Abstract
Eukaryotic cells regulate the progression and integrity of DNA replication forks to maintain genomic stability and couple DNA synthesis to other processes. The budding yeast proteins Mrc1 and Tof1 associate with the putative MCM–Cdc45 helicase and limit progression of the replisome when nucleotides are depleted, and the checkpoint kinases Mec1 and Rad53 stabilize such stalled forks and prevent disassembly of the replisome. Forks also pause transiently during unperturbed chromosome replication, at sites where nonnucleosomal proteins bind DNA tightly. We describe a method for inducing prolonged pausing of forks at protein barriers assembled at unique sites on a yeast chromosome, allowing us to examine for the first time the effects of pausing upon replisome integrity. We show that paused forks maintain an intact replisome that contains Mrc1, Tof1, MCM–Cdc45, GINS, and DNA polymerases α and ε and that recruits the Rrm3 helicase. Surprisingly, pausing does not require Mrc1, although Tof1 and Csm3 are both important. In addition, the integrity of the paused forks does not require Mec1, Rad53, or recombination. We also show that paused forks at analogous barriers in the rDNA are regulated similarly. These data indicate that paused and stalled eukaryotic replisomes resemble each other but are regulated differently.
Keywords: DNA replication forks, Mrc1, Tof1, Csm3, checkpoint, recombination
Chromosomal DNA replication in eukaryotic cells is initiated from multiple origins on each chromosome, each of which is activated just once during the S phase of each cell cycle, to ensure that a single copy of the genome is made (Kearsey and Cotterill 2003; Diffley 2004; Stillman 2005). DNA replication forks are established at each origin and then move away as the parental DNA duplex is unwound by the action of DNA helicases. In order to preserve genomic stability and prevent lethal damage to the chromosomes during mitosis, it is crucial that progression of the two converging forks from each pair of adjacent origins continues until the forks meet each other and termination occurs. Despite this fact, individual forks can stop for a variety of reasons before termination, and eukaryotic cells have thus evolved a variety of mechanisms that regulate both the progression and stability of DNA replication forks.
Much of our understanding of the regulation of eukaryotic DNA replication forks has come from studies of budding yeast cells treated with the drug hydroxyurea (HU), which inhibits ribonucleotide reductase and thus causes forks to stall. The stalling of fork progression is partly a consequence of the reduced availability of dNTPs but also requires the two replisome components Mrc1 and Tof1 (Katou et al. 2003). In the absence of these proteins, the replisome progresses faster than it is able to synthesize DNA; it thus appears that individual replisomes play an active role in the stalling of forks in response to HU. The mechanism by which Mrc1 and Tof1 restrain the progression of forks is not understood, but they both interact with the Cdc45–MCM2–7 complex that is thought to act as the DNA helicase responsible for unwinding the parental DNA duplex at DNA replication forks (Katou et al. 2003; Pacek and Walter 2004; Shechter et al. 2004; Nedelcheva et al. 2005).
Mrc1 and Tof1 are also both important for the activation of a “checkpoint” response following nucleotide depletion by HU. The budding yeast kinases Mec1 and Rad53 block mitosis and also inhibit initiation events at later origins of DNA replication under such conditions. When cells lacking either Mrc1 or Tof1 are treated with HU, neither anaphase nor the firing of late origins of DNA replication is inhibited, showing that checkpoint activation is partially defective (Alcasabas et al. 2001; Foss 2001; Katou et al. 2003). It thus appears that Mrc1 and Tof1 share a similar role at HU-stalled forks.
Checkpoint activation is also essential in order to preserve the stability of HU-stalled DNA replication forks (Lopes et al. 2001) and thus prevent disassembly of the replisome (Cobb et al. 2003). In addition, both Mec1 and Rad53 are essential to maintain the integrity of DNA replication forks that stall upon encountering DNA damage induced by the alkylating agent methyl methanesulphonate (MMS) (Tercero and Diffley 2001); cells lacking either kinase are therefore exquisitely sensitive to the stalling of DNA replication forks by HU or MMS (Allen et al. 1994; Desany et al. 1998; Tercero and Diffley 2001).
DNA replication forks can also pause transiently during the normal process of chromosome replication, upon encountering sites where nonnucleosomal proteins are bound tightly to DNA (Greenfeder and Newlon 1992; Wang et al. 2001; Ivessa et al. 2002, 2003; Makovets et al. 2004). It has been estimated that there are >1000 such pause sites in the budding yeast genome (Ivessa et al. 2003), and they are thought to represent an important challenge to the maintenance of genomic stability, as there is evidence that such paused forks may break and that their subsequent repair by homologous recombination may provide a source of genetic variability (Keil and McWilliams 1993; Ivessa et al. 2000, 2002, 2003; Ahn et al. 2005; Lambert et al. 2005; Prado and Aguilera 2005).
