Abstract
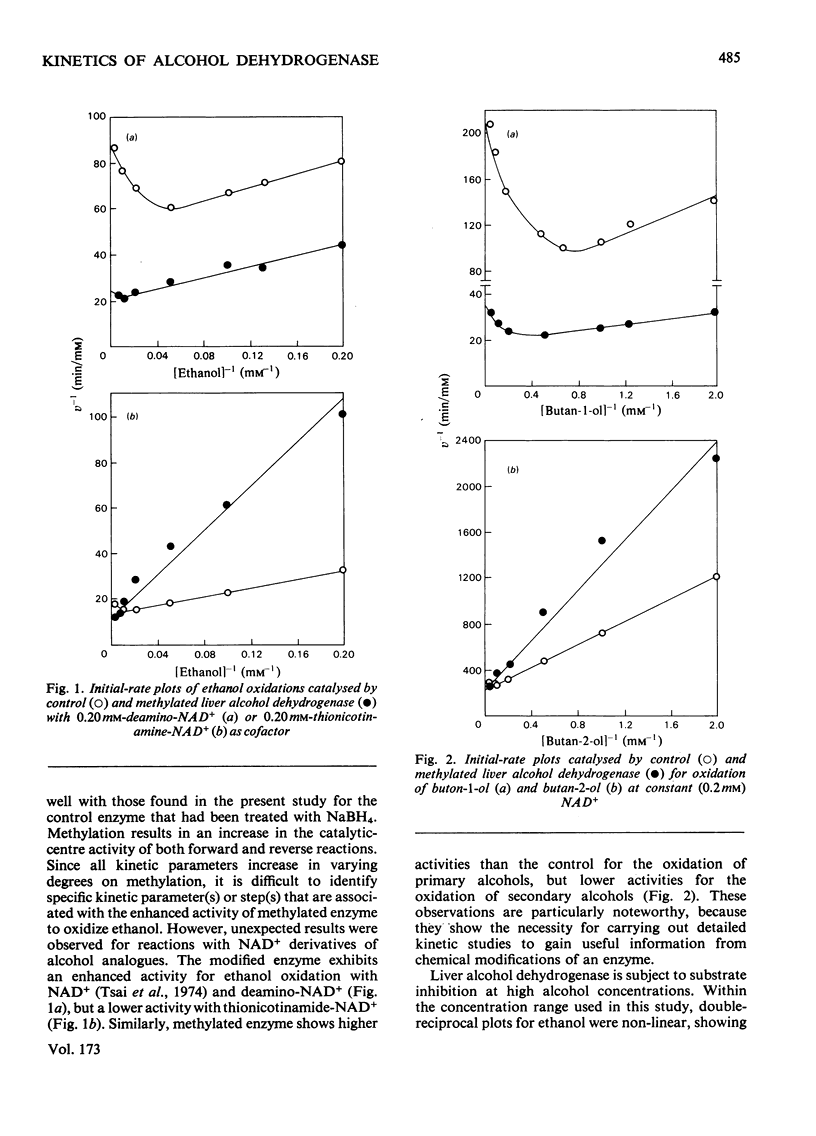
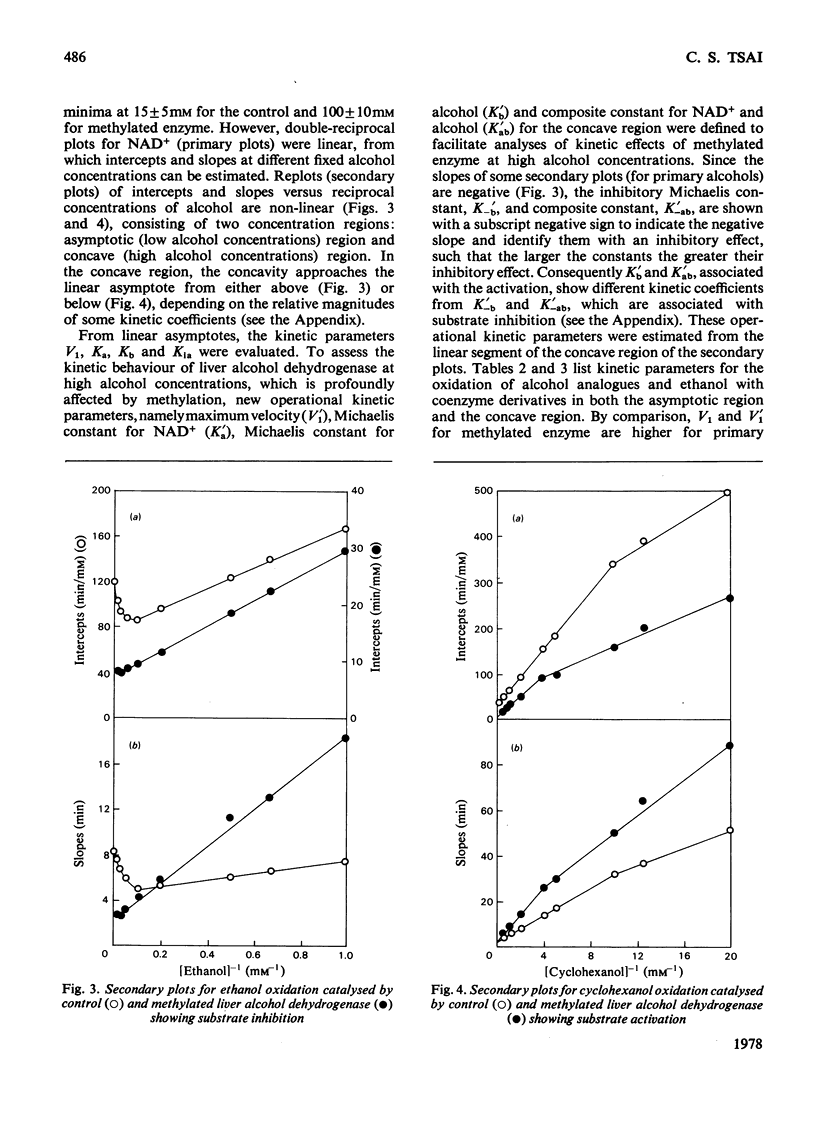
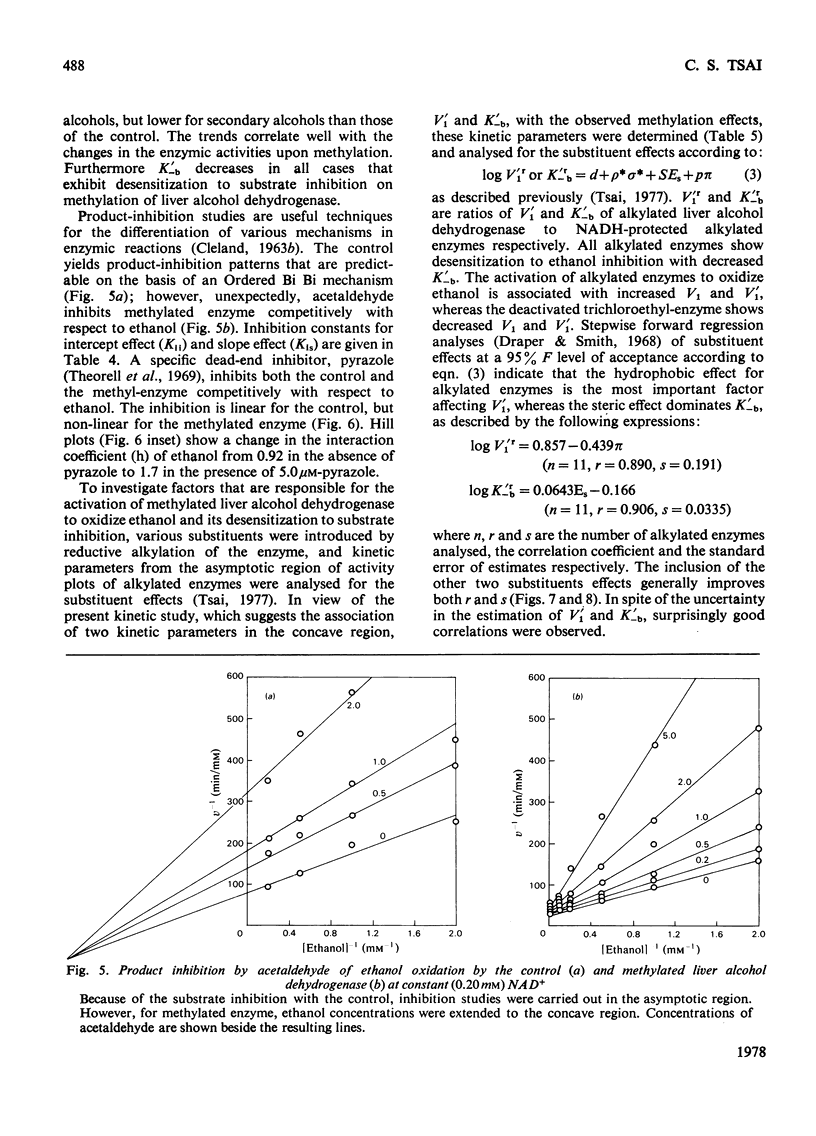
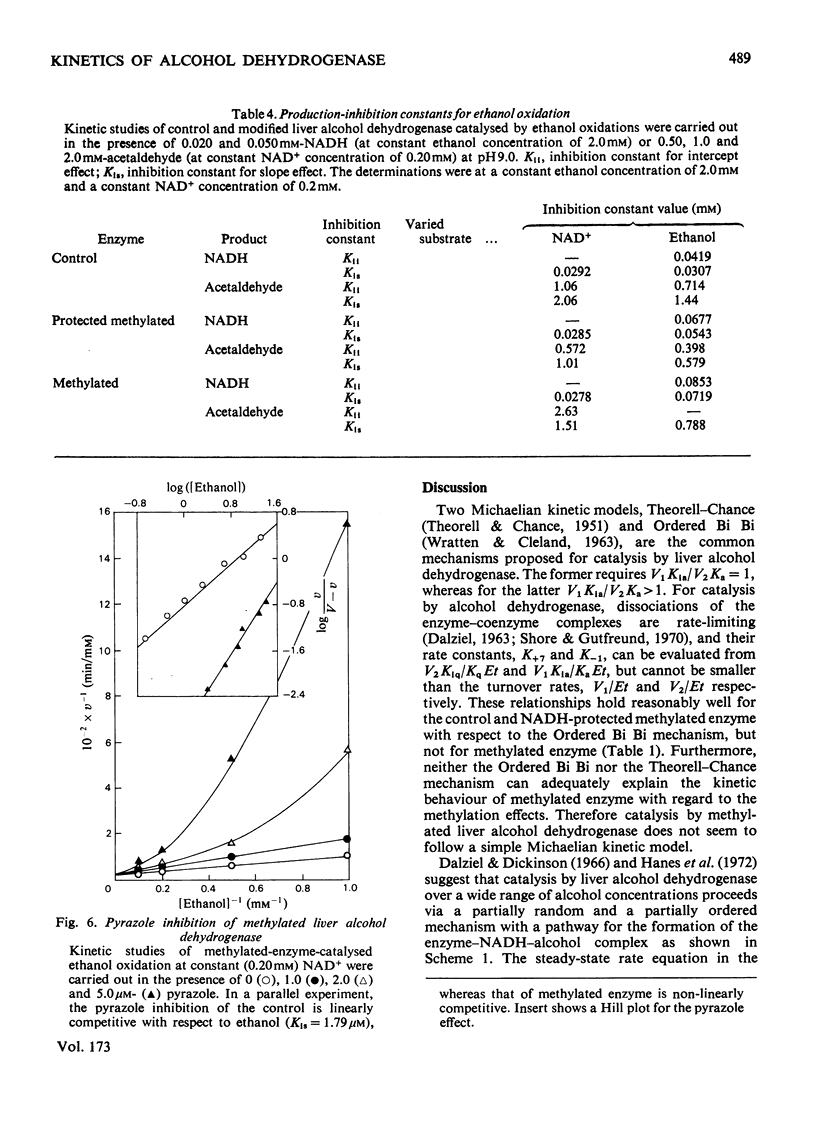
Reductive methylation of lysine residues activates liver alcohol dehydrogenase in the oxidation of primary alcohols, but decreases the activity of the enzyme towards secondary alcohols. The modification also desensitizes the dehydrogenase to substrate inhibition at high alcohol concentrations. Steady-state kinetic studies of methylated liver alcohol dehydrogenase over a wide range of alcohol concentrations suggest that alcohol oxidation proceeds via a random addition of coenzyme and substrate with a pathway for the formation of the productive enzyme-NADH-alcohol complex. To facilitate the analyses of the effects of methylation on liver alcohol dehydrogenase and factors affecting them, new operational kinetic parameters to describe the results at high substrate concentration were introduced. The changes in the dehydrogenase activity on alkylation were found to be associated with changes in the maximum velocities that are affected by the hydrophobicity of alkyl groups introduced at lysine residues. The desensitization of alkylated liver alcohol dehydrogenase to substrate inhibition is identified with a decrease in inhibitory Michaelis constants for alcohols and this is favoured by the steric effects of substituents at the lysine residues.