabilities are set as described above. In principle, we can apply any training algorithm, such as the Baum–Welch or Viterbi training algorithm (33), to learn the emission probabilities of each of the two characteristics and . Once we have determined the values of and for the states in the HMM, we can apply the Viterbi algorithm as previously described to partition the original region class into two region classes. We then decompose each of the two new region classes, stopping when the total number of region classes reaches a user specified limit; based on our experience a set of region classes of cardinality 40 works well in practice.
In practice, learning only the parameter with greater variance before applying the Viterbi algorithm and estimating the other parameter afterwards seems to give slightly better results than learning both parameters at once; this is due in large part to the greater ease in determining the initial parameters of the training algorithm in this case. Such an approach splits region classes more evenly, which is our chief goal in this process.
Seed indexing
Once we have the set of regions and region classes, we can proceed to build the index. Our algorithm must determine for each region which subset of the candidate set of seeds will be indexed. Rather than considering each region individually, we consider each region class individually and index the same subset of seeds for every region in the same region class. When it comes time to actually index the set of seeds that has been assigned to a region class, each region is indexed independently; i.e. we never index positions that cause a seed to span two regions. Our goal is to maximize the expected number of homologies in the multiple alignment matched to a homologue in a query.
We must first infer probable locations of homologies in the alignment, so that such positions can be indexed more aggressively. Our profile is partitioned into a set of regions, not homologies, and we therefore adjust our profile so that maximizing the expected number of regions matched to a homologue in the query approximates the number of homologies matched between the alignment and the query. The change that we must make concerns the length of regions; namely, very long regions are likely to contain more than one homology. In this case, there should be a reward for matching a long region to a homologue that is proportional to the expected number of homologies it contains. In accordance with the idea in (12) that 64 is a good characteristic length for homologies, we split regions of length longer than 64 into a set of contiguous regions of size 64; the last region is allowed to be slightly larger. This enables us to approximate the number of homologies matched within the original region by the number of new regions matched to a homologue.
There is only one technical problem with this approach: having many small regions creates many boundaries between the regions. As mentioned above, we do not index positions that cause seeds to span two regions, and, therefore, having many boundaries can decrease our sensitivity if we are not careful. We can circumvent this problem by allowing seeds to span region boundaries if the regions on both sides of the boundary are part of the same region class. Therefore, for the rest of this section we consider only the problem of maximizing the number of regions in the profile matched to a homologue in the query, with the understanding that this approximates the number of homologies matched between the alignment and the query.
We now begin by describing a general method for solving the seed assignment problem, and we then introduce a greedy approximation that we use in practice. To begin, we determine for each region class and for each number of seeds j the expected number of regions within that class matched to a homologue in the query if every position in that region class indexes the j candidate seeds that optimize the hitting probability for the region class. We will say that a region class is assigned j seeds if every region in that class indexes j seeds. This can be represented as a table with a row corresponding to each region class and a column corresponding to each possible subset size. Entry (i, j) represents the expected number of regions in region class i matched to a homologue if region class i is assigned j seeds.
We compute this table as follows. For a region class (, ), the probability that a region within that class matches a homologue is the probability that a homologue exists multiplied by the conditional probability that a set of seeds matches the region and its homologue, given that the homologue exists. The former probability can be approximated by , which gives the average probability that a typical region in the region class has a homologue in a query. The latter is the hitting probability, phit, which we can compute using dynamic programming. Algorithms for calculating this (18,20) require that we specify a region length over which the seeds will be indexed as well as the similarity level of the region. Because all regions are of approximate size 64, we can specify the same length for every region; furthermore, the characteristic of a region class approximates the similarity level of a typical region in the region class.
We fill in the first column of the table by computing for each region class i the optimal seed. This seed matches any region in the region class to its homologue with approximate conditional probability , given that the homologue exists; the seed therefore matches each region to a homologue with unconditional probability . If the region class has |C| regions, the seed is expected to match regions to their homologues; we fill in entry (i, 1) with this value. Next, we fill in successive columns using the greedy covering procedure described by (20). Briefly, to fill in column j for a region class i, we compute the optimal set of seeds of size j by augmenting the optimal set of seeds of size j − 1 with the seed that optimizes phit given that the previous j − 1 seeds do not match. This set of seeds matches with probability , so we fill in the table entry (i, j) with the value .
Once the table is full, we assign weights and values to each entry. The weight of entry (i, j) is the total length of all of the regions in region class i multiplied by j, the number of seeds corresponding to this entry; this represents the amount of budget we use by assigning j seeds to region class i. The value is the entry in the cell, which represents the expected number of regions in region class i matched to a homologue if j seeds are indexed. In this manner, each entry can be viewed as an object of a certain weight and value. Any set consisting of at most one object from each row in the table specifies a number of seeds to assign to each region class; if entry (i, j) is selected, then region class i is assigned j seeds. The sum of the weights of the objects is equal to the budget used by such an assignment, and the sum of the values of the objects is the expected number of regions matched. Therefore, such a set of objects that has maximum value and total weight less than our budget specifies the optimal set of seeds to index for each region class.
