Abstract
The relationship between human inherited genomic variations and phenotypic differences has been the focus of much research effort in recent years. These studies benefit from millions of single-nucleotide polymorphism (SNP) records available in public databases, such as dbSNP. The importance of identifying false dbSNP records increases with the growing role played by SNPs in linkage analysis for disease traits. In particular, the emerging understanding of the abundance of DNA and RNA editing calls for a careful distinction between inherited SNPs and somatic DNA and RNA modifications. In order to demonstrate that some of the SNP database records are actually somatic modification, we focus on one type of these modifications, namely A-to-I RNA editing, and present evidence for hundreds of dbSNP records that are actually editing sites. We provide a list of 102 RNA editing sites previously annotated in dbSNP database as SNPs, and experimentally validate seven of these. Interestingly, we show how dbSNP can serve as a starting point to look for new editing sites. Our results, for this particular type of RNA editing, demonstrate the need for a careful analysis of SNP databases in light of the increasing recognition of the significance of somatic sequence modifications.
INTRODUCTION
The genomes of different individuals typically differ in millions of nucleotides, mostly due to genetically inherited single-nucleotide polymorphisms (SNPs). SNPs are extensively studied in search of statistically significant associations between a particular allele of an SNP and certain phenotypes (usually diseases). SNPs associated with a phenotype can be used to pinpoint candidate causative genes, or as genetic markers that alter the risk for disease occurrence, outcome, response to specific treatments and side effects (1). The power of association studies is a function of the number of SNPs used and of their quality (i.e. the likelihood of the SNP locus actually being polymorphic in the population under study).
The largest depository of SNP is dbSNP (2), in which virtually all known SNPs are deposited. Most of the SNPs recorded in dbSNP were found in the course of sequencing the human genome, by algorithmic search for single nucleotide differences between aligned sequence reads of the genomic sequence. This approach has been successful in identifying common SNPs, namely those with a frequency >1–5%, in a diverse panel of individuals representative of different populations. This approach has concentrated on developing a dense map, with uniform coverage across the existing draft of the human genome (1). In addition, many other dbSNP records come from other origins and are of varying accuracy. Sources for erroneous SNP identifications include sequencing errors, mutations and duplications. A recent confirmation study has reported that a large fraction (>40%) of SNPs in these databases could not be confirmed, meaning that they are either of very low frequency, mis-mapped, or not polymorphic at all (3).
In addition, SNPs were identified using expressed data: aligning millions of available expressed sequence tags (ESTs), one can search clusters of ESTs for possible SNPs. Consistent variation between expressed sequences and the human genome was interpreted as genomic SNP, resulting in tens of thousands of dbSNP records in human (4–6). More recently, analyses of full-length human mRNAs have yielded more putative SNPs (7). These methods have yielded only tens of thousands of new SNPs, not a significant number compared with the millions of records in dbSNP. However, their importance lies in the fact that the resulting SNPs have an increased likelihood of residing in a coding region or untranslated region (UTR) of a gene. SNPs in these regions, or generally in regulatory and expressed regions, are considered much more important than those in non-functional regions (i.e. most of the SNPs) that are considered of low probability to contribute to phenotype. Large-scale EST searches for SNPs were also utilized in other organisms, such as rat (8) and Arabidopsis thaliana (9). This method is the most efficient method for the identification of SNPs in organisms that do not have a sequenced genome (10) and was employed to many organisms, e.g. the Bombyx mori silkworm (11).
Recently, much interest has been focused on enzymatic modification of DNA and RNA sequences (DNA/RNA editing), such as cytosine deamination of DNA by AID (12), cytosine deamination of RNA and DNA by the APOBEC family (13,14), and adenosine deamination of RNA by ADARs. It becomes clear that these are much more common than previously believed, but the full scope of these phenomena is yet to be exposed. The abundance of DNA/RNA editing raises the possibility that some of the observed sequence variations are actually DNA/RNA editing sites rather than genetically inherited SNPs. In the following, we explore this possibility in conjunction with one of the better-characterized types of such modification, namely A-to-I RNA editing.