The consequences of the pausing of eukaryotic DNA replication forks at protein–DNA barriers are much less well understood than is the case for HU-stalled forks, principally because forks only pause very transiently at most sites, so that a molecular analysis is much more complicated. Previous studies have not determined the kinetics of pausing at individual sites within the genome, and many fundamental questions remain to be answered. Most importantly, it is not known whether the replisome disassembles when a fork encounters a protein–DNA barrier or whether replisome proteins, including regulators such as Mrc1 and Tof1, remain stably associated with the fork, as is the case at HU-stalled forks. Previous work suggested that the MCM2–7 complex must be maintained at DNA replication forks in an uninterrupted fashion between initiation and termination, as removal of MCM proteins from HU-stalled forks blocks irreversibly the recovery of DNA synthesis (Labib et al. 2000), but it has previously not been possible to test whether the putative MCM helicase does indeed remain associated with paused forks, as would be predicted by this model. It would also be interesting, for example, to examine whether proteins such as the primase associated with DNA polymerase α remain at the paused fork, as lagging strand synthesis is completed rapidly after pausing occurs.
It appears, however, that the progression of paused forks is actively regulated in a somewhat analogous fashion to HU-stalled forks, as the fission yeast homolog of Tof1 is required, together the associated Swi3 protein, for forks to pause at the mating-type locus and in the rDNA (Dalgaard and Klar 2000; Krings and Bastia 2004). This suggests that the progression of paused and stalled forks may be restrained by the same mechanism, although the role of Mrc1 at paused forks has not been tested directly.
It is presently unclear whether checkpoint kinases play a role in preserving the stability of paused forks in budding yeast. Both Mec1 and Rad53 are essential for viability even in the absence of replication stress, and cells lacking Mec1 accumulate double-strand breaks in DNA and are unable to complete chromosome replication (Zhao et al. 2001; Cha and Kleckner 2002). The lethal effects of deleting the MEC1 or RAD53 genes can be suppressed by overexpressing the large subunit of ribonucleotide reductase (Desany et al. 1998) or by additionally deleting the gene encoding the Sml1 protein, which is an inhibitor of ribonucleotide reductase (Zhao et al. 1998; Chabes et al. 1999). Previous studies of budding yeast have not determined whether DNA replication forks still pause at protein barriers under such conditions.
The rDNA in budding yeast contains an efficient replication fork barrier sequence (RFB) that actively blocks the progression of DNA replication forks when bound tightly by a specific protein called Fob1 (Fork block 1) (Brewer and Fangman 1988; Linskens and Huberman 1988; Kobayashi and Horiuchi 1996; Kobayashi 2003; Mohanty and Bastia 2004). As shown in Figure 1A (panel i), each of the ∼200 rDNA repeats contains a potential origin of bidirectional DNA replication. Initiation occurs within a limited number of the repeats during S phase, and the rightward fork is free to progress into the adjacent unit, in the same direction as transcription. The leftward fork passes through a 5S rRNA gene without opposing transcription, but then pauses at the RFB (Brewer and Fangman 1988; Linskens and Huberman 1988). The barrier only blocks the progression of leftward forks, so that replication of the rDNA repeats occurs principally in the same direction as the highly active transcription by RNA PolI.
Figure 1.
Using the Fob1–RFB system to pause specific DNA replication forks on chromosome 3. (A, panel i) Map of two consecutive repeats of the rDNA on chromosome 12 of budding yeast. The positions of the replication fork barrier (RFB), origin of DNA replication (ARS), 35S rRNA (35S), and 5S rRNA (5S) genes are indicated. (Panel ii) Insertion of RFB sequences on chromosome 3 to block the forks from ARS305 and ARS306. The distances indicated are measured from the left end of chromosome 3; “B” and “S” mark the positions of BclI and SalI restriction enzyme sites used for the two-dimensional DNA gels. See Materials and Methods and Supplementary Figure 1 for more information. (Panel iii) Using two-dimensional DNA gels to study DNA replication intermediates; see text for details. The two spots below the Y-arc in the left panel correspond to other sites in the genome that are recognized weakly by the chosen probe. (B) DNA replication forks do not pause at the RFBs on chromosome 3 in the absence of Fob1. (C, panel i) In cells expressing Fob1, the forks from ARS305 and ARS306 pause for an extended period at the two RFBs on chromosome 3. (Panel ii) The histograms show a quantification of paused forks at the indicated times (calculated as described in Materials and Methods).