This problem can be solved exactly in time linear in terms of the size of the database by a solution closely related to that of the Knapsack Problem (39). In practice, we use a different algorithm to obtain an approximate solution that is both faster and allows us to avoid computing the entire table of values, which is inefficient since we typically index for many region classes a small subset of the candidate seeds. We define the density of an object as its value divided by its weight and select objects in order of decreasing density, disallowing more than one object from a given row.
In detail, we proceed as follows. In order to choose the set of objects, we begin with a candidate list of objects corresponding only to the first column in the table. At each step in the algorithm, we examine the candidate list and select the object of highest density. If the object we choose corresponds to cell (i, j), then we remove it from the candidate list and add the object corresponding to cell (i, j + 1) to the candidate list. Rather than setting the value of the new object to be the value in cell (i, j + 1) of the table, though, we set its value to be the difference between the values in the cell (i, j + 1) and cell (i, j). Similarly, rather than setting its weight to the total length of all regions in region class i multiplied by the size of the seed set, we set its weight to be merely the total length of the regions.
Viewing this step in terms of seed selection, choosing object (i, j) corresponds to choosing to index j seeds for each region in region class i. The value of the object added to the candidate set corresponds to the additional number of regions matched to a homologue if j + 1 rather than j seeds are indexed for region class i, and its weight is the amount of budget used by indexing one additional seed. This object can only be added after object (i, j) is added because the first j seeds must be indexed before seed j + 1 is indexed. If an object cannot be added because its weight is too high, we remove it from consideration and try to add the object with the next highest density. We stop this process right before we exceed our budget.
The quality of this approximation depends on the relative weights and values of the objects. For instance, if all the objects are all of similar weight, the algorithm will perform well since it is exact in the case that all objects have the same weight. Similarly, if all objects except those with small values can be chosen without exceeding our budget, the greedy algorithm will perform well because it is not that important which subset of low valued objects it chooses to fill out the budget. In our case, we attempt to partition the region classes into roughly equal sizes, which, in practice, is mostly successful. When some region classes are very large, they typically contain regions with many gaps and consequently have low hitting probabilities. Therefore, in practice either objects of similar weight fill our entire budget, or we are faced with choosing between the objects of low values. In both cases our algorithm works adequately.
RESULTS
Assessment
We evaluated our algorithm on real biological data in an attempt to assess its performance. For this purpose, we implemented Typhon, a fully functional local alignment tool that incorporates our indexing algorithm into a framework supporting BLAST-like un-gapped and gapped extensions as well as other options such as extensions of pairs of seeds on the same diagonal (11). Typhon applies our indexing algorithm to a database and then scans a query sequence against the database, looking up every pattern in the candidate set at every position in the query. A beta version of Typhon is available at http://typhon.stanford.edu; this website also contains supplementary information and additional experimental results. We stress that our focus in this paper is on the indexing algorithm; any seed-based alignment tool, such as PatternHunter (12) or BLAST (11), can incorporate the method used in Typhon. Once the index is built, the local aligner implementation is orthogonal.
Past performance evaluations of spaced seed local alignment techniques have focused on sensitivity with respect to the Smith–Waterman algorithm (18). The implication is that the performance of an algorithm is directly correlated with the number of high scoring local alignments that it finds. We believe that this testing methodology is imperfect because it does not attempt to evaluate whether alignments are truly homologous. For example, it is expected to give too much importance to transposable elements and other repeats in a genome. A core principle of Typhon is to incorporate conservation information, which likely indicates homology, to improve alignment algorithms. Therefore, it is desirable to evaluate Typhon based on how many true homologies it can detect between a database and a query.
One means of testing this is to use known homologous annotations of a database and a query for evaluation purposes; however, there is a dearth of such data, with exons being an exception. Unfortunately, increasing our budget does not allow us to align more exons when considering query species closely related to our database, as even low budgets capture almost all exons capable of being aligned.
Therefore, we tested Typhon as follows. We first constructed a multiple alignment database using LAGAN (24) and post-processed it using the refinement techniques of MUSCLE (40). We then chose three species as query species and globally aligned each to the database using LAGAN. Parts of the alignment to which the query aligns without gaps are likely to be true homologies; while this is not always the case, we expect it to be a reasonable approximation of the set of homologies present between the database and the query. To prune obviously bad alignments, we scored all potential un-gapped alignments and kept only those with positive scores; to score alignments, we constructed the consensus sequence from the database and scored the query against that sequence. While this pruning step may in principle remove some alignments that score low overall but have segments that score high, almost all alignments removed in practice are genuinely low scoring and are potential misalignments (data not shown). We kept each remaining un-gapped alignment as a hypothetical homologous alignment (HHA).