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CLELAND W. W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. I. Nomenclature and rate equations. Biochim Biophys Acta. 1963 Jan 8;67:104–137. doi: 10.1016/0006-3002(63)91800-6. [DOI] [PubMed] [Google Scholar]
- CLELAND W. W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. III. Prediction of initial velocity and inhibition patterns by inspection. Biochim Biophys Acta. 1963 Feb 12;67:188–196. doi: 10.1016/0006-3002(63)91816-x. [DOI] [PubMed] [Google Scholar]
- Chen S. S., Engel P. C. Horse liver alcohol dehydrogenase. A study of the essential lysine residue. Biochem J. 1975 Sep;149(3):627–635. doi: 10.1042/bj1490627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALZIEL K. KINETIC STUDIES OF LIVER ALCOHOL DEHYDROGENASE AND PH EFFECTS WITH COENZYME PREPARATIONS OF HIGH PURITY. J Biol Chem. 1963 Aug;238:2850–2858. [PubMed] [Google Scholar]
- Dalziel K., Dickinson F. M. The kinetics and mechanism of liver alcohol dehydrogenase with primary and secondary alcohols as substrates. Biochem J. 1966 Jul;100(1):34–46. doi: 10.1042/bj1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degani Y., Veronese F. M., Smith E. L. Nicotinamide adenine dinucleotide-specific glutamate dehydrogenase of Neurospora. II. Selective chemical reactivity of amino and sulfhydryl groups. J Biol Chem. 1974 Dec 25;249(24):7929–7935. [PubMed] [Google Scholar]