The highly repetitive nature of the rDNA impedes the analysis of individual paused DNA replication forks, and each leftward fork at the RFB is generally resolved by the arrival of a rightward fork from the preceding repeat (Brewer and Fangman 1988; Linskens and Huberman 1988). We describe a system that exploits the efficiency of the Fob1–RFB to allow extended but finite pausing of specific DNA replication forks that originate from adjacent origins on chromosome 3 of budding yeast. By studying the composition and regulation of such individual paused eukaryotic replisomes, we identify similarities with, but also important differences from, their counterparts at HU-stalled forks.
Results
A model system for studying paused eukaryotic replisomes
Our initial aim was to create an experimental system with which to study the fate of eukaryotic replisomes when DNA replication forks pause transiently at protein–DNA barriers during the process of chromosome replication. We wanted to study forks from the earliest origins so that generation of the paused forks would not be affected if we subsequently used strains or conditions that activate checkpoint kinases, which inhibit the firing of later origins. Previous studies determined the location and timing of origins of DNA replication throughout the budding yeast genome (Raghuraman et al. 2001), and we used this information to design a strain in which two early forks would pause for an extended but finite period of time at unique sites, without being resolved by the arrival of forks from a neighboring replicon. The two origins ARS305 and ARS306 on chromosome 3 are separated by 36 kb and are among the earliest and most active origins in the genome. In almost every cell cycle, a rightward fork from ARS305 and a leftward fork from ARS306 enter the intervening region with similar timing and subsequently meet toward the center of the region (Reynolds et al. 1989; Raghuraman et al. 2001). RFB sequences from the rDNA were inserted within this region, ∼10 kb from each origin, so that the forks from both ARS305 and ARS306 would pause with similar kinetics, without being resolved by a fork from elsewhere in the genome (see Fig. 1A, panel ii; Supplementary Fig. 1). We used a yeast strain in which expression of the FOB1 gene was controlled by the regulatable GAL1,10 promoter, so that we could switch-off FOB1 and thus grow cells without activating the RFBs between ARS305 and ARS306, synchronize cells in the G1 phase of the cell cycle, and then rapidly induce FOB1 to activate the barriers before examining the immediate consequences in the subsequent round of chromosome replication.
We used two-dimensional (2D) “neutral-neutral” DNA gels (Brewer and Fangman 1987) to study DNA replication intermediates isolated from samples taken every 15 min after releasing cells from G1 arrest. After digestion of genomic DNA with appropriate restriction enzymes, both unreplicated and fully-replicated linear DNA fragments corresponding to a particular locus migrate in an identical fashion in both dimensions, producing a prominent spot in the bottom right of the gel (Fig. 1A, panel iii). In contrast, restriction fragments that contain replication intermediates are retarded in the first dimension by virtue of their greater size, and are also retarded in the second dimension due to their abnormal shape. Passive replication of the region produces a characteristic “Y-arc” beginning at the spot corresponding to linear DNA and ending at a point equivalent to a restriction fragment that is just less than fully replicated (Fig. 1A, panel iii, left). Pausing of the fork at a specific site produces an accumulation of molecules with the same size and shape, corresponding to a distinctive spot on the Y-arc (Fig. 1A, panel iii, middle, spot labeled RFB).
Initially we grew cells in the absence of Fob1 throughout the experiment and examined replication of the two barrier sites together with an intervening locus (Fig. 1B). We observed simple Y-arcs at all three sites, peaking between 30 and 45 min after release from G1 arrest. This shows that the two forks from ARS305 and ARS306 replicate the region without pausing significantly at the RFB sequences in the absence of the Fob1 protein.
We then released cells from G1 arrest after first switching-on expression of Fob1. As shown in Figure 1C, a strong spot appeared on the Y-arc for each of the two forks, at a position corresponding to the RFB sequence within the restriction fragment (Fig. 1C, panel i, top and bottom). The rightward fork from ARS305 and the leftward fork from ARS306 arrive at the corresponding RFB with very similar kinetics and then pause for an extended period relative to the time required to replicate the rest of the genome (Fig. 1C, panel ii; Supplementary Fig. 2). Pausing of the two forks delays replication of the site between the two barriers, although Y-arcs are eventually observed at later times (Fig. 1C, panel i, middle), indicating that DNA synthesis does subsequently resume at the paused forks, and cells are thus able to grow well in the continued presence of active barriers on chromosome 3 (Supplementary Fig. 3).