A-to-I RNA editing is the modification of adenosine to inosine in precursor messenger RNAs, catalyzed by members of the double-stranded-RNA (dsRNA) specific ADAR family (15). ADAR-mediated RNA editing is essential for the development and normal life of both invertebrates and vertebrates (16–18). Altered editing patterns were associated with inflammation (19), epilepsy (20), depression (21), amyotrophic lateral sclerosis (22) and malignant gliomas (23). In a few known examples, editing changes the translated protein and its functionality. However, this may not be the primary role of ADARs, as most documented editing events occur within UTRs and other non-coding regions (24). These editing events may affect splicing, RNA localization, RNA stability and translation (25), but full understanding of the role of editing in these regions is yet elusive. Several groups have recently reported the identification of abundant A-to-I editing in human, affecting thousands of genes (26–29). Most of these editing sites reside in Alu elements within UTRs. Alu elements are short interspersed elements, typically 300 nt long, which account for >10% of the human genome (30). The abundance of A-to-I RNA editing sites and the fact that the EST signature of an SNP is virtually the same as the EST signature of an editing site naturally lead to the hypothesis that some of the SNPs predicted by EST data are actually RNA editing sites. In the following, we describe an initial search for editing sites that were deposited in dbSNP as SNPs. We find over a hundred such sites and claim that the actual number is much higher.
MATERIALS AND METHODS
Experimental protocol
Total RNA and genomic DNA (gDNA) were isolated simultaneously from the same tissue sample using TriZol reagent (Invitrogen, Carlsbad, CA). We used tumor and normal samples of lung and oral cavity carcinoma.
The total RNA underwent oligo(dT)-primed reverse transcription using M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA and gDNA (at 20 ng) were used as templates for PCRs. We aimed at high sequencing quality and thus amplified rather short genomic sequences (∼200 nt). The amplified regions chosen for validation were selected only if the fragment to be amplified maps to the genome at a single site. PCRs were carried out using Abgene ReddyMix™ kit (Takara Bio, Shiga, Japan) using the primers and annealing conditions as detailed in the following. PCR fragments were purified from agarose gel using QIAquick Gel Extraction Kit (Qiagen) followed by sequencing using ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
We have used build 119 (January 2004) of dbSNP.
RESULTS
dbSNP (build 119) consists of a total of 6 134 414 non-redundant human RefSNP clusters. Most of these were validated by comparing DNA of different individuals, but for 30 879 clusters the only evidence of polymorphism is mismatches between DNA and expressed data (expressed SNPs). A total of 5 672 327 of the SNPs (92.5%) are a simple single-nucleotide substitution, including virtually all expressed SNPs (30 774; 99.7%).
However, these mismatches between DNA and RNA that were interpreted as expressed SNPs can potentially be not a result of an SNP but rather a signature of DNA or RNA editing. In particular, sequences undergoing A-to-I RNA editing will read G instead of the genomic A, and this could be erroneously identified as an A/G SNP. Although the expressed SNPs are only a small fraction (0.5%) of the total number of SNPs, they are a significant fraction (12%) of SNPs in coding sequences, including 13% of the non-synonym SNPs. Thus, curation of this subset of SNPs is of great importance. In order to test the possibility of editing sites incorrectly reported as SNPs, we checked for over-representation of A/G-expressed SNPs within Alu repetitive elements, in which A-to-I RNA editing is enhanced (26–29).