- Dove M. J., Tsai C. S. Kinetic effect of some aliphatic amines on yeast alcohol dehydrogenase. Can J Biochem. 1976 May;54(5):432–437. doi: 10.1139/o76-062. [DOI] [PubMed] [Google Scholar]
- Dworschack R., Tarr G., Plapp B. V. Identification of the lysine residue modified during the activation of acetimidylation of horse liver alcohol dehydrogenase. Biochemistry. 1975 Jan 28;14(2):200–203. doi: 10.1021/bi00673a002. [DOI] [PubMed] [Google Scholar]
- Fan C. C., Plaut G. W. Functional groups of diphosphopyridine nucleotide linked isocitrate dehydrogenase from bovine heart. II. Studies of an active amino group by reaction with aldehydes. Biochemistry. 1974 Jan 1;13(1):52–59. doi: 10.1021/bi00698a009. [DOI] [PubMed] [Google Scholar]
- Hanes C. S., Bronskill P. M., Gurr P. A., Wong J. T. Kinetic mechanism for the major isoenzyme of horse liver alcohol dehydrogenase. Can J Biochem. 1972 Dec;50(12):1385–1413. doi: 10.1139/o72-182. [DOI] [PubMed] [Google Scholar]
- Jörnvall H. Reactive lysine residues in horse liver alcohol dehydrogenase. Biochem Biophys Res Commun. 1973 Apr 16;51(4):1069–1076. doi: 10.1016/0006-291x(73)90036-3. [DOI] [PubMed] [Google Scholar]
- McKinley-McKee J. S., Morris D. L. The lysines in liver alcohol dehydrogenase. Chemical modification with pyridoxal 5'-phosphate and methyl picolinimidate. Eur J Biochem. 1972 Jun 23;28(1):1–11. doi: 10.1111/j.1432-1033.1972.tb01877.x. [DOI] [PubMed] [Google Scholar]
