Abstract
4E-BP is an important regulator of overall translation levels in cells. By binding eIF4E, 4E-BP impairs recruitment of the 40S ribosomal subunit to the cap structure present at the 5′-end of all eukaryotic cellular mRNAs. 4E-BP activity is controlled by TOR (Target of Rapamycin). 4E-BP has been studied extensively in cell culture; however, the biological role of 4E-BP in developing organisms is unclear to date. Since TOR has been shown to control tissue growth during animal development, 4E-BP has also been assumed to serve as a growth regulator. Here, we study the relevance of 4E-BP function for organismal development, and present evidence for an alternate view. We show that 4E-BP strongly affects fat metabolism in Drosophila. We suggest that 4E-BP works as a metabolic brake that is activated under conditions of environmental stress to control fat metabolism. 4E-BP mutants lack this regulation, reducing their ability to survive under unfavorable conditions.
Keywords: Translation, metabolism, fat, triglycerides, stress, rapamycin
The cellular activity and mechanism of action of 4E-BP as a translation inhibitor have been extensively studied. 4E-BP can bind eIF4E, and block its normal function of recruiting the initiation complex (containing the 40S ribosomal subunit) to the m7GpppX cap structure present at the 5′-end of all eukaryotic cellular mRNAs (for review, see Gingras et al. 1999b). In the absence of this cap-dependent recruitment, most cellular mRNAs are poorly translated (Both et al. 1975; Muthukrishnan et al. 1975). Therefore, when 4E-BP is active, overall translation levels are strongly dampened. 4E-BP activity is regulated through phosphorylation by the protein kinase TOR (Hay and Sonenberg 2004). 4E-BP is phosphorylated in a complex manner on a large number of sites (Gingras et al. 2001). Briefly, when TOR activity is low, 4E-BP is hypophosphorylated allowing it to bind efficiently to eIF4E and block translation. When TOR activity increases, it phosphorylates 4E-BP, causing its affinity for eIF4E to drop and allowing cap-dependent translation to occur. TOR activity is regulated by growth factor signaling (especially insulin/PI3K), by amino acid availability, and by the energy state of the cell (for review, see Hay and Sonenberg 2004). Recently, several reports have indicated that 4E-BP activity is also regulated at the transcriptional level: 4E-BP transcription has been reported to be inhibited by the stress-dependent kinases ERK and p38 (Rolli-Derkinderen et al. 2003) and positively regulated by the forkhead transcription factor FOXO (Junger et al. 2003; Puig et al. 2003).
The in vivo relevance of 4E-BP's activity remains a topic of conjecture, since almost all studies on 4E-BP have been done in cell culture. These studies have explained how 4E-BP functions molecularly, but not under what circumstances a living organism uses 4E-BP activity, nor what the consequences are for the whole animal when 4E-BP activity is lacking. A knockout mouse for one of the three mammalian 4E-BP proteins, 4E-BP1, has been described. These mice are viable and have elevated metabolism and reduced adipose tissue (Tsukiyama-Kohara et al. 2001). These phenotypes are surprising, given that 4E-BP blocks a core component of the cell translation machinery. Since mice have three 4E-BP proteins, it is unclear whether this is a derived function for 4E-BP1 or whether it is an ancestral function for all 4E-BPs. Alternatively, 4E-BP has also been proposed to control the rate of growth of tissues during development (Miron et al. 2001; Johnston and Gallant 2002). This conjecture was made since cellular translation is an important part of tissue growth, and since 4E-BP's main regulator, TOR, controls tissue growth during development. This conjecture, however, has not been thoroughly tested. Since Drosophila has only one 4E-BP gene, we use this model system to study the ancestral function of 4E-BP at the organismal level.