We evaluated the performance of Typhon by locally aligning each query to the database and determining how many HHAs were overlapped by at least one resulting local alignment. Besides approximating the number of homologies detected, this performance metric is relevant to global alignment algorithms incorporating an anchoring step (23,25,28,41), as a high number of potentially true anchors helps such aligners. We tested un-gapped extensions as well as gapped extensions for evaluation. The relative performance of the algorithms in all of the tests we ran was unaffected by the extension method used; since un-gapped extensions are somewhat easier to analyze, we report only those results.
The query sequences we used were the CFTR regions of mouse, pig and fugu obtained from (42). For each query we tested how many HHAs were identified, and we tested how many exons were identified for fugu as well. The sequences included in the multiple alignment were also obtained from (42) and consisted of baboon, cat, chicken, chimp, cow, dog, human, mouse, pig and rat; we removed mouse and rat when testing mouse and pig when testing pig. For a phylogenetic tree, we used the tree given in (A. Siepel, 2002, http://www.cse.ucsc.edu/classes/cmps242/Fall02/projects/proj02.html).
We compared the performance of the Typhon indexing algorithm to the existing scheme that simply indexes every position in the database with the same set of seeds (12,17,18,20). We refer to this algorithm as STANDARD. We did not test Typhon against existing algorithms such as PatternHunter (12) or Wu-BLAST (Gish, W., 1996–2004) because, such tests introduce many uncontrolled experimental variables. For instance, each incorporates optimizations that are entirely orthogonal to the indexing algorithm used, which is our chief focus in this paper. Because the code base of STANDARD is identical to that of Typhon, the only variation in our experiments was due to the seeding method used.
Note that when indexing a multiple alignment, STANDARD indexes the consensus sequence, rather than simply indexing the sequence of a certain species. We show below that STANDARD is significantly more sensitive when indexing a multiple alignment than when indexing the closest species in the alignment to the query, which is by itself a significant reason to index multiple alignments rather than individual sequences.
Sensitivity comparison
We tested Typhon versus STANDARD on three query species for varying values of the budget B as described above; the database used in these tests was the full multiple alignment. We used patterns of weight 10 and span 18 as well as patterns of weight 11 and span 19; these choices for pattern weights are based on the tests presented in past research on spaced seeds (12,17–20). To handle low complexity regions and repeats, we pre-processed the query using DUST (R. L. Tatusov and D. J. Lipman, unpublished data) and discarded from the index those k-mers occurring more frequently than 5 SD above the mean. We chose this discarding technique because we found that it significantly speeds up searches with no loss in sensitivity. We used a candidate set of seeds of size 15 for Typhon and designed the set of seeds using Mandala's greedy covering algorithm (20). When partitioning the profile into a set of region classes, Typhon used 40 region classes. Complete specification of the parameters we used for scoring alignments can be found as supplementary information on our website.
For a seed based local aligner, the method used to score alignments is orthogonal to the seeding method. However, the choice can have an impact on performance, particularly in the case of aligning a sequence to a multiple alignment. We focus here on results obtained using consensus scoring, which constructs from the multiple alignment the consensus sequence and then scores hits by standard pairwise alignment scoring. The advantage of this approach is that it allows a comparison of Typhon to sequence to sequence aligners; because the scoring is done between the two sequences, we can use Karlin–Altschul statistics to evaluate the local alignments (43).
The number of HHAs identified by Typhon and STANDARD as the parameter B is varied is shown for each species in Table 1; we also show the number of exons identified for fugu. Note that Typhon permits non-integral values for B. The number of seed extensions is shown as part of our supplementary results to confirm that Typhon obtains no advantage over STANDARD by performing more seed extensions; both algorithms perform roughly the same number of extensions, which as expected scales linearly with B. The total number of alignments found is not shown but was also roughly the same for each method for a given B. This is because many alignments found by STANDARD are potentially poor multiple alignments. For example, many alignments for fugu are gapped in half of the species in the database. Such alignments can score reasonably well with consensus scoring but are unlikely to be truly homologous alignments. Indeed, if sum of pairs scoring is used instead, which is more common when performing multiple alignments, Typhon finds significantly more local alignments than STANDARD. Results are shown as supplementary information on our website.
Table 1.