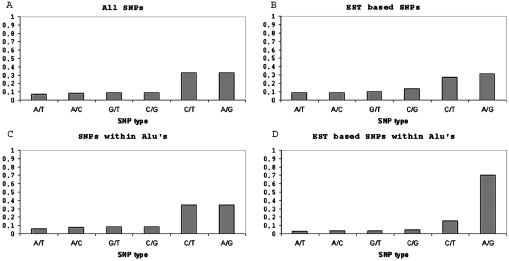
Figure 1 shows the distribution of the different types of simple substitution SNPs. A/G SNPs account for 33% of all single substitution SNPs, and for 35% of single substitution SNPs within Alu repeats. In contrast, A/G-expressed SNPs are highly over-represented in Alu repeats, whereas only 27% of all expressed single-substitution SNPs are of type A/G; 70% of these that reside within an Alu repeat are A/G SNPs (P-value < 10−100). Although in most cases the mismatch type of the expressed SNPs is defined according to the RNA sequence, the annotation of the SNPs from genomic data does not distinguish between strands. Therefore, it might be necessary to look at the statistics of A/G and C/T SNPs combined. These types of SNPs account for 66% of all single substitution SNPs, and for 69% of single-substitution SNPs within Alu repeats. In contrast, A/G- and C/T-expressed SNPs are highly over-represented in Alu repeats, whereas only 59% of all expressed single-substitution SNPs are of type A/G or C/T; 86% of these that reside within an Alu repeat are SNPs of these types (P-value < 10−35). This over-representation of A/G- and C/T-expressed SNPs within Alu elements suggests that ∼20% of the expressed SNPs of these types within Alu elements are actually not SNPs but rather the result of RNA editing.
Figure 1.
Distributions of the different types of simple substitution SNPs. (A) All SNPs; (B) SNPs inferred from expressed data only; (C) SNPs within Alu repetitive elements; (D) SNPs within Alu elements inferred from expressed data only. The enrichment of A/G SNPs in the last panel is attributed to editing sites within Alu elements that were previously interpreted as SNPs.
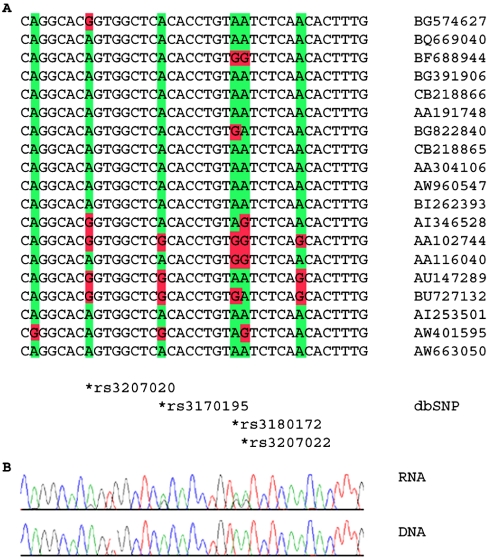
How can one distinguish between an A-to-I editing site and an SNP? There are a number of characteristics of editing that can be used for this purpose: (i) A-to-I editing occurs in dsRNA regions; (ii) A-to-I editing occurs mainly within Alu repeats; (iii) A-to-I editing sites tend to cluster and show a combinatorial nature: different sequences will be edited in different subsets of the cluster. For example, the genomic locus shown in Figure 2 includes five different expressed SNPs that we suspect to be editing sites (we manage to validate four of them in our specimen). The different transcripts presented in the figure exhibit nine different combinations (out of the possible 25 = 32) of adenosines and guanosines in these five sites. Such a combinatorial behavior is not expected for SNPs, since the short distance between the sites does not allow for many recombinations. If one would assume this diversity to follow from genomic diversity, such a large number of haplotypes would require assuming the existence of at least four recombination sites between the five editing sites. However, it is unlikely to have so many recombination sites within such a short genomic region.
Figure 2.
Editing sites in the ribosomal protein S19 (RPS19) locus, previously identified as SNPs. (A) Some of the publicly available expressed sequences that cover this gene, together with the corresponding genomic sequence. The locations of the dbSNP SNP records are indicated at the bottom. The editing location is highlighted in green for non-edited sequences and in red for edited sequences. (B) Experimental results: sequencing matching human DNA and cDNA RNA sequences. Editing is characterized by a trace of guanosine (black) in the cDNA RNA sequence, where the DNA sequence exhibits only adenosine signals (green). We note that the results show that rs3207020, not found in our set, is also an editing site rather than an SNP.
