Abstract
Rrs1p, a ribosomal protein L11-binding protein, has an essential role in biogenesis of 60S ribosomal subunits. We obtained conditionally synthetic lethal allele with the rrs1-5 mutation and determined that the mutation is in REX1, which encodes an exonuclease. The highly conserved leucine at 305 was substituted with tryptophan in rex1-1. The rex1-1 allele resulted in 3′-extended 5S rRNA. Polysome analysis revealed that rex1-1 and rrs1-5 caused a synergistic defect in the assembly of 60S ribosomal subunits. In vivo and in vitro binding assays indicate that Rrs1p interacts with the ribosomal protein L5–5S rRNA complex. The rrs1-5 mutation weakens the interaction between Rrs1p with both L5 and L11. These data suggest that the assembly of L5–5S rRNA on 60S ribosomal subunits coordinates with assembly of L11 via Rrs1p.
INTRODUCTION
Eukaryotic ribosomes are synthesized mainly in the nucleolus [for reviews see (1–4)]. Yeast ribosomes consist of 4 rRNAs and 78 ribosomal proteins (RPs). All four rRNAs are encoded by a 9.1 kb rDNA unit, which is tandemly repeated 100–200 times on chromosome XII. Three rRNAs (25S, 18S and 5.8S) are transcribed as a long 35S precursor by RNA polymerase I, whereas 5S rRNA is transcribed by RNA polymerase III as a slightly longer precursor with the 3′-extension (12 nt in Saccharomyces cerevisiae). The 35S pre-rRNA associated with trans-acting factors and RPs forms a large ribonucleoprotein particle, which is subsequently converted into pre-40S and pre-60S ribosomal subunits by cleavage of the pre-rRNA. It has been demonstrated that a number of trans-acting proteins are involved in processing of pre-rRNAs and assembly of ribosomal subunits. However, little is known about how RPs are properly recruited into pre-ribosomal subunits.
Yeast cells consume a large amount of energy in ribosome synthesis, which accounts for 80–90% of total transcription (5). Therefore, ribosome synthesis is regulated in response to environmental changes. In S.cerevisiae, a secretory defect leads to a significant repression of ribosome synthesis (6), suggesting coupling between plasma membrane and ribosome synthesis. RRS1 was identified in a screen for mutations that failed to repress RP genes resulting from a secretion block (7). We demonstrated that Rrs1p is essential for growth, localized in the nucleus with enrichment into the nucleolus, and required for ribosome biogenesis, especially for maturation of 25S rRNA and the assembly of 60S ribosomal subunits (7). Rrs1p depletion leads to the accumulation of 27SB pre-rRNA, suggesting that Rrs1p is required for the processing of 27SB into mature 25S rRNA (8). We also demonstrated that normal function of Rrs1p is required for export of 60S ribosomal subunits from the nucleolus to the cytoplasm (9). Furthermore, we isolated RPL11A encoding ribosomal protein L11 in yeast two-hybrid screening using RRS1 as bait [(10), for a nomenclature of RPs, see (11)]. Ribosomal protein L11 is necessary for the assembly of 60S ribosomal subunits and is localized near the top surface of the central protuberance, where the 60S subunit potentially contacts the 40S subunit (12). We proposed that Rrs1p has a role to recruit L11 to pre-60S subunits. However, it remains unclear how Rrs1p functions in assembly of 60S ribosomal subunits. In order to learn more detailed functions of Rrs1p, in this paper, we have obtained a conditionally synthetic lethal allele with the rrs1-5 mutation and determined that the mutation is in REX1/RNH70/YGR276c. The mutant cells have 3′-extended 5S rRNA as shown previously in the rex1Δ mutant (13). It is known that 5S rRNA is recruited by ribosomal protein L5. Our results suggest that the assembly of L5-5S rRNA on 60S ribosomal subunits coordinates with the assembly of L11. Interestingly, the Escherichia coli homologue of L11 is a 5S rRNA-binding protein. We propose a model for the assembly process of the 60S ribosomal subunit.
MATERIALS AND METHODS
Yeast strains, media and a library
The yeast strains used in this study are listed in Table 1. The conditional rrs1 allele, rrs1-5, was generated by random-PCR mutagenesis of RRS1 (9). Strain 4795-408 (MATa ade2 ade3 leu2 ura3 his7 can1 sap3; a gift from Dr L. Hartwell) was crossed twice with W303-background strain and KM426 (MATa ade2 ade3 leu2 ura3 trp1 can1 rrs1Δ::HIS3 rrs1-5-TRP1 integrated at RRS1 YCp50-RRS1-ADE3) was obtained as a parental strain for mutant screening. Yeast cells were grown in YPD (yeast extract, polypeptone and glucose) rich medium, synthetic complete medium containing 2% glucose (SC) or SC dropout medium, depending on the plasmid markers. A library consisting of partial Sau3A fragments of S.cerevisiae genomic DNA inserted into single-copy yeast vector YCp50, was provided by Dr M. D. Rose (14). Standard techniques were used for yeast manipulation (15).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303 | MATa/MATα his3-11,15/his3-11,15 ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 can1-100/can1-100 | R. Rothstein |
| W303a | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 | R. Rothstein |
| KM148 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1Δ::LEU2 pRS313-HA-RRS1 (ARS/CEN HA-RRS1 HIS3) | (10) |
| KM421 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rrs1Δ::HIS3 rrs1-5-TRP1 integrated at RRS1 | This study |