Sensitivity comparison of indexing algorithms
| Budget | w = 10 | w = 11 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fugu | Mouse HHAs | Pig HHAs | Fugu | Mouse HHAs | Pig HHAs | |||||||||||
| HHAs | Exons | HHAs | Exons | |||||||||||||
| S | T | S | T | S | T | S | T | S | T | S | T | S | T | S | T | |
| 1 | 56 | 68 | 57 | 74 | 2020 | 2137 | 6006 | 5998 | 48 | 64 | 46 | 69 | 1856 | 2060 | 5930 | 5949 |
| 1.5 | – | 69 | – | 75 | – | 2240 | – | 6049 | – | 65 | – | 71 | – | 2158 | – | 6027 |
| 2 | 61 | 69 | 65 | 76 | 2190 | 2259 | 6074 | 6079 | 51 | 65 | 52 | 72 | 2089 | 2208 | 6044 | 6034 |
| 3 | 66 | 69 | 70 | 77 | 2258 | 2295 | 6089 | 6086 | 56 | 67 | 61 | 74 | 2186 | 2248 | 6067 | 6079 |
| 5 | 68 | 69 | 72 | 77 | 2302 | 2325 | 6104 | 6096 | 61 | 67 | 67 | 75 | 2256 | 2286 | 6083 | 6084 |
| 7 | 68 | 69 | 75 | 77 | 2324 | 2339 | 6121 | 6098 | 63 | 67 | 70 | 75 | 2283 | 2312 | 6105 | 6089 |
| 10 | 69 | 69 | 76 | 77 | 2338 | 2346 | 6135 | 6115 | 66 | 67 | 74 | 75 | 2301 | 2320 | 6118 | 6092 |
Shown here are sensitivity results for mouse, fugu and pig using an alignment of baboon, cat, chicken, chimp, cow, dog, human, mouse, pig and rat as a database. For STANDARD and Typhon the number of HHAs overlapped by an alignment is shown for various values of B, the average number of seeds indexed at each position. The columns labeled S show results for STANDARD while the columns labeled T show results for Typhon. For each test the best performing method is shown in bold; we tested seed weights of 10 and 11. HHAs were generated as described in the text by eliminating corresponding annotations with consensus scores <0 There were in total 10 801 HHAs that could potentially be identified for mouse, 852 for fugu and 11 873 for pig. There were 116 exons that could be identified for fugu.
The results indicate that Typhon achieves the highest gains in sensitivity for distant species; it performs best for fugu, where it is more sensitive using a seed weight of 11 for low budgets than STANDARD using a seed weight of 10. It is more sensitive than STANDARD for mouse, although not as dramatically, and it performs the worst on pig, where it performs roughly the same as STANDARD. This suggests that Typhon is most effective for queries that are far away from each species in the multiple alignment. Because pig has a close relative in cow present in the database, the advantage of Typhon is diminished; similarly, its advantage for fugu is greatest because fugu has no close relatives in the alignment.
The reason that Typhon is less sensitive for queries with a close relative in the database is two-fold. First, the close relative is likely to dominate the other species in the multiple alignment, so that the extra information in the multiple alignment that Typhon can use is minimal. Second, indexing multiple patterns does not yield a large benefit because one pattern finds most of the true matches; e.g. in STANDARD, B = 10 identifies 15% more HHAs than B = 1 for mouse but only 2% more for pig. There is simply not much room for improvement; Typhon typically achieves sensitivity gains by skipping a region and reallocating its budget, but for close species skipping a region is very costly while budget reallocation yields minimal gains.
We note that for all species, Typhon's performance advantage is largest when values of B are small. This is important because large values of B are less useful for large-scale genome searches, since searches run slower and sensitivity improvements diminish as B increases. An example of this can be seen by examining the data for fugu; with a seed weight of 11 and budget of 1, Typhon is almost as sensitive as STANDARD using a seed weight of 10 and budget of 3. Despite similar sensitivities, we would expect Typhon to perform ∼12 times fewer seed extensions in this case.
Although consensus scoring allows easy comparison to existing pairwise local aligners, many multiple aligners use sum-of-pairs scoring to score alignments. We therefore also tested Typhon using sum-of-pairs scoring. Complete details of these tests can be found as supplementary information on our website, but we summarize here that with respect to HHAs, Typhon increases its advantage over STANDARD for mouse and fugu queries, while the relative performance of the two methods remains roughly the same with pig. In addition, as mentioned above, Typhon finds significantly more alignments in total than STANDARD. This is encouraging, as high sum of pairs scores indicate similarity to many species, unlike high consensus scores. Complete analysis of scoring schemes in a local aligner is a topic for further research, but our results indicate that Typhon is effective using either consensus, sum or pairs scoring.
Finally we briefly mention an example of how seeds are distributed among region classes. With mouse as a query and a budget of 2, Typhon allocates between 0 and 5 seeds to each region. There is a high correlation between and the number of seeds allocated, while the value has secondary effects. In some cases, lower values of cause more seeds to be allocated even in the face of lower ; once region classes with high values receive a moderate number of seeds, allocating additional seeds leads to diminishing returns. A plot of the number of seeds allocated for each region class is shown as supplementary information on our website.