Results and Discussion
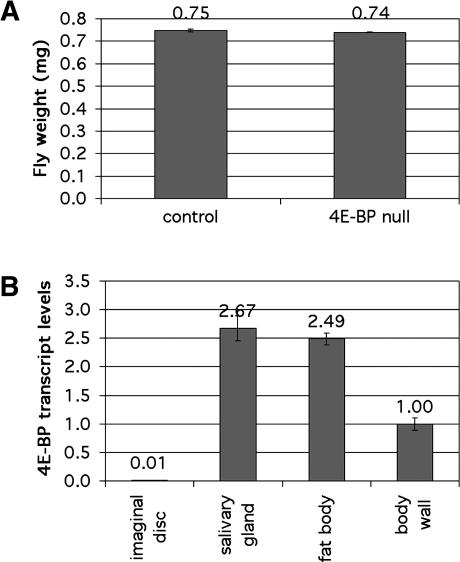
To assess the contribution of 4E-BP toward controlling tissue growth in Drosophila, we measured the size of adult flies of the previously described 4E-BP-null mutant Thor2, which is viable and fertile (Bernal and Kimbrell 2000; Bernal et al. 2004). This allele deletes part of the promoter region, the translation start site, and a substantial part of the coding region. We confirmed by real-time quantitative RT–PCR that this allele produces no detectable 4E-BP transcript and by immunoblotting that no detectable protein is produced. If under normal growth conditions 4E-BP functions to limit tissue growth, we would expect 4E-BP mutant flies to be larger than normal flies. Instead, we found that 4E-BP mutant flies are comparable in body weight to control flies (Fig. 1A). The developmental rate of 4E-BP mutant and control flies was also comparable (data not shown). This suggests that 4E-BP does not normally play a role in tissue growth control. Using quantitative real-time RT–PCR, we found that 4E-BP transcript levels in the wing, haltere, and leg imaginal discs were <1% those found in other larval tissues such as the salivary gland or the body wall (Fig. 1B). In agreement with this, no 4E-BP protein could be detected in imaginal disc extracts, whereas it could easily be detected in other larval tissues (data not shown). This suggests that despite being expressed abundantly in larval tissues, 4E-BP might have little effect on body size in part because it is expressed only at low levels in imaginal tissues, which form the external structures of the adult body. However, this seems unlikely because we observed that even ectopic overexpression of 4E-BP under GAL4 control was not able to reduce growth of imaginal tissues (in agreement with Miron et al. 2001). To test whether this is due to excess TOR activity, which can inactivate 4E-BP via phosphorylation, we generated transgenic flies that express under UAS control either wild-type 4E-BP or a 4E-BP form with two threonine-to-alanine substitutions (T37A, T46A), 4E-BP(AA). Abolishing the possibility of phosphorylation on these two threonine residues by TOR has been shown to render mammalian 4E-BP constitutively active in binding and inhibiting eIF4E (Gingras et al. 1999a). We found that ectopic overexpression of either 4E-BP(WT) or 4E-BP(AA) in the wing disc did not cause any reduction in final wing size, suggesting that 4E-BP cannot affect final tissue size even when constitutively active. (Expression levels for the two different forms were comparable to each other and to the level of endogenous 4E-BP protein found in larval tissues [data not shown].) These results indicate that 4E-BP does not control tissue growth during Drosophila development.
Figure 1.
4E-BP mutant flies are normal in size. (A) Newly hatched L1 larvae of genotypes w1118 (“control”) and Thor2 (“4E-BP null”) were seeded in vials at a density of 50/vial and grown at 25°C. Male adult flies were then aged 3 d and weighed in batches of 40. Average fly weights for control and 4E-BP-null flies were 0.75 and 0.74 mg, respectively. Error bars indicate standard deviations of 0.005 and 0.002 mg/fly, respectively. (B) Quantitative real-time RT–PCR was performed on RNA extracted from indicated tissues of wandering third instar larvae. The values shown are 4E-BP transcript levels normalized to rp49 transcript levels. Error bars show standard deviation on the assay done in triplicate.