| KM426 | MATaade2-1 ade3 ura3 leu2 trp1-1 can1 rrs1Δ::HIS3 rrs1-5-TRP1 integrated at RRS1 YCp50-RRS1-ADE3 | This study |
| KM427 | MATα his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rex1-1 | This study |
| KM428 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rex1-1 rrs1Δ::HIS3 rrs1-5-TRP1 integrated at RRS1 | This study |
| KM429 | MATα his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rex1Δ::LEU2 | This study |
| KM430 | MATahis3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rex1Δ::LEU2 rrs1Δ::HIS3 rrs1-5-TRP1 integrated at RRS1 | This study |
| L40 | MATahis3Δ200 ade2 lys2-801am trp1-901 leu2-3,112 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ | R. Sternglanz |
Plasmid construction
YCp50 (CEN URA3)-RRS1-ADE3 was constructed as follows. The 1.6 kb EcoRI–SacI fragment containing RRS1 was cloned into the same sites of YCp50 to generate YCp50-RRS1 [pAT-35; (7)]. The 5.0 kb BamHI–SalI fragment of pDK255 (16) containing ADE3 was cloned into the same sites of pUC19 and the 5.0 kb SacI–SalI fragment of the generated plasmid was cloned into YCp50-RRS1 to make YCp50-RRS1-ADE3. The RRS1 fragment in pRS313 (9) was cloned as a SacI–EcoRI fragment into pRS304 to generate pRS304-RRS1. The RRS1 fragment in pRS304 was cloned as a SacI–XhoI fragment into pRS315 (CEN LEU2) to generate pRS315-RRS1. pRS315-rrs1-5 was similarly constructed. The plasmids for two-hybrid system were constructed by PCR cloning into pBTM116 or pACT2 as described previously (10,17). The plasmid expressing L5-myc (YCplac22-L5-myc) was generated by PCR cloning of an SphI–SalI fragment carrying RPL5 and its upstream promoter region (primers: 5′-TGGGCATGCTCAATACTTTAATAAAATCCAATG and 5′-TTTGTCGACTTGTTGACCAGCCAAAGCAGC) into the CTF vector (provided by Dr D. Kornitzer), YCPlac22 (CEN TRP) containing 9× myc, stop codon and TDH2 terminator, digested with the same enzymes. pGEX-4T-RPL5 and pMAL-C2-RRS1, which encode glutathione S-transferase (GST)-L5 and maltose-binding protein (MBP)-Rrs1p, respectively, were obtained by inserting fragments generated by PCR into the vectors. PCR was performed with KODplus (TOYOBO, Japan) and the entire PCR products were sequenced, and the structures of all plasmids were confirmed by restriction site analysis.
Isolation of mutants
To obtain mutated alleles that cause synthetic lethality with the rrs1-5 allele, 9.2 × 104 cells of strain KM426 containing the plasmid YCp50-RRS1-ADE3 were plated on YPD and subsequently treated with UV at 25–30 J/m2 (viability 20–61%). Plates were incubated at 32°C for 6 days. Colonies showing a red non-sectoring phenotype were isolated and checked for whether they could not grow on 5-fluoroorotic acid (5-FOA) medium at 32°C. Sixteen selected colonies were subsequently transformed with pRS315-RRS1 or pRS315-rrs1-5 to test whether pRS315-RRS1, and not pRS315-rrs1-5 could replace YCp50-RRS1-ADE3 on SC plate containing 5-FOA, and one mutant was obtained. After crossing the mutant with the RRS1 or rrs1-5 strain, tetrad analysis revealed that the allele is derived from single mutation of genomic DNA.
Cloning and sequencing of the REX1 gene
The mutated allele of the chromosomal gene was isolated by PCR. DNA fragments including the open reading frame (ORF) of REX1 were amplified by PCR using a set of primers (5′-CCGTTCTTAAGAGAATGTCAAAG and 5′-AGTAAGGAATCATGGAGGTATGA) and total chromosomal DNA that was isolated from wild-type and m1 mutant cells. The PCR was performed in duplicate and the products of each reaction were independently sequenced.
Gene disruption
A deletion–insertion mutation of REX1 was constructed in the diploid W303. The REX1 gene was subcloned into pUC19 as a 3.1 kb EcoRI–KpnI fragment. NcoI fragments (221–507 amino acids) were deleted from the REX1 gene and a 2.2 kb long SmaI–HpaI fragment containing the LEU2 gene from YEp351 was inserted at SmaI site of the REX1 gene. The resulting plasmid was excised with SacI as a 4.0 kb fragment and used to transform the diploid W303 strain. Leu+ transformants were obtained and correct integration of the rex1::LEU2 gene at the homologous locus was confirmed by PCR. A correct integrant was sporulated and tetrads were dissected to obtain the rex1Δ strain.
Polysome analysis
Yeast crude cell extracts were overlaid on top of 11 ml of a 7–47% (w/v) sucrose gradient and centrifuged for 3.4 h at 35 000 r.p.m. at 4°C in a Hitachi RPS40T rotor as described previously (18). Gradients were collected by pumping up using a peristaltic pump and monitored at 254 nm.
[methyl-3H]Methionine pulse–chase and northern blot analyses
Processing of pre-rRNA was analyzed by [methyl-3H]methionine pulse–chase as described previously (19,20). Total RNA was separated on a denaturing 8% polyacrylamide/8 M urea gel and visualized by ethidium bromide staining or transferred to a Nytran membrane by electroblotting. Northern hybridyzation was carried out using a 32P-labeled oligonucleotide probe, 5′-GGTAGATATGGCCGCAACC for 5S rRNA.
Two-hybrid assays
Two kinds of plasmid for production of lexA binding domain-fusion proteins and Gal4p activation domain-fusion proteins were co-transformed into yeast L40 strain cells as described previously (17). Leu+ Trp+ transformants were selected and 5-fold serial dilutions of the cell cultures were stamped on SC–Leu, Trp, His plates containing 1 mM 3-amino-1, 2, 4-triazole and incubated at 30°C for 3 days unless indicated.