The above characteristics were used in a recently published algorithm to search for RNA editing (26). Here, we used the set of putative editing sites (predicted accuracy > 95%, experimental validation of a random subset shows accuracy of ∼90%) and aligned each predicted editing site against the database of expressed SNPs using the BLAST algorithm. We retained only alignments 90 nt or longer with identity levels higher than 95%. We found 562 expressed SNPs that were mapped on predicted A-to-I editing sites, a list of which is given in Supplementary Table 1. As expected for editing sites, these 562 sites tend to cluster and belong to only 197 different genomic loci. However, as most of these SNPs are located within Alu elements, only 102 of these SNPs have an unambiguous mapping onto the genome in dbSNP. The list of these 102 SNPs is given in Supplementary Table 2. Given the extremely low false-positive rate of the RNA editing database, we expect only a few of these 102 sites to be SNPs after all. For each dbSNP record, the RefSeq sequence onto which the SNP is mapped (if any) and the location within the RefSeq sequence are given. In addition, it is indicated whether the SNP resides within an Alu repeat. Out of the 102 SNPs, 56 are mapped onto a RefSeq sequence—37 of which (66%) are mapped to the UTR of the RefSeq and the remaining 19 (34%) are located within introns of the RefSeq sequence (coming either from splice variants not represented in the RefSeq database, or from pre-mRNA sequences). None of the 102 SNPs is mapped onto RefSeq coding sequences. A total of 96 out of the 102 SNPs in the table (94%) are located within Alu repeats.
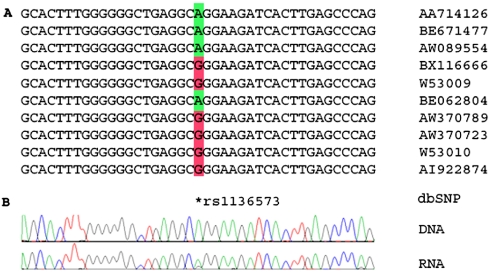
In order to validate our results, we chose four transcripts that contain SNPs from the list of 102 candidates and are relatively easy to sequence, having a long, unique, flanking region out of the Alu in the same exon. We then sequenced PCR products of matching DNA and RNA samples in a number of tissues. The occurrence of editing was determined by the presence of an unambiguous trace of guanosine in positions for which the genomic DNA from the same sample clearly indicated the presence of an adenosine (Figures 2 and 3). All sites tested have been shown to be editing sites and not SNPs or somatic mutations. One of the amplified transcripts included more than 1 SNP in our list, and thus we validated 7 out of the predicted 102 (dbSNP ID numbers: rs1136573, rs3170195, rs3180172, rs3207022, rs3180175, rs3192564 and rs1057026). In addition, these experiments have yielded one more false SNP not present in our list: rs3207020. The results for two of these transcripts are presented in Figures 2 and 3.
Figure 3.
An editing site in the eukaryotic translation initiation factor (eIF3k) locus, previously identified as SNPs. (A) Some of the publicly available expressed sequences, which cover this gene, together with the corresponding genomic sequence. The location of the dbSNP SNP record is indicated at the bottom. The editing location is highlighted in green for non-edited sequences and in red for edited sequences. (B) Experimental results: sequencing matching human DNA and cDNA RNA sequences from the same source. Editing is characterized by a trace of guanosine (black) in the cDNA RNA sequence, where the DNA sequence exhibits only adenosine signals (green).
DISCUSSION
The above analysis relies on a previously published RNA editing database (26). This database consist of more than 12 000 putative editing sites, but the actual number of editing sites in the human genome is probably much higher. Recently, it is was shown by direct sequencing of 3 Mb of human brain cDNA that the average editing rate within intronic and intergenic regions is ∼1:1000 bp, raising the total number of potential editing sites in the genome to over a million.