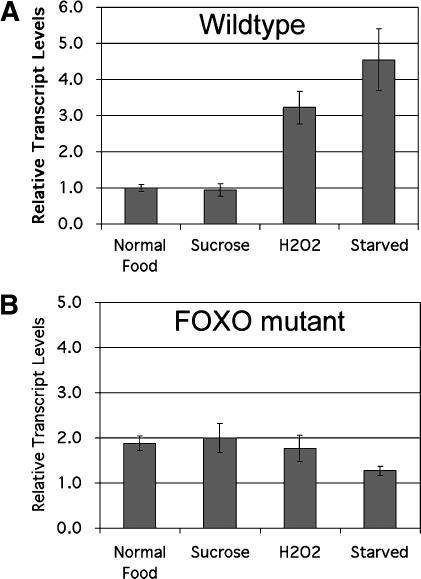
To date, the only defect reported for 4E-BP-null flies is an impaired innate immune response to wounding or bacterial infection (Bernal and Kimbrell 2000). This prompted us to examine whether 4E-BP is involved in other stress responses. We first asked whether 4E-BP expression is induced under oxidative stress or starvation. Quantitative RT–PCR was performed on RNA from whole flies grown under identical, controlled conditions and then placed for 1 d on (1) normal food, (2) medium containing sucrose but no amino acids, (3) medium supplemented with hydrogen peroxide, or (4) medium lacking all nutrients (Fig. 2A). We found that 4E-BP transcript levels are strongly up-regulated under these environmental stress conditions, in agreement with previous microarray experiments (Zou et al. 2000; Zinke et al. 2002). These results were statistically significant (t-test = 0.02 comparing “H2O2” or “starved” to normal food conditions). Since it has been shown that 4E-BP is a direct target of transcriptional regulation by FOXO (Junger et al. 2003; Puig et al. 2003), we asked whether this 4E-BP expression induction is a FOXO-dependent response. We performed quantitative RT–PCR on FOXO-null mutant flies and found that the basal level of 4E-BP was somewhat elevated, but that it was not further increased by starvation or oxidative stress (Fig. 2B). These data indicate that under conditions of environmental stress, in particular starvation, 4E-BP expression is induced and that this occurs in part via FOXO.
Figure 2.
Stress induction of 4E-BP expression is FOXO-dependent. Quantitative real-time RT–PCR on RNA from whole adult control w1118 flies (A) or FOXO21/25 mutant flies (B). Larvae were grown under controlled conditions. Adult males were aged for 3 d and then placed in vials containing 0.8% agarose in PBS (“starved”) or supplemented with 10% sucrose or 10%sucrose + 5% H2O2. The values shown are 4E-BP transcript levels normalized to rp49 transcript levels. Error bars show standard deviation on the assay done in triplicate.
We next asked whether 4E-BP mutant flies have an impaired response to nutrient deprivation. 4E-BP mutant and control flies were grown under identical conditions, aged 3 d, and then deprived of all sources of energy. 4E-BP mutants died faster than control flies under these conditions (Fig. 3). At 37 h of starvation, 92% of control flies were alive, whereas only 38% of 4E-BP mutant flies were still alive. This difference was statistically highly significant (t-test < 0.001). A possible explanation for this finding is that the 4E-BP mutant flies started out with smaller energy reserves than control flies. Flies store energy predominantly as triglycerides in the fat body (Van der Horst 2003). 4E-BP-null flies did not prove to have lower triglyceride levels than control flies under the standard nutritional conditions used here; if anything, fat levels were slightly higher than in wild-type flies (Fig. 4A). An alternative possibility is that 4E-BP-null flies do not reduce their metabolism appropriately when starved and thus burn their reserves too quickly. A time-course analysis of triglyceride levels in 4E-BP-null and control flies during starvation supports this possibility (Fig. 4B). Although the triglyceride stores of both 4E-BP mutant and control flies dropped during starvation, the triglyceride stores of 4E-BP mutants dropped more rapidly. This was consistent in independently repeated experiments. By 24 h of starvation, the difference in fat content was statistically significant (t-test < 0.05). At 36 h of starvation, the fat content of 4E-BP mutants reached almost zero, whereas control flies still had a substantial amount of fat left (t-test < 0.01). The time at which the flies had no fat left coincides with when they started dying (Fig. 3). We performed two control experiments to verify that this phenotype is due specifically to a lack of 4E-BP function. First, we repeated this experiment with control flies, 4E-BP-null flies, and 4E-BP-null flies in which 4E-BP activity was restored by ubiquitous expression of 4E-BP under Gal4 control (Fig. 4C). We used 30 h of starvation because at this time point the phenotype is robust and clearly evident. After 30 h of starvation, control flies had >25% of their original fat content, whereas 4E-BP-null flies had only half as much, indicating an increased burn rate as shown in Figure 4B. The 4E-BP-null mutants in which 4E-BP activity was restored using a transgene were rescued, retaining >25% of their original fat, as the control flies (Fig. 4C). Second, we tested a fly strain in which the P-element used to generate the 4E-BP-null strain was instead precisely excised (termed “revertant strain”) (Fig. 4D). Again, the 4E-BP-null flies had an accelerated fat burn rate compared with control, whereas the revertant strain exhibited a fat burn rate similar to that of the control. Thus, the accelerated fat loss in the mutant can be attributed to the absence of 4E-BP activity. We conclude that although 4E-BP mutant flies start out with at least the same amount of fat as normal flies, they are impaired in their ability to reduce their energy burn rate under conditions of nutrient deprivation.