Analysis of database types
Suppose that we have the genomes of several mammals and we have just sequenced a new species. A common next step might be to align the new species to the sequences we already have. Currently, one way of doing so might be to use a local aligner to align the newly sequenced species to the human sequence, due to the importance and sequence quality of the human genome. Another method might use the closest available relative of the query as a database. However, if we have a reference alignment of the already sequenced mammals, it is worthwhile to ask if we can improve sensitivity by aligning the new species directly to the multiple alignment. In other words, do multiple alignment databases allow search sensitivities greater than those permitted by sequence databases, irrespective of the indexing algorithm used?
To address this question, we tested aligning each query sequence to human as well as to its closest relative using STANDARD; for mouse and fugu it happens that the closest relative in our alignment is human. Because the human sequence is much shorter than the full alignment, the alignment database we used for comparison consisted of the full alignment projected to include only columns un-gapped in human. We tested STANDARD and Typhon on this projected database using consensus scoring to allow direct comparisons to the method that indexes only a sequence. The results are shown in Table 2.
Table 2.
Sensitivity comparison of databases
| Budget | Fugu | Mouse HHAs | Pig HHAs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HHAs | Exons | ||||||||||||
| H | S | T | H | S | T | H | S | T | H | C | S | T | |
| w = 10 | |||||||||||||
| 1 | 57 | 58 | 61 | 54 | 56 | 65 | 2771 | 3151 | 3298 | 7166 | 8651 | 8709 | 8709 |
| 1.5 | – | – | 69 | – | – | 71 | – | – | 3473 | – | – | – | 8715 |
| 2 | 64 | 63 | 71 | 64 | 64 | 74 | 3045 | 3450 | 3528 | 7361 | 8721 | 8751 | 8749 |
| 3 | 69 | 69 | 72 | 69 | 70 | 75 | 3140 | 3573 | 3630 | 7439 | 8748 | 8760 | 8761 |
| 5 | 71 | 71 | 72 | 70 | 72 | 77 | 3262 | 3661 | 3691 | 7499 | 8765 | 8771 | 8772 |
| 7 | 71 | 71 | 72 | 72 | 75 | 77 | 3303 | 3708 | 3719 | 7520 | 8771 | 8778 | 8775 |
| 10 | 72 | 72 | 72 | 74 | 76 | 77 | 3328 | 3727 | 3735 | 7554 | 8775 | 8789 | 8790 |
| w = 11 | |||||||||||||
| 1 | 48 | 49 | 56 | 48 | 47 | 58 | 2522 | 2909 | 3062 | 6755 | 8492 | 8665 | 8656 |
| 1.5 | – | – | 65 | – | – | 64 | – | – | 3297 | – | – | – | 8666 |
| 2 | 52 | 52 | 65 | 54 | 52 | 69 | 2850 | 3286 | 3379 | 6976 | 8576 | 8734 | 8731 |
| 3 | 58 | 59 | 68 | 61 | 61 | 73 | 3013 | 3431 | 3497 | 7145 | 8646 | 8750 | 8749 |
| 5 | 64 | 64 | 69 | 66 | 67 | 75 | 3131 | 3545 | 3591 | 7355 | 8723 | 8759 | 8759 |
| 7 | 66 | 66 | 70 | 68 | 70 | 75 | 3215 | 3610 | 3644 | 7382 | 8735 | 8775 | 8771 |
| 10 | 69 | 69 | 70 | 71 | 74 | 75 | 3271 | 3665 | 3688 | 7405 | 8739 | 8782 | 8773 |
For mouse, fugu and pig, we examined the relative performances of sequence and multiple alignment databases. The multiple alignment used consisted of baboon, cat, chicken, chimp, cow, dog, human, mouse, pig and rat. The sequence database consisted solely of the closest sequence in the alignment to the query, and the multiple alignment database consisted of the multiple alignment projected to remove columns gapped in the closest sequence. Columns labeled H present results using STANDARD with human as the database, C present results using STANDARD with cow as the database, S present results using STANDARD with the projected alignment as the database, and T present results using Typhon with the projected alignment as the database. The testing methodology was the same as that used for tests comparing sensitivity results on the full alignment. There were in total 10 678 HHAs that could potentially be identified for mouse, 848 for fugu and 11 094 for pig.
The first observation we make is that the two methods that index the multiple alignment are indeed more sensitive than the method that indexes only a sequence. This suggests that, regardless of the indexing method used, using a multiple alignment as a database can boost search sensitivity.
We next observe that, in these tests, Typhon maintains most of its performance advantage over STANDARD, particularly in the case of small budgets. Some degradation is expected because the filtering of columns that are gapped in human should have some overlap with the filtering performed by Typhon.