Accordingly, the number of erroneously assigned EST-based SNPs is probably much higher than the 102 putative sites we found. Indeed, during our experimental validation procedure we found more sites, which were previously annotated as expressed SNPs but actually are editing sites, e.g. the SNP rs3207020 (Figure 2).
The above results demonstrate the effect of one particular type of sequence modification on dbSNP. Similarly, other types of RNA editing in the human transcriptome, such as the C-to-U RNA editing of apoB transcripts by APOBEC-1 (apolipoprotein B mRNA editing catalytic polypeptide 1), could result in erroneously identified SNPs. There are probably many more substrates for this enzyme family than the only one known target, since other members of the family have yet unknown targets (31,32). The possibility of editing events of these types being recorded as EST-based SNPs should be taken into account in future analyses using dbSNP.
Furthermore, dbSNP might be helpful as a starting point for searching new editing targets. Indeed, in a recent work (33) we proposed an algorithm to find novel A-to-I editing sites within the coding sequence and employed it to find four new proteins affected by editing: BLCAP, FLNA, CYFIP2 and IGFBP7. Interestingly, all of the new editing sites found were previously recorded as SNPs in dbSNP (dbSNP IDs: BLCAP, rs11557677; FLNA, rs3179473; CYFIP2, rs3207362; IGFBP7, rs1133243 and rs11555284), even though this fact was not used at all in any stage of the algorithm. All of these presumed SNPs have no evidence for genomic polymorphisms and were included in dbSNP based solely on expressed data. We thus conclude that the erroneously recorded expressed SNPs could serve as a powerful tool in future studies screening for RNA editing sites.
On the other hand, for careful genotyping analyses, one might want to be on the safe side and ignore all SNPs of expressed origin (or at least remove all A/G and C/T SNPs). A less drastic solution would be to use the known properties of editing sites (e.g. they tend to cluster, to appear in dsRNAs and in Alu repeats) and remove only the expressed SNPs that satisfy these properties. Such measures would prevent focusing linkage studies on false SNPs, allowing the finding of more associations between certain disease phenotypes and true SNPs. These considerations are especially important for correct definition of haplotype blocks, which requires accurate sets of SNPs.
DNA editing mechanisms have also attracted much interest recently. Programmed introduction of uracil into DNA is induced by AID through targeted cytosine deamination, thus triggering multiple pathways for somatic modification of antibody genes. The resulting U:G lesion can then be repaired and replicated over, yielding C-to-T and G-to-A transition mutations (34). Similarly, APOBEC3G can edit not only infectious viral DNA, but also endogenous retroelements: it inhibits retrotransposition of IAP and MusD elements in mouse by inducing G-to-A hypermutations in their DNA copies (13). One should bear in mind that most editing enzymes in human have yet no known endogenous target, suggesting that many more editing events are yet to be revealed (14). These DNA editing events could also be misinterpreted for SNPs.
The identification of DNA editing sites among the SNPs poses even a bigger challenge. These sites are modified on the genomic level; therefore, the experimental distinction between these and regular SNPs requires sequencing of DNA from different tissues of the same individual to show that the modification is tissue dependent. From a bioinformatic point of view, better characterization of these sites is yet required in order to design and conduct a systematic search for DNA editing sites. The extensive activity in this emerging field promises to provide such information in the coming years.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Sergey Nemzer, Lital Singer, Shaul Zevin and Compugen's LEADS team for technical assistance, and Harold Smith for many helpful comments on the manuscript. The work of E.Y.L. was performed in partial fulfillment of the requirements for a PhD degree from the Sackler Faculty of Medicine, Tel Aviv University, Israel. E.E. is supported by an Alon fellowship at Tel-Aviv University. Funding to pay the Open Access publication charges for this article was provided by Sheba Cancer Research Center, Tel-Hashomer Israel.