Immunoprecipitation, western and northern analyses
Yeast cells were grown in SC selective media to a mid-log phase, collected by centrifugation, washed twice with ice-cold IP buffer [50 mM Tris–HCl, (pH7.5), 1 mM EDTA, 10% glycerol, 30 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol (DTT), 1 mM phenylmethane sulfonyl fluoride, 1 μg/ml leupeptin and 1 μg/ml pepstatin A], and resuspended in 100 μl of IP buffer. The cells were broken with glass beads by using multi-beads shocker (Yasui Kikai) at 4°C. The homogenates were centrifuged twice at 15 000 r.p.m. for 15 min at 4°C in a micro-centrifuge rotor. The supernatants were incubated with the anti-hemagglutinin (anti-HA, 12CA5; Roche) or anti-myc (9E10; COVANCE) mouse monoclonal antibodies and protein A-Sepharose beads (Sigma) overnight (for western blotting) or for 1.5 h (for northern blotting) at 4°C and then precipitated by centrifugation. The immunoprecipitates for western blotting were washed five times with IP buffer, fractionated by SDS–PAGE, transferred onto Hybond ECL membrane (Amersham Biosciences), and probed separately with the anti-HA or anti-myc antibodies. Horseradish peroxidase-conjugated sheep anti-mouse IgG (NA931; Amersham Biosciences) was used as the secondary antibody. Signals were visualized by Enhanced Chemiluminescence (Amersham Biosciences), according to the manufacturer's instructions. The expression of binding domain (BD)- and activation domain (AD)-fusion proteins was detected by using anti-lexA binding protein antibodies [anti-LexA (2–12): sc-7544; Santa Cruz] and anti-HA antibodies, respectively. RNA was prepared by phenol/chloroform extraction from the immunoprecipitates followed by ethanol precipitation, and was resolved on a denaturing gel before northern blotting.
In vitro interaction of Rrs1p with L5
Expression of GST and MBP fusion proteins in E.coli BL21 was induced by the addition of 0.1 mM IPTG to a culture medium. The cells were cultured for 2 h at 37°C except for GST-L5 expression, for which cells were cultured for 2 h at 25°C. GST and MBP fusion proteins were affinity-purified with glutathione–Sepharose 4B (Amersham Biosciences) and amylose resin beads (New England BioLabs), respectively, according to the manufacturer's instructions. Protein concentration was determined by SDS–PAGE followed by Coomassie blue staining by using BSA as a control. To examine the interaction of the purified proteins in vitro, 75 pmol of GST-L5 was incubated with 75 pmol of MBP-Rrs1p immobilized on amylose resin in 150 μl of reaction mixture [20 mM Tris–HCl (pH 7.5) and 1 mM DTT] overnight at 4°C. After six washes of the resin with the buffer containing 20 mM Tris–HCl (pH 7.5), 10% glycerol, 137 mM NaCl and 1% NP-40, the associated proteins were eluted with 30 μl of elution buffer [20 mM Tris–HCl (pH 7.5), 1 mM DTT and 1% maltose] and analyzed by SDS–PAGE followed by immunoblotting with rabbit anti-GST antibodies (kindly provided by Dr A. Kikuchi) and rabbit anti-MBP antibodies (T. Okada and K. Mizuta, unpublished data). Horseradish peroxidase-conjugate donkey anti-rabbit IgG (NA934; Amersham Biosciences) was used as the secondary antibody. Signals were visualized by Enhanced Chemiluminescence (Amersham Biosciences), according to the manufacturer's instructions.
RESULTS
Isolation of a yeast mutant that exhibits a synthetic growth defect with rrs1-5
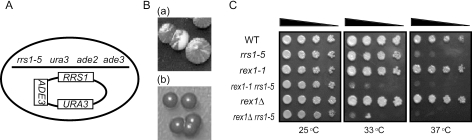
We previously showed that RRS1 encodes a nuclear protein essential for 25S rRNA maturation and 60S ribosomal subunit assembly in S.cerevisiae (7). In order to obtain more details about the function of Rrs1p, we isolated temperature-sensitive rrs1 mutants by random-PCR mutagenesis of RRS1 (9). Here, we tried to isolate alleles that exhibit a synthetic growth defect with rrs1. We selected one of the mutated rrs1 alleles, rrs1-5, because it results in weak temperature sensitivity owing to only one amino acid substitution of L65P. The rrs1-5 cells are not able to grow at 37°C, but grow at 35.5°C on YPD medium, whereas rrs1-84 and rrs1-124, which we previously used for analysis of Rrs1p functions, exhibit severe growth defects at 35.5°C (9). In this study, we screened mutants based on plasmid dependency for growth by both a red and white colony sectoring assay and counter-selection on a medium containing 5-FOA (Figure 1A and B). Parent strain cells were mutagenized by UV irradiation to 20–61% survival. Mutations that conferred synthetic lethality with rrs1-5 at 32°C were identified as red, non-sectoring colonies on plates under non-selective conditions for the plasmid, because growth is dependent on RRS1 in the ADE3-containing plasmid. Among 9.2 × 104 colonies screened, 64 exhibited this phenotype. Among them, 16 colonies were unable to grow on medium containing 5-FOA, a drug that kills cells harboring the URA3 gene. To confirm the synthetic growth defect with rrs1-5, the LEU2-based plasmid containing the RRS1 gene or the rrs1-5 gene was transformed. In only one strain, the plasmid containing the RRS1 gene, but not the rrs1-5 gene could replace the YCp50-RRS1-ADE3 plasmid at 32°C by plasmid shuffling. As the mutant strain without the RRS1 plasmid was obtained on a plate containing 5-FOA at 25°C, it was temporarily designated as m1 mutant. Genetic analysis revealed that a recessive mutation in the genome resulted in the synthetic growth defect in the m1 mutant (data not shown).
Figure 1.