Figure 3.
4E-BP mutant flies have impaired survival under starvation conditions. Control w1118 (solid line) and 4E-BP-null flies (dashed line) were raised under controlled conditions, aged 3 d, and starved; 20 males per vial (0.8% agarose/PBS). Error bars show standard deviation on the assay done in triplicate.
Figure 4.
4E-BP mutant flies burn their fat stores more quickly than control flies. (A) Total body triglycerides and total body protein were measured after controlled growth and aging. Plotted values are triglyceride levels normalized to protein levels. Batches of eight males were assayed, and the experiment was done in quadruplicate. Average values were 4.3 and 4.6 μg triglycerides/μg protein for control and 4E-BP-null flies, respectively. Error bars show standard error. (B) Total triglycerides were measured for flies treated as in A. Batches of eight males were measured in duplicate. Values show triglyceride levels as a percentage of prestarvation levels ± standard error. (C,D) Flies of indicated genotypes were grown under controlled conditions and aged for 3 d. Total body triglycerides were measured for half of the flies immediately, and for the other half after 30 h of starvation on PBS/agarose. Three batches of eight males were used for each measurement. Error bars show standard deviation.
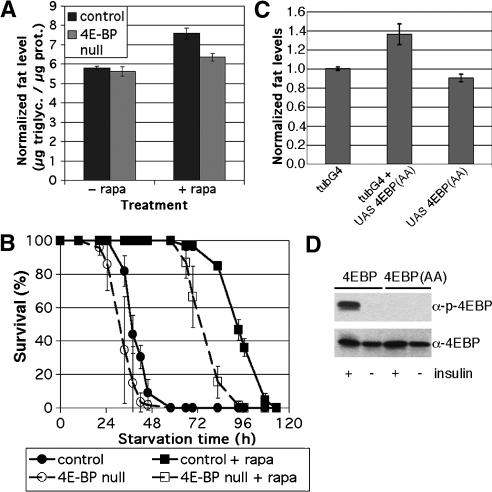
Since 4E-BP mutants are hypersensitive to nutrient deprivation, we wanted to perform the converse experiment, to ask whether increasing 4E-BP activity could increase fat accumulation. We increased 4E-BP activity systemically by pharmacologically blocking TOR with the specific inhibitor rapamycin. Since TOR also has other targets (including ribosomal protein S6 kinase) (Hay and Sonenberg 2004), we assessed how much of rapamycin's activity is 4E-BP-mediated by comparing the effect of rapamycin administration on control flies and 4E-BP mutants. Wild-type flies treated with rapamycin in normal food for 3 d accumulated more fat than untreated flies (Fig. 5A). This effect was statistically significant (t-test < 0.02), and much of it was dependent on the activity of 4E-BP, since the 4E-BP mutants did not accumulate as much excess fat in response to rapamycin (Fig. 5A). The difference between the response of wild-type and 4E-BP mutant flies represents the fat accumulation caused specifically by increased 4E-BP activity (t-test < 0.03). Therefore, artificially increasing 4E-BP activity systemically causes the fly to become fatter. Accordingly, survival of control flies under conditions of nutrient deprivation was strongly extended by rapamycin administration (Fig. 5B, solid black lines). This enhancement was in part due to 4E-BP activity as 4E-BP mutants did not respond as strongly (Fig. 5B, dashed lines)—the half-life of control flies was extended by 55 h, whereas the half-life of 4E-BP mutants was extended by 43 h. As with the fat measurement experiments, this shows that the effect of rapamycin was in part due to increased 4E-BP activity and in part due to other TOR targets.