Conflict of interest statement. None declared.
REFERENCES
- 1.Taylor J.G., Choi E.H., Foster C.B., Chanock S.J. Using genetic variation to study human disease. Trends Mol. Med. 2001;7:507–512. doi: 10.1016/s1471-4914(01)02183-9. [DOI] [PubMed] [Google Scholar]
- 2.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang R., Duan J., Windemuth A., Stephens J.C., Judson R., Xu C. Genome-wide evaluation of the public SNP databases. Pharmacogenomics. 2003;4:779–789. doi: 10.1517/phgs.4.6.779.22821. [DOI] [PubMed] [Google Scholar]
- 4.Buetow K.H., Edmonson M.N., Cassidy A.B. Reliable identification of large numbers of candidate SNPs from public EST data. Nature Genet. 1999;21:323–325. doi: 10.1038/6851. [DOI] [PubMed] [Google Scholar]
- 5.Picoult-Newberg L., Ideker T.E., Pohl M.G., Taylor S.L., Donaldson M.A., Nickerson D.A., Boyce-Jacino M. Mining SNPs from EST databases. Genome Res. 1999;9:167–174. [PMC free article] [PubMed] [Google Scholar]
- 6.Irizarry K., Kustanovich V., Li C., Brown N., Nelson S., Wong W., Lee C.J. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nature Genet. 2000;26:233–236. doi: 10.1038/79981. [DOI] [PubMed] [Google Scholar]
- 7.Furey T.S., Diekhans M., Lu Y., Graves T.A., Oddy L., Randall-Maher J., Hillier L.W., Wilson R.K., Haussler D. Analysis of human mRNAs with the reference genome sequence reveals potential errors, polymorphisms, and RNA editing. Genome Res. 2004;14:2034–2040. doi: 10.1101/gr.2467904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guryev V., Berezikov E., Malik R., Plasterk R.H., Cuppen E. Single nucleotide polymorphisms associated with rat expressed sequences. Genome Res. 2004;14:1438–1443. doi: 10.1101/gr.2154304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid K.J., Sorensen T.R., Stracke R., Torjek O., Altmann T., Mitchell-Olds T., Weisshaar B. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 2003;13:1250–1257. doi: 10.1101/gr.728603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevreux B., Pfisterer T., Drescher B., Driesel A.J., Muller W.E., Wetter T., Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng T.C., Xia Q.Y., Qian J.F., Liu C., Lin Y., Zha X.F., Xiang Z.H. Mining single nucleotide polymorphisms from EST data of silkworm, Bombyx mori, inbred strain Dazao. Insect. Biochem. Mol. Biol. 2004;34:523–530. doi: 10.1016/j.ibmb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E.coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 13.Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A.J., Heidmann T., Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 14.Wedekind J.E., Dance G.S., Sowden M.P., Smith H.C. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 15.Polson A.G., Crain P.F., Pomerantz S.C., McCloskey J.A., Bass B.L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991;30:11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- 16.Palladino M.J., Keegan L.P., O'Connell M.A., Reenan R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q., Khillan J., Gadue P., Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 19.Patterson J.B., Samuel C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brusa R., Zimmermann F., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 21.Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 22.Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 23.Maas S., Patt S., Schrey M., Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl Acad. Sci. USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morse D.P., Aruscavage P.J., Bass B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl Acad. Sci. USA. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 27.Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA Editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blow M., Futreal P.A., Wooster R., Stratton M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 32.Begum N.A., Kinoshita K., Kakazu N., Muramatsu M., Nagaoka H., Shinkura R., Biniszkiewicz D., Boyer L.A., Jaenisch R., Honjo T. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- 33.Levanon E.Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nussenzweig M.C., Alt F.W. Antibody diversity: one enzyme to rule them all. Nature Med. 2004;10:1304–1305. doi: 10.1038/nm1204-1304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.