Isolation of a mutant exhibiting a synthetic growth defect with rrs1-5. (A) Schematic representation of the strain for screening mutants. (B) Non-sectoring phenotype of a mutant. Parent strain cells (a) and isolated mutant cells (b) were streaked on YPD media and incubated at 32°C for 6 days. (C) Synthetic growth defect of rrs1-5 with rex1-1 and rex1Δ. W303a (WT), KM421 (rrs1-5), KM427 (rex1-1), KM428 (rex1-1 rrs1-5), KM429 (rex1Δ) and KM430 (rex1Δ rrs1-5) cells were cultured in SC medium at 25°C overnight. The culture was diluted to OD600 = 0.75 and 5-fold serial dilutions were stamped onto YPD media. The cells were cultured at 25, 33 or 37°C.
REX1/RNH70 is mutated in the m1 mutant
In order to isolate a gene complementing the growth defect of the m1 mutant, the strain was transformed with a library of yeast genomic DNA constructed in YCp50 (14) and cultured at 33°C. Of 1.4 × 104 Ura+ transformants, six colonies grew well at 33°C, but not at 37°C on YPD medium, which is similar to the temperature sensitivity of rrs1-5. One of the six plasmids that were isolated from the colonies complemented the temperature sensitivity of the m1 mutant cells, whereas the others not. A partial DNA sequence of the plasmid revealed that the insert DNA corresponds to chromosome VII from 1037596 to 1048620, including YGR271c-A, YGR272c, YGR273c, TAF1, RTT102, REX1, YGR277c and CWC22. Subcloning revealed that the complementing activity was contained in REX1/RNH70/YGR276c (data not shown). In order to confirm that the mutation of REX1 is responsible for the synergistic temperature sensitivity of the m1 mutant, DNA fragments containing the REX1 gene were amplified by PCR using chromosomal DNA extracted from wild-type and the m1 mutant cells. The DNA fragment from wild-type cells, but not that of the m1 mutant cells, could complement the temperature sensitivity of the m1 mutant cells (data not shown). This result confirms that REX1 is mutated in the m1 mutant cells and is responsible for the synthetic defect with rrs1-5.
REX1 encodes an exonuclease consisting of 553 amino acid residues. DNA sequence analysis revealed that the m1 mutant had a point mutation in REX1, a T-to-G conversion within codon 305, which changed a TTG leucine codon to a TGG tryptophan codon. We named the allele rex1-1. The plasmid containing rex1-1 constructed by site-directed mutagenesis could not complement the temperature sensitivity of the m1 mutant cells (data not shown), confirming that rex1-1 is responsible for the synthetic growth defect with rrs1-5. By searching the protein database at DDBJ using the BLAST program, we found many similar sequences from various organisms. Schizosaccharomyces pombe putative exonuclease (NP_594627.1) has similarity to Rex1p throughout the whole sequence. The restricted region from around (196–225) to around (377–384) amino acids in Rex1p is similar to various sequences such as Caenorhabditis elegans exonuclease (NP_504838.1), Drosophila melanogaster LD30051p (AAN71248.1), Mus musculus similar to RIKEN cDNA 1700021p10 (XP_14328.2) and Homo sapiens exonuclease GOR (NP_258439.1). An alignment of the conserved region (225–377 amino acids in Rex1p) reveals that leucine 305, the mutated amino acid in rex1-1, is conserved in the middle of this region (data not shown).
REX1 is not required for cell growth when RRS1 functions normally
After crossing the m1 mutant with a wild-type strain, we obtained a rex1-1 RRS1 strain. This strain grows well at 25, 33 and 37°C whereas the rex1-1 rrs1-5 strain shows a severe temperature sensitivity (Figure 1C). As the rex1-1 mutation exhibited synthetic sick phenotype with rrs1-5, we determined the effect of the rex1-null mutation on the synthetic phenotype. A disruptant of REX1, rex1Δ grows well at 25, 33 or 37°C similar to rex1-1 and rex1Δ has a similar synthetic growth defect with rrs1-5 (Figure 1C). Furthermore, rex1Δ exhibits a synthetic growth defect with other rrs1 alleles, rrs1-84 and rrs1-124 (data not shown). These data indicate that REX1 is not required for cell growth, but normal function of REX1 is required in the background of rrs1.
rex1-1 leads to extended 5S rRNA
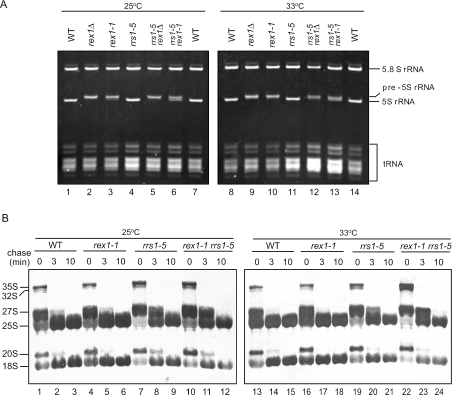
Previously it was reported that Rex1p has a role in maturation of 5S rRNA and that rex1Δ results in accumulation of3′-extended 5S rRNA by ∼3 nt (13). Consistent with this observation, the rex1Δ strain accumulates 5S rRNA that is longer than the corresponding RNA in wild type (Figure 2A). The rex1-1 strain also accumulates extended 5S rRNA with the same length as in the rex1Δ strain (Figure 2A). On the other hand, 5.8S rRNA is normally produced in both the rex1-1 and rex1Δ strains (Figure 2A). In the rex1-1 rrs1-5 strain, extended 5S rRNA appears to be partially trimmed compared with that in the rex1-1 strain and the rex1Δ strain at both 25 and 33°C (Figure 2A). On the other hand, in the rex1Δ rrs1-5 strain cells, such trimming is not detected, and 5S rRNA appears to have similar mobility to that in the rex1Δ strain (Figure 2A). These results indicate that the rex1-1 strain cells have very weak activity to trim the 3′-extension of 5S rRNA, which is detected only in the background of rrs1-5.