- Plapp B. V., Brooks R. L., Shore J. D. Horse liver alcohol dehydrogenase. Amino groups and rate-limiting steps in catalysis. J Biol Chem. 1973 May 25;248(10):3470–3475. [PubMed] [Google Scholar]
- Plapp B. V. Enhancement of the activity of horse liver alcohol dehydrogenase by modification of amino groups at the active sites. J Biol Chem. 1970 Apr 10;245(7):1727–1735. [PubMed] [Google Scholar]
- Shore J. D., Gutfreund H. Transients in the reactions of liver alcohol dehydrogenase. Biochemistry. 1970 Nov 24;9(24):4655–4659. doi: 10.1021/bi00826a006. [DOI] [PubMed] [Google Scholar]
- Theorell H., Yonetani T., Sjöberg B. On the effects of some heterocyclic compounds on the enzymic activity of liver alcohol dehydrogenase. Acta Chem Scand. 1969;23(1):255–260. doi: 10.3891/acta.chem.scand.23-0255. [DOI] [PubMed] [Google Scholar]
- Tsai C. S., Tsai Y. H., Lauzon G., Cheng S. T. Structure and activity of methylated horse liver alcohol dehydrogenase. Biochemistry. 1974 Jan 29;13(3):440–443. doi: 10.1021/bi00700a007. [DOI] [PubMed] [Google Scholar]
- WRATTEN C. C., CLELAND W. W. PRODUCT INHIBITION STUDIES ON YEAST AND LIVER ALCOHOL DEHYDROGENASES. Biochemistry. 1963 Sep-Oct;2:935–941. doi: 10.1021/bi00905a007. [DOI] [PubMed] [Google Scholar]