Figure 5.
4E-BP regulates fat content. (A) w1118 (“control”) or Thor2 (“4E-BP null”) male flies were raised under controlled conditions and aged for 3 d on normal food or normal food supplemented with 50 μM rapamycin. Total body triglycerides and protein were measured, and the ratio of triglycerides to protein is shown. Measurements were done on batches of eight male flies in triplicate. Error bars show standard error. Average values are 5.8 μg triglycerides/μg protein (control flies, normal food), 5.6 μg triglycerides/μg protein (4E-BP-null flies, normal food), 7.5 μg triglycerides/μg protein (control flies on rapamycin), or 6.4 μg triglycerides/μg protein (4E-BP-null flies on rapamycin). (B) Control or 4E-BP-null male flies were raised under controlled conditions and aged for 3 d on normal food or normal food supplemented with 50 μM rapamycin. Flies were then starved on 0.8% agarose/PBS either supplemented with 50 μM rapamycin or not. Batches of 20 males were assayed in triplicate. Error bars show standard deviation. (C) Flies of various genotypes were grown and aged for 3 d under controlled conditions. Total body triglycerides and protein were measured for each, and the ratio of triglycerides to protein is shown. Three batches of eight males were used for each. (D) Immunoblots of S2 cells transfected to express 4E-BP or the double-alanine mutant 4E-BP(AA). Cells were treated with insulin or not as indicated to induce Tor activity. (Lower panel) Probed with antibody to 4E-BP to show relative loading. (Upper panel) Probed with antibody specific to the phosphorylated form of 4E-BP.
These findings indicate that Drosophila does not fully turn off TOR activity, even though doing so would extend life span under conditions of nutrient deprivation. It can be expected that flies are subject to periods of nutrient deprivation in the natural environment, yet they do not apparently make use of the full extent of TOR regulation that might be beneficial under the circumstances. TOR clearly has other important functions, thus this may reflect an optimized balance between resistance to starvation and other TOR functions. In this context, the finding that manipulation of insulin pathway activity can extend life span under normal nutritional conditions (Clancy et al. 2001; Giannakou et al. 2004; Hwangbo et al. 2004) raises the question of whether rapamycin or related compounds might have similar effects on life span in mammals.
As an independent means to confirm the finding that systemic 4E-BP activity causes the animal to accumulate fat, we made use of the constitutively active 4E-BP(AA) mutant protein. The insensitivity of the 4E-BP(AA) mutant protein to insulin-induced phosphorylation was confirmed in S2 cells (Fig. 5D). Flies expressing the 4E-BP(AA) mutant protein under control of a ubiquitous Gal4 driver had elevated body fat compared with control flies, both when normalized to total body protein (Fig. 5C) and in absolute levels (data not shown). This is the same construct that gives no tissue growth phenotype when expressed in the wing (see above). Furthermore, we observed no altered total body fat levels if expression of the 4E-BP(AA) mutant protein was limited to the adipose tissue itself (using pplGal4) (data not shown). Thus systemically activating 4E-BP results in a net increase in fat accumulation in the adipose tissue.
Our findings provide evidence that 4E-BP plays an important role as a regulator of metabolism. Increasing 4E-BP activity within the context of a whole living animal increases fat accumulation. Conversely, reduced 4E-BP activity leads to an increased rate of fat burn. Furthermore, we have shown that either eliminating, or ectopically increasing 4E-BP activity does not lead to abnormal tissue growth. These results parallel those found in mice mutant for one of the three mammalian 4E-BP proteins, 4E-BP1, suggesting that the ancestral activity of 4E-BP is, indeed, that of a metabolic regulator. We show that 4E-BP expression is induced under various stress conditions including starvation. We favor the simple model that 4E-BP functions as a “metabolic brake,” through its regulation of overall cellular translation, which accounts for a large fraction of total cellular energy consumption. Although this brake is not applied during normal development to limit growth, the 4E-BP brake is applied under stress conditions, including starvation, to control the animal's metabolic rate and thus the rate at which it consumes its fat reserves. In support of the notion that 4E-BP's effect on fat metabolism is mediated by eIF4E, it has recently been shown that another modulator of eIF4E activity, Lk6, also affects total body fat levels (Arquier et al. 2005; Reiling et al. 2005). An alternate model is that the activity of 4E-BP is somehow specific to fat metabolism. Our finding that systemic activation of 4E-BP makes the animal fat, but that adipose-tissue specific activation of 4E-BP does not, would seem to support the argument against this possibility.