Figure 2.
(A) Extended 5S rRNA in rex1-1 and rex1Δ. W303a (WT), KM429 (rex1Δ), KM427 (rex1-1), KM421 (rrs1-5), KM430 (rex1Δ rrs1-5) and KM428 (rex1-1 rrs1-5) strain cells were grown at 25°C and one-half of the cultures were shifted to 33°C and maintained at that temperature for 5 h. Total RNA (10 μg each) prepared from the cell extract was separated on an 8% polyacrylamide/8 M urea gel. RNA was stained with ethidium bromide. (B) [methyl-3H]Methionine pulse–chase analysis. W303a (WT), KM427 (rex1-1), KM421 (rrs1-5) and KM428 (rex1-1 rrs1-5) strain cells were grown in SC–Met medium at 25°C and one-half of the cultures were shifted to 33°C and maintained at that temperature for 5 h. Each culture was pulsed with [methyl-3H]methionine (10 μCi/ml) for 3 min and chased with non-radioactive methionine (500 μg/ml). Samples were taken at the time of addition of non-radioactive methionine (t = 0) and after a chase time of 3 or 10 min to prepare total RNA. RNA (8 μg) was analyzed by electrophoresis and blotted onto Nytran membrane. The membrane was sprayed with En3Hance (PerkinElmer Life Sciences) and exposed to a film.
In order to examine whether rex1-1 affects processing of 35S pre-rRNA, we performed a [methyl-3H]methionine pulse–chase analysis (newly synthesized pre-rRNA is methylated immediately). Figure 2B shows that rex1-1 does not cause a defect in maturation of 25S or 18S rRNA (lanes 1–6 and 13–18). This suggests that the 3′-extended 5S rRNA has little effect on processing of 35S pre-rRNA to mature RNAs. On the other hand, rrs1-5 results in a slight defect in maturation of 25S rRNA at both 33 and 25°C (Figure 2B, lanes 7–9 and 19–21). In the rex1-1 rrs1-5 mutant, more accumulation of the 35S and 32S pre-rRNAs and slower processing of 27S to 25S were observed at 33°C (Figure 2B, lanes 22–24) compared with the rrs1-5 mutant (Figure 2B, lanes 19–21).
rex1-1 and rrs1-5 show a synthetic defect in ribosome biogenesis
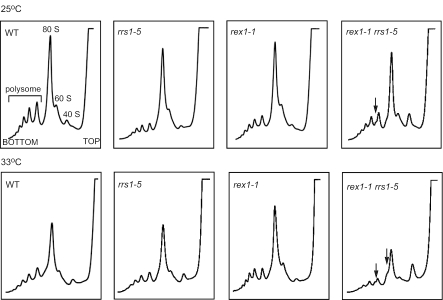
As both Rex1p and Rrs1p are involved in biogenesis of 60S ribosomal subunits, we analyzed polysome profiles by sucrose density gradient ultracentrifugation (Figure 3). Five hours after shifting the rex1-1 rrs1-5 strain cells from 25 to 33°C, 40S subunits accumulated, 60S subunits, 80S monosomes and polysomes decreased and half-mer polysomes, which contain 43S initiation complexes stalled at the AUG start codon, appeared. This result indicates that the rex1-1 rrs1-5 strain is defective in biogenesis of 60S ribosomal subunits at 33°C. Even at 25°C, half-mer polysomes appear in the rex1-1 rrs1-5 strain. However, both the rex1-1 strain and the rrs1-5 strain appear to produce 60S subunits at 25 and at 33°C. This is because rrs1-5 is a weak temperature sensitive allele, although it causes a defect in biogenesis of 60S ribosomal subunits at 37°C (data not shown). These results indicate that rex1-1 and rrs1-5 cause a synthetic defect in assembly of 60S ribosomal subunits. Disruption of REX1 exhibited similar effects on polysome profiles to that of rex1-1; the rex1Δ rrs1-5 strain was defective in biogenesis of 60S ribosomal subunits at 33°C, whereas the rex1Δ strain appears to produce 60S subunits normally (data not shown).
Figure 3.
A synthetic defect of rex1-1 and rrs1-5 in biogenesis of 60S ribosomal subunits. W303a (WT), KM421 (rrs1-5), KM427 (rex1-1) and KM428 (rex1-1 rrs1-5) strain cells were grown at 25°C overnight. One-half of the cultures were shifted to 33°C and maintained at that temperature for 5 h. Cell extracts were used for polysome analysis by sucrose density gradient ultracentrifugation. Ribosomal profiles were determined by OD254 measurement of the gradient (7–47%) fractions. Arrows indicate half-mer polysomes.
Rrs1p interacts with L5/5S rRNA
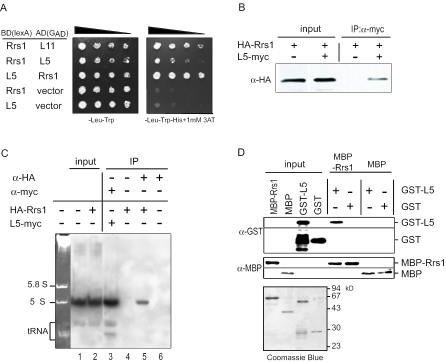
During the cloning of the gene that complements the temperature sensitivity of the m1 mutant, RPL5 encoding ribosomal protein L5 was isolated as a weak suppressor (data not shown). The suppressor effect is assumed to be due to increased expression of L5. The m1 mutant had no mutated nucleotide in the ORF of the RPL5 gene (data not shown). Since L5 is known as a 5S rRNA-binding protein, we examined whether Rrs1p interacts with L5/5S rRNA. In a yeast two-hybrid system, interaction between Rrs1p and L5 is indicated by growth on a plate without histidine containing 3-amino-1,2,4-triazole (Figure 4A) and by increased β-galactosidase activity (data not shown). To further investigate the interaction between Rrs1p and L5/5S rRNA, we performed immunoprecipitation followed by western and northern analyses. As shown in Figure 4B, HA-Rrs1p co-purified with L5-myc. Northern analysis following immunoprecipitation revealed that 5S rRNA co-purified with HA-Rrs1p (Figure 4C). These results indicate that Rrs1p interacts with the L5–5S rRNA complex. To further confirm a direct interaction, we performed an in vitro pull-down experiment. GST-L5, GST, MBP-Rrs1p and MBP were produced in E.coli and purified. GST-L5 or GST was incubated with MBP-Rrs1p or MBP immobilized on amylose resin. GST-L5 was precipitated with MBP-Rrs1p but not with MBP and GST was not precipitated with MBP-Rrs1p (Figure 4D). The result indicates that Rrs1p binds directly to L5.