Pausing of forks at the Fob1–RFB does not cause replisome disassembly
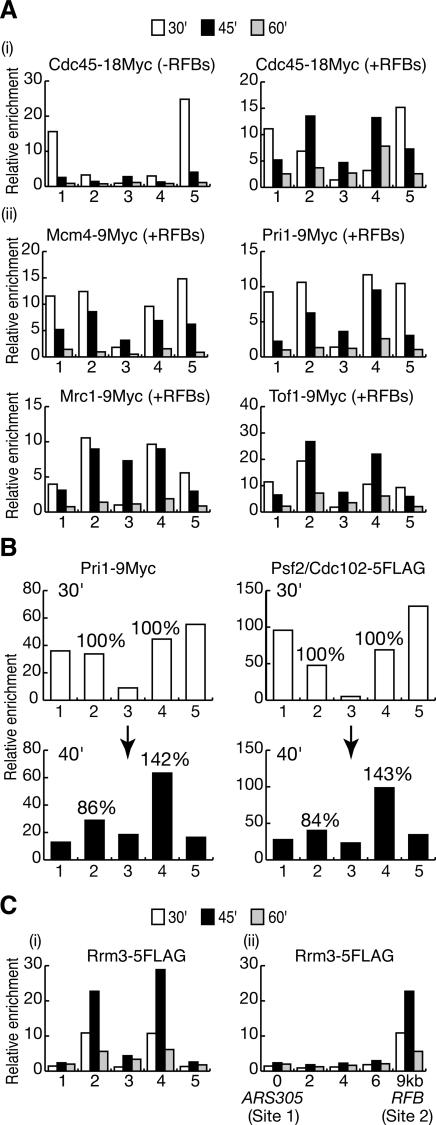
The preceding experiments show that the introduction of two RFBs between ARS305 and ARS306 generates an ideal system with which to study the protein composition and stability of paused eukaryotic replisomes, as the two forks pause for an extended but finite period at unique sites. We could thus examine the kinetics with which the replisome arrives at the RFB and is either maintained or disassembled, by using cultures of cells that are replicating their chromosomes synchronously. We used chromatin immunoprecipitation (ChIP) to examine the composition of the replisome under such conditions, by treating cells with formaldehyde before preparing an extract that was sonicated to shear chromosomal DNA into fragments of several hundred base pairs; we then isolated the protein of interest by immunoprecipitation. We used a “real-time” version of the polymerase chain reaction (PCR) to determine quantitatively the association of replisome proteins with specific DNA sequences in vivo (see Materials and Methods), and initially examined five sites on chromosome 3: ARS305, a site very close to where the first RFB was inserted, a site between the two barriers, a site very close to the second RFB, and ARS306 (sites 1–5 in Fig. 2A; Supplementary Fig. 4). In all the subsequent experiments, cells were grown under identical conditions to those described above for the experiment shown in Figure 1C.
Figure 2.
Pausing of forks at the RFBs does not cause the replisome to disassemble. (A) ChIP was used to study the localization of proteins at the five sites indicated. Further details can be found in Supplementary Figure 4. (B) Cells were synchronized in the G1 phase of the cell cycle before expressing GAL-FOB1-9MYC for 45 min, maintaining the G1 arrest throughout. The Fob1-9Myc protein was detected specifically at the two RFBs introduced on chromosome 3. (C [panel i], D) The localization of Pol2-9Myc or Psf2/Cdc102-9Myc was examined at the same five sites, as cells entered synchronously into the S phase of the cell cycle in the presence (+RFBs) or absence (–RFBs) of the Fob1–RFBs on chromosome 3; see text for details. (C, panel ii) The region between ARS305 and the corresponding RFB was also examined with higher resolution.
To confirm that we were indeed able to detect proteins present at the RFB sites on chromosome 3, we used ChIP to examine the putative association of the Fob1 protein at the five sites mentioned above, and observed a strong enrichment in the Fob1 immunoprecipitates of the DNA sequences close to each of the two RFBs introduced on chromosome 3 (Fig. 2B, sites 2 and 4). Although the resolution of ChIP is limited to several hundred nucleotides, this experiment confirmed that we could use our assay to detect proteins that accumulate at the RFB sites on chromosome 3, where DNA replication forks pause during the process of chromosome replication.