TOR signaling can differentially regulate both tissue growth and fat metabolism, the relative strength of these two activities depending on context (e.g., adipose tissue vs. imaginal tissue). The activity of S6K has profound effects on cellular and tissue growth (Thomas 2002), whereas we show here that 4E-BP has insignificant effects on tissue growth but profound effects on metabolism. Thus, the relative expression of S6K and 4E-BP in various tissues may help explain why TOR activity can have different phenotypic outputs. Our findings also suggest the possibility that control of 4E-BP activity could provide a means to influence fat metabolism and thus might be of clinical use in treating obesity or diabetes.
Materials and methods
Constructs, fly strains and reagents
The 4E-BP-null mutant strain Thor2 and the revertant strain Thor1rev1were kindly provided by D. Kimbrell (University of California, Davis) (Bernal and Kimbrell 2000; Bernal et al. 2004). DNA encoding UAS-4E-BP-HA was kindly provided by N. Sonenberg (McGill University, Montreal, Quebec, Canada) (Miron et al. 2003). This DNA was injected to create transgenic flies. FOXO21 and FOXO25 mutant flies were previously described (Junger et al. 2003). Rapamycin was obtained from Sigma (catalog #R0395). A 2 mM solution of rapamycin/ethanol was placed on the surface of normal fly food or agarose/PBS and allowed to soak in. The anti-d4EBP antibody is as in Miron et al. (2001). The antiphospho-T37/46-4EBP antibody is from Cell Signaling.
Triglyceride measurement
Newly hatched L1 larvae were seeded in vials at a density of 50/vial and grown at 25°C. Adult flies were then aged 3 d and placed into Eppendorf tubes in batches of eight (for males) or six (for females). The tubes were either placed on ice for immediate triglyceride quantification or frozen at –80°C for later processing. To process tubes that were frozen at –80°C, they were then placed on dry ice until homogenization. This significantly reduced error as triglycerides in thawed but unhomogenized flies got degraded over time. Flies were then homogenized in 1 mL of 0.05% Tween-20 + 2× protease inhibitor (Roche Complete protease inhibitor cocktail, catalog #1-836-145) in H2O for 40 sec at 5000 rpm using a homogenizer with a ceramide cylinder, and placed on ice (not dry ice, to avoid freeze/thaw cycles). The homogenates were then spun at 5000 rpm on a tabletop centrifuge for 1 min, and 500 μL of supernatant was transferred to a fresh tube, avoiding debris. This supernatant was then spun at 14,000 rpm for 3 min in a tabletop centrifuge at 4°C. To measure triglycerides, 80 μL of the resulting supernatant was combined with 1 mL of Triglyceride Reagent (Thermo Electron Corp. catalog #981786) and incubated for 10 min at 37°C; the OD520 was measured and compared with a standardization curve. To measure protein levels, 100 μL of the final supernatant was combined with 700 μL of H2O and 200 μL of Bio-Rad Protein Assay Reagent (catalog #500-0006), incubated at room temperature for 3 min, and the OD595 was then measured and compared with a standardization curve.
Quantitative RT–PCR
Total RNA was obtained from larval tissues or adult flies by homogenization in TRIzol reagent (Invitrogen catalog #15596-018). Five micrograms of total RNA from each sample was used to generate first-strand cDNA using the SuperScript II kit (Invitrogen 11904-018) using the poly(dT) method. Quantitative RT–PCR was then done on the Applied Biosystems ABI Prism 7000 Sequence Detection System using the SYBR Green PCR master mix (ABI catalog #4309155). All assays were done in triplicate and normalized to rp49 levels, and errors were propagated in all calculations.