Figure 4.
Interaction of Rrs1p with ribosomal protein L5. (A) Rrs1p has a two-hybrid interaction with L5. Protein–protein interactions were analyzed by yeast two-hybrid assay. (B) HA-Rrs1p is co-immunoprecipitated with L5-myc. KM148 strain cells expressing HA-Rrs1p with or without YCplac22-L5-myc expressing L5-myc. The cell extracts were immunoprecipitated with the anti-myc antibody, and the immunoprecipitates were probed with the anti-HA antibodies. (C) 5S rRNA is co-immunoprecipitated with Rrs1p. KM148 strain cells expressing HA-Rrs1p (lanes 2, 4 and 5) and W303a with (lane 3) or without (lanes 1 and 6) YCplac22-L5-myc expressing L5-myc were cultured. The cell extracts were immunoprecipitated with the anti-myc antibodies (lane 3) or anti-HA antibodies (lanes 5 and 6) and the immunoprecipitates were used for northern blotting analysis to detect 5S rRNA. Ethidium bromide staining of RNA (2.5 μg) extracted from wild type cells is shown at left. (D) Direct interaction of Rrs1p with L5. GST-L5 or GST was incubated with MBP-Rrs1p or MBP immobilized on amylose resin. The resin was precipitated by centrifugation, and the precipitates were probed with the anti-GST and anti-MBP antibodies. Coomassie brilliant blue stainings of MBP-Rrs1p, MBP, GST-L5 and GST used in this experiment are shown below.
The rrs1-5 mutation in Rrs1p weakens the interaction of Rrs1p with both L5 and L11
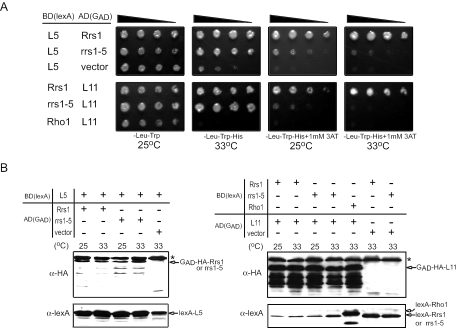
In order to test the effect of the L65P mutation of Rrs1p on L5 and L11 interactions, we created the L65P mutation in the lexA BD-Rrs1p and Gal4p AD-Rrs1p. The apparent strength of interaction was changed depending on the BD-fusion protein and AD-fusion protein; interaction of AD-Rrs1p with BD-L5 appears to be stronger than that of BD-Rrs1p with AD-L5, whereas interaction of BD-Rrs1p and AD-L11 appears to be stronger than that of AD-Rrs1p and BD-L11. Thus, to examine the effect of L65P mutation in Rrs1p, two sets of interactions, AD-Rrs1p/BD-L5 and BD-Rrs1p/AD-L11, were determined. As shown in Figure 5A, the L65P mutation in AD-Rrs1p and BD-Rrs1p almost lost interaction with BD-L5 and AD-L11, respectively. This effect was not caused by a deference in the expression level of AD- or BD-fusion proteins; as shown in Figure 5B, the mutation of Rrs1p had little effect on the expression level of AD-Rrs1p or BD-Rrs1p and furthermore, expression level of BD-L5 in the presence of AD-Rrs1p or AD-L11 in the presence of BD-Rrs1p was not affected by the mutation of L65P in Rrs1p. The results suggest that leucine at 65 in Rrs1p is critical for Rrs1p-L5 and Rrs1p-L11 interactions.
Figure 5.
Effect of L65P mutation in Rrs1p on the interaction with ribosomal proteins, L5 and L11. (A) Protein–protein interactions were analyzed by yeast two-hybrid assay. BD-Rho1 was used as a negative control. (B) Expression of BD- and AD-fusion proteins was detected in western blotting analysis. Asterisks indicate non-specific bands.
DISCUSSION
We demonstrated previously that Rrs1p has important functions in 25S rRNA maturation and 60S ribosomal subunit assembly (7) and is also required for nuclear transport of 60S ribosomal subunits (9). We proposed that Rrs1p has a function to recruit L11 into the pre-60S ribosomal particle (8). In this paper, we have isolated the rex1-1 allele that causes a synthetic growth defect with rrs1-5. Rex1p was first described as Rnh70p, a 70 kDa protein that co-purified with RNase H activity. However, Rex1p/Rnh70p has no sequence similarity to known RNase H proteins and rex1/rnh70 mutants do not show reduced RNase H activity (21,22). Although Rex1p functions in various steps of RNA processing, Rex1p appears to be a unique enzyme for 5S rRNA trimming because rex1Δ results in extended 5S rRNA (13). rRNA (5S) is transcribed as a precursor by RNA polymerase III. The 5′ end of the mature 5S rRNA corresponds to that of the primary transcript, whereas the 3′ end is processed from a pre-5S rRNA that is extended by 12 nt. However, Rex1p and Rex2p function redundantly in 5.8S rRNA maturation and Rex1p, Rex2p and Rex3p are redundant for the processing of U5 snRNA and RNA subunit of RNase P. We show that the rex1-1 mutation causes extended 5S rRNA similar to a rex1-null mutation, without any effect on 5.8S, 18S and 25S rRNA maturation. The results indicate that the synergistic defect was caused by the rrs1-5 mutation and 3′-extended 5S rRNA. The rex1-1 mutation does not exhibit a synthetic defect with the rrs1-5 allele at 25°C, although at this temperature the rex1-1 allele has a defect in pre-5S rRNA processing. This is because the rrs1-5 mutant is temperature sensitive and less defective at 25°C than at 33°C. In the rex1-1 rrs1-5 strain cells, but not in the rex1Δ rrs1-5 strain cells, a smearing band showing extended 5S rRNA is detected at both 25 and 33°C (Figure 2A). This suggests that in the background of rrs1-5, extended 5S rRNA is not assembled into the pre-ribosome particle and has a chance to be trimmed by a weak activity of Rex1 that is encoded by rex1-1.