For mouse and pig, the number of identified HHAs in these tests is larger than the number identified in the tests involving the full alignment. This appears to be due to many annotations that are far apart in the full alignment but are joined in the projected alignment. We refrain from stating that if this indicates that it is better to index the projected alignment than the full alignment, as it may be an artifact of the manner in which the alignment is constructed or the quality of the sequences in the alignment. At any rate, an examination of the results for fugu shows that the projected alignment does not always uncover more HHAs.
Performance comparison
To ensure that Typhon is a practical algorithm, we ran some tests to measure its running time relative to STANDARD. The running time consists of two components: time spent building the index and time spent scanning the index. To measure both of these, we recorded the CPU time spent building and scanning the index for the tests using mouse as a query with consensus scoring. We ran two tests with seed weight of 10; one used the human sequence and the projected alignment as databases, and the other used the full alignment as a database. The tests were run on a 2.8 GHz Pentium 4 processor with 2 GB of RAM.
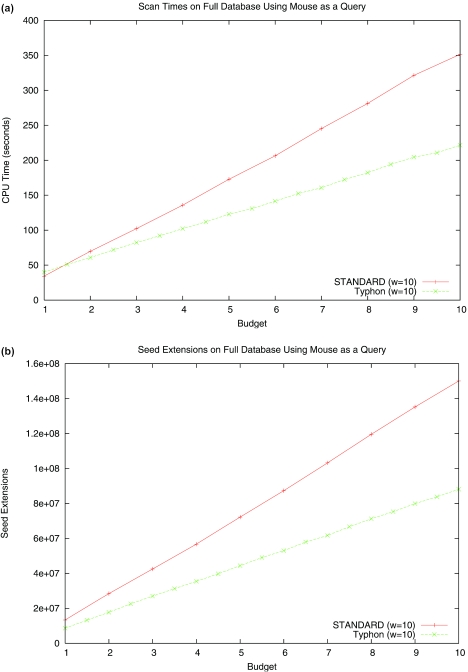
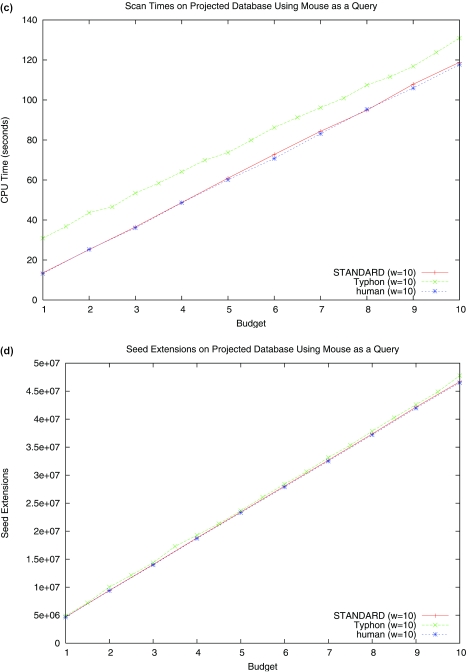
The results of these tests are shown in Figure 6. In the case of the full alignment Typhon runs faster, and in the case of the smaller database STANDARD performs better. As one might expect, these differences are based almost entirely on the different number of seed extensions performed by each algorithm; this is also evident from Figure 6. Running times are therefore extremely data dependent and difficult to gauge accurately from simple tests. Our chief point here is that Typhon does not incur a major performance overhead relative to STANDARD. We also stress that Typhon is still a beta version implementation; careful optimizations are almost certainly possible and will most likely improve the performance of Typhon. In addition, as noted in (20), the overhead of scanning multiple seeds permits substantial parallelization, which may eliminate much of the overhead Typhon faces.
Figure 6.
Running time and seed extension comparison of indexing algorithms. The performance of both Typhon and STANDARD is highly data dependent. Above are plots of scan times as database size is varied. In all tests using alignment databases the alignment consisted of baboon, cat, chicken, chimp, cow, dog, human and pig. We used mouse as a query and seed weights of 10. Tests were run on a 2.8 GHz Pentium 4 processor with 2 GB of RAM. (a) CPU time spent scanning the index and (b) seed extensions performed when using the full alignment (4.2 Mbp) as an index are shown, as well as (c) CPU time spent scanning the index and (d) seed extensions performed when using the projected alignment (1.8 Mbp) as an index. Shown also is performance while scanning a database consisting of solely the human sequence, which is the same length as the projected alignment.
Typhon does take longer to build the index than STANDARD. Our tests showed that the running times of both algorithms scale roughly linearly with the size of the database, with Typhon running ∼3 to 4 times slower. However, for large sequences, build times are typically much less than scan times, which are quadratic in the size of the sequences. Furthermore, it is likely that the cost of building the index can be in some cases amortized over many query sequences, just as can be the cost of designing optimal spaced seeds (12). We note that our index is more query specific than standard spaced seed indexes; such indexes are optimized only for a specific similarity level while our indexes are optimized for a given phylogenetic tree. However, if many queries admit the same tree structure and only alter branch lengths, which will often happen if all are distant from the species in the database (e.g. chicken and fugu queries to mammalian genomes), our index is less restricted. In such cases only the absolute values of pid change; this alters the index to a much lesser extent than do cases in which the relative values of pid or ppresent change across regions.