Acknowledgments
We thank Vladimir Benes for help with quantitative RT–PCR and Ann Mari Voie for fly injections.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.341505.
References
- Arquier N., Bourouis, M., Colombani, J., and Leopold, P. 2005. Drosophila Lk6 Kinase Controls Phosphorylation of Eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr. Biol. 15: 19–23. [DOI] [PubMed] [Google Scholar]
- Bernal A. and Kimbrell, D.A. 2000. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc. Natl. Acad. Sci. 97: 6019–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A. Schoenfeld, R., Kleinhesselink, K., and Kimbrell, D.A. 2004. Loss of Thor, the single 4E-BP gene of Drosophila, does not result in lethality. Dros. Inf. Serv. 87: 81–84. [Google Scholar]
- Both G.W., Banerjee, A.K., and Shatkin, A.J. 1975. Methylation-dependent translation of viral messenger RNAs in vitro. Proc. Natl. Acad. Sci. 72: 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D.J., Gems, D., Harshman, L.G., Oldham, S., Stocker, H., Hafen, E., Leevers, S.J., and Partridge, L. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106. [DOI] [PubMed] [Google Scholar]
- Giannakou M.E., Goss, M., Junger, M.A., Hafen, E., Leevers, S.J., and Partridge, L. 2004. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Gygi, S.P., Raught, B., Polakiewicz, R.D., Abraham, R.T., Hoekstra, M.F., Aebersold, R., and Sonenberg, N. 1999a. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes & Dev. 13: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught, B., and Sonenberg, N. 1999b. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15: 807–826. [DOI] [PubMed] [Google Scholar]
- Hay N. and Sonenberg, N. 2004. Upstream and downstream of mTOR. Genes & Dev. 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- Hwangbo D.S., Gersham, B., Tu, M.P., Palmer, M., and Tatar, M. 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566. [DOI] [PubMed] [Google Scholar]
- Johnston L.A. and Gallant, P. 2002. Control of growth and organ size in Drosophila. Bioessays 24: 54–64. [DOI] [PubMed] [Google Scholar]
- Junger M.A., Rintelen, F., Stocker, H., Wasserman, J.D., Vegh, M., Radimerski, T., Greenberg, M.E., and Hafen, E. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M., Verdu, J., Lachance, P.E., Birnbaum, M.J., Lasko, P.F., and Sonenberg, N. 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3: 596–601. [DOI] [PubMed] [Google Scholar]
- Miron M., Lasko, P., and Sonenberg, N. 2003. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol. Cell. Biol. 23: 9117–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S., Both, G.W., Furuichi, Y., and Shatkin, A.J. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255: 33–37. [DOI] [PubMed] [Google Scholar]
- Puig O., Marr, M.T., Ruhf, M.L., and Tjian, R. 2003. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes & Dev. 17: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J.H., Doepfner, K.T., Hafen, E., and Stocker, H. 2005. Diet-dependent effects of Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr. Biol. 15: 24–30. [DOI] [PubMed] [Google Scholar]
- Rolli-Derkinderen M., Machavoine, F., Baraban, J.M., Grolleau, A., Beretta, L., and Dy, M. 2003. ERK and p38 inhibit the expression of 4E-BP1 repressor of translation through induction of Egr-1. J. Biol. Chem. 278: 18859–18867. [DOI] [PubMed] [Google Scholar]
- Thomas G. 2002. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 35: 305–313. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Poulin, F., Kohara, M., DeMaria, C.T., Cheng, A., Wu, Z., Gingras, A.C., Katsume, A., Elchebly, M., Spiegelman, B.M., et al. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7: 1128–1132. [DOI] [PubMed] [Google Scholar]
- Van der Horst D.J. 2003. Insect adipokinetic hormones: Release and integration of flight energy metabolism. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136: 217–226. [DOI] [PubMed] [Google Scholar]
- Zinke I., Schutz, C.S., Katzenberger, J.D., Bauer, M., and Pankratz, M.J. 2002. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 21: 6162–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Meadows, S., Sharp, L., Jan, L.Y., and Jan, Y.N. 2000. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. 97: 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]