Rex1p is a unique exonuclease for 5S rRNA trimming. However, both the rex1Δ strain and the rex1-1 strain are indistinguishable from wild type in growth in spite of accumulating 3′-extended 5S rRNA. This indicates that 3′-extended 5S rRNA can be normally assembled into the 60S ribosomal subunits. Nevertheless, 3′-extended 5S rRNA causes significant growth defect under the rrs1-5 background. The result suggests that normal function of Rrs1p is required for the assembly of 5S rRNA into the 60S ribosomal subunit as well as for the assembly of L11. RPL5 encoding a ribosomal protein L5 was cloned as a weak suppressor of rrs1-5 rex1-1. It was suggested that L5 protects nascent 5S rRNA and recruits 5S rRNA to ribosomal particles (23). Since it appears that extended 5S rRNA is not degraded in rex1-1 or rex1Δ strains, it is likely that excess L5 suppresses the rex1-1 mutation by recruiting 5S rRNA. Either rpl11aΔ or rpl11bΔ, which results in a decreased level of L11, did not cause a synthetic growth defect with rrs1-5, suggesting that a decreased level of a component of ribosome does not cause a synthetic defect with rrs1 (T. Okada and K. Mizuta, unpublished data). Our results suggest that the recruitment of L11 and that of L5/5S rRNA are highly coordinated and that leucine at 65 in Rrs1p is critical for Rrs1p–L5 and Rrs1p–L11 interactions. We performed tandem affinity purification and detected both L11 and L5 in the fraction purified with Rrs1p-TAP (K. Ehara and K. Mizuta, unpublished data). It remains to be elucidated if both L5/5S rRNA and L11 are recruited into pre-ribosomal particles at the same time, or if assembly of one component followed by conformational rearrangement of the particle is required for assembly of the other component.
In eukaryotes, 5S rRNA tightly binds to L5 (the eukaryotic orthologue of E.coli L18) and the L5–5S rRNA complex is assembled into ribosomes, whereas in bacteria, 5S rRNA is assembled into ribosomes as a complex with three RPs, L5, L18 and L25 [for reviews see (24,25)]. In S.cerevisiae, both L11 (the orthologue of E.coli L5) and L5 (the orthologue of E.coli L18) are localized near the top surface of the central protuberance of the ribosomal large subunit (12,26). E.coli L5 homologues have similar lengths; E.coli L5, H.marismortui L5, S.cerevisiae L11 consists of 178, 176 and 173 amino acids, respectively. In contrast, comparison of the amino acid sequences of the E.coli L18 homologues reveals significant difference in their lengths; E.coli L18, H.marismortui L18, S.cerevisiae L5 consists of 117, 186 and 297 amino acids, respectively. In this paper, we show genetic and physical interactions among Rrs1p, L11-binding protein and L5-5S rRNA. Our results suggest that evolutionary changes of bacterial L18 confer the ability to recruit 5S rRNA to S.cerevisiae L5 and that the coordinated function of E.coli L5 and L18 in 5S rRNA recruitment is somehow conserved in S.cerevisiae L11 and L5 via Rrs1p (Figure 6).
Figure 6.
Model for assembly of 5S rRNA into the subunit in budding yeast and E.coli. In budding yeast, Rrs1p might recruit both ribosomal L11 and 5S rRNA (5S in the figure)-L5 into the 60S subunit. Yeast L11 and L5 are the orthologues of E.coli L5 and L18, respectively.
It was demonstrated previously that in 5S rRNA mutants, a decrease in the amount of 5S RNA was paralleled by a decrease in the amount of 60S subunits due to a specific defect in the processing of the 27SB rRNA. It was proposed that 5S RNA is recruited by pre-ribosomal particles containing the 27SB precursor and that its binding allows processing to proceed at a normal rate (27). Such a mechanism could ensure that all newly formed mature 60S subunits contain equal amounts of the three rRNAs and the RPs (27). We demonstrated previously that the depletion of Rrs1p leads to the accumulation of 27SBS and 27SBL intermediates whereas level of 27SA2 intermediate remained constant (8). The result indicates that Rrs1p is required for the processing from 27SB to 25S rRNA. These results suggest that L5–5S rRNA and L11 are recruited by Rrs1p to pre-60S subunits at the same step of the processing pathway and the recruitment is necessary for the further efficient processing.