We finally note that the space requirements of Typhon are identical to STANDARD except for an overhead for each pattern in the candidate set. This overhead is present due to the need for a lookup table for each such pattern; the size of this table is independent of database size. Full performance results are available as supplementary information on our website.
DISCUSSION
In this paper we have argued in favor of using a multiple alignment to improve seeded local alignment techniques. Our results indicate that not only can multiple alignment databases increase search sensitivity irrespective of the search algorithm used, but also they permit improved search algorithms. We have presented an algorithm encapsulated in Typhon, which is a step in this direction.
We believe that our results are particularly relevant given the current trends in comparative genomics. In the near future, a common task will be to search a database of genomes, such as those consisting of mammals or flies, with a query set of sequences of interest. Such queries may contain a newly sequenced mammalian genome or a set of fly genes and may not have a close relative in the database. Our results indicate that instead of indexing a single representative species from the database, it is more effective to directly query an alignment of the database species. Even if our dynamic indexing algorithm is not employed, reconstructing the consensus sequence of the multiple alignment at the point where the query's immediate ancestor meets the phylogenetic tree is likely to improve sensitivity.
As our results show, by indexing a multiple alignment we can obtain sensitivities comparable to existing sequence local aligners while performing many fewer seed extensions. This combined improvement of the sensitivity/speed trade-off can be quite significant, especially across a whole-genome search where local alignment extensions consume the majority of the running time. For instance, suppose we use the sequence of mouse as a query mammal and the sequence of rat is not available in our database. If we use a Typhon-generated alignment database consisting of on average two weight-11 seeds per position, we can perform searches in no more than half the time than can an algorithm that searches a human database consisting of any number of weight-10 seeds per position; this performance improvement comes at no cost with respect to sensitivity.
Because our index is specific to a given query species, our method is most applicable in the case where query sequences are large, such as a newly sequenced whole-genome. In particular, if the tree consisting of the query species and the species in the database changes, we must rebuild the index for optimal performance. However, as mentioned above, if two species cause the tree to change only with respect to branch lengths but not structurally, our algorithm must only reconsider the similarity level between the query and the index. This situation can be common and affects our index much less severely than do cases in which the entire tree structure changes. In future work, it will be useful to examine the exact degree to which the index is affected by non-structural changes in the phylogenetic tree.
Our work also brings to light several directions for future research. Most notably, our method for constructing the probabilistic profile can be formalized and expanded upon. The interplay of the probabilities ppresent and pid decoded from the multiple alignment can have dramatic effects on performance, and improvements in obtaining this information will almost certainly improve the results of our algorithm further. It will be interesting to explore with simulations or real data how these values will change if the species in the multiple alignment are varied. Adjacent to these issues is the question of how best to deal with possible errors in the multiple alignment that may mislead Typhon; it may be possible to detect and correct for such problems with a more flexible algorithm.
In addition, many opportunities exist for algorithmic improvements. First of all, more care in choosing the candidate set of spaced seeds may result in increased sensitivity. It is possible that using higher-order models for seed evaluation as previously suggested (19) may confer a benefit, especially since the multiple alignment may yield information useful in constructing a model of sequence similarity. In addition, it may be beneficial to use the multiple alignment to identify conserved positions in the alignment and treat those differently than less conserved positions; vector seeds (22) may be relevant here due to their ability to weight different positions in a seed differently. This would allow seeds to change on a position-by-position basis, as opposed to a region-by-region basis; an intermediate idea might be to allow the set of candidate seeds to vary on a region-by-region basis, rather than fixing the candidate set for all regions. Finally, our method of indexing a multiple alignment by using only consensus characters is restrictive. For instance, if a position has equal probabilities of 2 nucleotides, it might be beneficial for multiple seeds indexed at that position to incorporate different nucleotides.
Based on the results of this paper, we believe that a complete examination of all issues relevant to the task of indexing a multiple alignment will be fruitful. At this stage in time, our results indicate that Typhon is an attractive alternative to existing local aligners that index sequence databases. Furthermore, the opportunity for future algorithmic developments appears large. We feel that as more sequencing data becomes available, tools for querying multiple alignment databases will become increasingly important.