Rrs1p is highly conserved in eukaryotes and is 36.8% identical to its human homologue (7). Several lines of evidence show that the mammalian homologues of Rrs1p, L5 and L11 play important roles in various kinds of cellular responses in higher eukaryotes: (i) The expression of RRS1 mRNA is stimulated very early in the disease cascade in Hdh CAG knock-in mice, Huntington's disease model mice (28). It was also demonstrated that RRS1 mRNA in human brains is elevated in Huntington's disease. (ii) Human homologues of L5 and L11 can interact with HDM2 and inhibit its function; thus, resulting in stabilization and activation of the p53 tumor suppressor, similar to ARF (29,30). (iii) L5 and L11 also interact with fragile X mental retardation 1 gene (FMR1) product. The Drosophila homologues of L5 and L11 were identified as components of a dFMR1-associated complex (31). (iv) RRS1 is induced by influenza virus replication, whereas viral replication results in the downregulation of many cellular mRNAs (32). Studies on Rrs1p will shed a light on research of human diseases.
Acknowledgments
We thank Dr John L. Woolford, Jr for critical reading of the manuscript, Dr Akira Kikuchi for the antibodies, Dr Leland Hartwell for yeast strain and Dr Mark D. Rose for yeast genomic DNA library. This research was supported by Grants-in-aid for Scientific Research from Japan Society for the Promotion of Science and from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a research grant from the Naito Foundation to K.M. C.S. is the recipient of a JSPS Research Fellowship (DC2). Funding to pay the Open Access publication charges for this article was provided by JSPS, MEXT, MEXT of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Woolford J.L., Jr, Warner J.R. The ribosome and its synthesis. In: Broach J.R., Pringle J.R., Jones E.W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 2.Kressler D., Linder P., de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Gen. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 5.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 6.Mizuta K., Warner J.R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuno A., Miyoshi K., Tsujii R., Miyakawa T., Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita D., Miyoshi K., Matsui Y., Toh-e A., Shinkawa H., Miyakawa T., Mizuta K. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27SB Pre-rRNA to 25S rRNA and the 60S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28780–28786. doi: 10.1074/jbc.M203399200. [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi K., Shirai C., Horigome C., Takenami K., Kawasaki J., Mizuta K. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60 S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004;565:106–110. doi: 10.1016/j.febslet.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi K., Tsujii R., Yoshida H., Maki Y., Wada A., Matsui Y., Toh-e A., Mizuta K. Normal assembly of 60S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:18334–18339. doi: 10.1074/jbc.M201667200. [DOI] [PubMed] [Google Scholar]
- 11.Mager W.H., Planta R.J., Ballesta J.G., Lee J.C., Mizuta K., Suzuki K., Warner J.R., Woolford J.L., Jr A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsay Y.F., Shankweiler G., Lake J., Woolford J.L., Jr Localization of Saccharomyces cerevisiae ribosomal protein L16 on the surface of 60 S ribosomal subunits by immunoelectron microscopy. J. Biol. Chem. 1994;269:7579–7586. [PubMed] [Google Scholar]
- 13.van Hoof A., Lennertz P., Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose M.D., Novick P., Thomas J.H., Botstein D., Fink G.R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- 16.Koshland D., Kent J.C., Hartwell L.H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 17.Tsujii R., Miyoshi K., Tsuno A., Matsui Y., Toh-e A., Miyakawa T., Mizuta K. Ebp2p, yeast homologue of a human protein that interacts with Epstein–Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells. 2000;5:543–553. doi: 10.1046/j.1365-2443.2000.00346.x. [DOI] [PubMed] [Google Scholar]
- 18.Shirai C., Takai T., Nariai M., Horigome C., Mizuta K. Ebp2p, the yeast homolog of Epstein–Barr virus nuclear antigen 1-binding protein 2, interacts with factors of both the 60S and 40S ribosomal subunit assembly. J. Biol. Chem. 2004;279:25353–25358. doi: 10.1074/jbc.M403338200. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi K., Miyakawa T., Mizuta K. Repression of rRNA synthesis due to a secretory defect requires the C-terminal silencing domain of Rap1p in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:3297–3303. doi: 10.1093/nar/29.16.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi K., Shirai C., Mizuta K. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1969–1973. doi: 10.1093/nar/gkg278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank P., Braunshofer-Reiter C., Karwan A., Grimm R., Wintersberger U. Purification of Saccharomyces cerevisiae RNase H(70) and identification of the corresponding gene. FEBS Lett. 1999;45:251–256. doi: 10.1016/s0014-5793(99)00512-8. [DOI] [PubMed] [Google Scholar]
- 22.Qiu J., Qian Y., Frank P., Wintersberger U., Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshmukh M., Tsay Y.F., Paulovich A.G., Woolford J.L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spierer P., Zimmermann R.A. Stoichiometry, cooperativity, and stability of interactions between 5S RNA and proteins L5, L18, and L25 from the 50S ribosomal subunit of Escherichia coli. Biochemistry. 1978;17:2474–2479. doi: 10.1021/bi00606a002. [DOI] [PubMed] [Google Scholar]
- 25.Szymanski M., Barciszewska M.Z., Erdmann V.A., Barciszewski J. 5S rRNA:structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rotenberg M.O., Moritz M., Woolford J.L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 27.Dechampesme A.M., Koroleva O., Leger-Silvestre I., Gas N., Camier S. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol. 1999;145:1369–1380. doi: 10.1083/jcb.145.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fossale E., Wheeler V.C., Vrbenac V., Lebel L.A., Teed A., Mysore J.S., Gusella J.F., MacDonald M.E., Persichetti F. Identification of a presymptomatic molecular phenotype in Hdh CAG knock-in mice. Hum. Mol. Genet. 2002;11:2233–2241. doi: 10.1093/hmg/11.19.2233. [DOI] [PubMed] [Google Scholar]
- 29.Lohrum M.A., Ludwig R.L., Kubbutat M.H., Hanlon M., Vousden K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Wolf G.W., Bhat K., Jin A., Allio T., Burkhart W.A., Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishizuka A., Siomi M.C., Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiss G.K., An M.C., Bumgarner R.E., Hammersmark E., Cunningham D., Katze M.G. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 2001;75:4321–4331. doi: 10.1128/JVI.75.9.4321-4331.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]