Acknowledgments
We thank George Asimenos and Antal Novak for helpful discussions and aid with code design, as well as Arend Sidow for helpful discussions. We also thank two anonymous referees for comments that resulted in an improved manuscript. This work was supported in part by NIH grant U01-HG003162. J.F. was supported in part by an SGF fellowship. S.B. acknowledges support from the NSF CAREER Award and the Alfred P. Sloan Fellowship. Funding to pay the Open Access publication charges for this article was provided by NIH grant U01-HG003162.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cooper G.M., Brudno M., Stone E.A., Dubchak I., Batzoglou S., Sidow A. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pevzner P., Tesler G. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 2003;13:37–45. doi: 10.1101/gr.757503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller D. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Hillier L.W., Miller W., Birney E., Warren W., Harsison R.C., Ponting C.P., Bork P., Burt D.W., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 7.Sharan R., Ideker T., Kelley B.P., Shamir R., Karp R.M. Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data. Proceedings of the Eighth Annual International Conference on Computational Molecular Biology (RECOMB); San Diego, CA. New York, NY: ACM Press; 2004. pp. 282–289. [DOI] [PubMed] [Google Scholar]
- 8.The ENCODE Project Consortium. The ENCODE Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 9.Lipman D.J., Pearson W.R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden T.L., Schaeffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma B., Tromp J., Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- 13.Kent W.J. BLAT–The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 15.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown D.G., Li M., Ma B. A tutorial of recent developments in the seeding of local alignment. J. Bioinform. Comput. Biol. 2004;2:819–842. doi: 10.1142/s0219720004000983. [DOI] [PubMed] [Google Scholar]
- 17.Buhler J., Keich U., Sun Y. Designing seeds for similarity search in genomic DNA. Proceedings of the Seventh Annual International Conference on Computational Molecular Biology (RECOMB); Berlin, Germany. New York, NY: ACM Press; 2003. pp. 67–75. [Google Scholar]
- 18.Li M., Ma B., Kisman D., Tromp J. PatternHunter II: highly sensitive and fast homology search. J. Bioinform. Comput. Biol. 2003;2:417–439. doi: 10.1142/s0219720004000661. [DOI] [PubMed] [Google Scholar]
- 19.Brejova B., Brown D.G., Vinar T. Optimal spaced seeds for homologous coding regions. J. Bioinform. Comput. Biol. 2004;1:595–610. doi: 10.1142/s0219720004000326. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y., Buhler J. Designing multiple simultaneous seeds for DNA similarity search. Proceedings of the Eighth Annual International Conference on Computational Molecular Biology (RECOMB); San Diego, CA. New York, NY: ACM Press; 2004. pp. 76–84. [DOI] [PubMed] [Google Scholar]
- 21.Choi K.P., Zhang L. Sensitivity analysis and efficient method for identifying optimal spaced seeds. Journal of Computer and System Sciences. 2004;68:22–40. [Google Scholar]
- 22.Brejova B., Brown D.G., Vinar T. Vector seeds: An extension to spaced seeds allows substantial improvements in sensitivity and specificity. Proceedings of WABI (Workshop on Algorithms in Bioinformatics); Budapest, Hungary: Springer-Verlag; 2003. pp. 39–54. [Google Scholar]
- 23.Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S. NISC Comparative Sequencing Program. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray N., Pachter L. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raphael B., Zhi D., Tang H., Pevzner P. A novel method for multiple alignment of sequences with repeated and shuffled elements. Genome Res. 2004;14:2336–2346. doi: 10.1101/gr.2657504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loots G.G., Ovcharenko I. rVista 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batzoglou S., Pachter L., Mesirov J.P., Berger B., Lander E.S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 30.Simon A., Stone E.A., Sidow A. Inference of functional regions in proteins by quantification of evolutionary constraints. Proc. Natl Acad. Sci. USA. 2002;99:2912–2917. doi: 10.1073/pnas.042692299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gribskov M., McLachlan A.M., Eisenberg D. Profile Analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durbin R., Eddy S., Krogh A., Mitchison G. Biological Sequence Analysis. 1st edn. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 36.Li J., Miller W. Significance of inter-species matches when evolutionary rate varies. Proceedings of the Sixth Annual International Conference on Computational Biology (RECOMB); Washington, DC, USA. New York, NY: ACM Press; 2002. pp. 216–224. [Google Scholar]
- 37.Boggess A., Narcowich F.J. First Course in Wavelets with Fourier Analysis. 1st edn. Upper Saddle River, NJ: Prentice Hall; 2001. [Google Scholar]
- 38.MacQueen J.B. Some methods for classification and analysis of multivariate observations. Berkeley Symposium on Mathematical Statistics and Probability; Berkeley, CA. Berkeley, CA: University of California Press; 1967. pp. 281–297. [Google Scholar]
- 39.Kleinberg J., Tardos E. Algorithm Design. 1st edn. San Francisco, CA: Addison Wesley; 2005. [Google Scholar]
- 40.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delcher A.L., Phillippy A., Carlton J., Salzberg S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., et al. Comparative analyses of multi-species sequencing from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 43.Karlin A., Altschul S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]