We thus proceeded to examine the association of replisome proteins with the same sites at different points during the S phase of the cell cycle. We started by examining Pol2, the catalytic subunit of DNA polymerase ε, which has previously been shown to remain associated with the stalled replisome at DNA replication forks when cells are treated with HU (Aparicio et al. 1997, 1999; Tanaka and Nasmyth 1998; Masumoto et al. 2000; Cobb et al. 2003). In a strain lacking RFB sequences on chromosome 3, Pol2 was strongly enriched at ARS305 and ARS306 30 min after release from G1 arrest (Fig. 2C, panel i, –RFBs, sites 1 and 5) and could also be observed ∼10 kb from each origin (sites 2 and 4), consistent with its presence at active DNA replication forks. Fifteen minutes later, association of Pol2 with the above sites was greatly reduced, and instead, the protein could be detected toward the middle of the region (site 3), as replication was completed.
We then examined Pol2 in a strain with active RFBs on chromosome 3. Thirty minutes after release from G1 arrest, the association of Pol2 with ARS305 and ARS306 was similar to that seen in the previous experiment (Fig. 2C, panel i, +RFBs, sites 1 and 5). Strikingly, however, enrichment of the RFB-proximal sequences was greatly increased in the Pol2 immunoprecipitates (sites 2 and 4). Moreover, we observed that these sequences were still strongly enriched in the Pol2 immunoprecipitates after 45 min, in contrast to the strain lacking RFBs. By 60 min, the association of Pol2 with the RFB-proximal sequences was largely but not completely diminished.
We also examined with higher resolution the 10-kb region to the right of ARS305 (Fig. 2C, panel ii). At 30 min after release from G1 arrest, Pol2 was detected throughout this region regardless of the presence or absence of RFBs on chromosome 3; 15 min later, however, only the sequence next to the RFB insertion site was strongly associated with Pol2, and this was dependent upon presence of the RFB. Taken together, these experiments indicate that Pol2 remains associated with the forks from ARS305 and ARS306 that pause at the two RFBs on chromosome 3.
We next examined the GINS complex, which plays an essential though poorly characterized role during chromosome replication and has been shown to associate with stalled forks after HU treatment (Kanemaki et al. 2003; Kubota et al. 2003; Takayama et al. 2003). We used ChIP to determine the association of the GINS subunit Psf2/Cdc102 with the region between ARS305 and ARS306 during S phase. As shown in Figure 2D, GINS behaves in a very similar fashion to Pol2, associating 30 min after release from G1 arrest with both of the origins and also with DNA replication forks (Fig. 2D, –RFBs and +RFBs), and then remaining associated with the two paused DNA replication forks (Fig. 2D, +RFBs 45 min), until replication of the region resumes and is completed (Fig. 2D, +RFBs 60 min). We conclude, therefore, that GINS remains associated with paused DNA replication forks.
Previous studies of forks that stall after depletion of nucleotides showed that the putative MCM–Cdc45 helicase complex remains part of the stalled replisome together with the two associated proteins, Mrc1 and Tof1 (Aparicio et al. 1997; Katou et al. 2003; Osborn and Elledge 2003). As shown in Figure 3A, panels i,ii, Mcm4, Cdc45, Mrc1, and Tof1 are all retained at the site of the paused forks originating from ARS305 and ARS306. We also examined the Pri1 protein that forms part of the DNA polymerase α-primase complex, and we found that this also associates with the paused DNA replication forks (Fig. 3A, panel ii), just as previously shown for HU-stalled forks (Tanaka and Nasmyth 1998; Aparicio et al. 1999; Masumoto et al. 2000; Cobb et al. 2003; Lucca et al. 2004), despite the rapid completion of lagging-strand synthesis behind the pause site (Lucchini and Sogo 1994; Gruber et al. 2000).
Figure 3.
Other components of the paused replisome. (A, panel i) Cdc45 accumulates during S phase at the sites of the paused forks (+RFBs) but is more transiently associated with the same sites in the absence of pausing (–RFBs). (Panel ii) Mcm4, Mrc1, Tof1, and Pri1 all remain associated with the forks that pause at the Fob1–RFBs on chromosome 3. (B) The localization of Pri1-9Myc and Cdc102-5Flag was determined in the same cells at the indicated times. The change in the specific enrichment of the RFB sequences is shown from 30 min (100%) to 40 min. (C) Rrm3 is specifically recruited to paused replisomes (panel i), and is not a stable component of active forks established at origins of replication (panel ii).
To provide stronger evidence that the various proteins described above all associate in a similar manner with the paused forks (despite small differences in kinetics seen from one experiment to another), we examined two different replisome components in the same cells. In the experiment shown in Figure 3B, both Pri1 and Psf2/Cdc102 associated in a similar manner with the two RFB sites 30 min after release from G1 arrest (Fig. 3B, 30 min, sites 2 and 4). Ten minutes later, the enrichment of the ARS305-proximal barrier in the Pri1 immunoprecipitate had reduced slightly to 86% of the value at 30 min, whereas the enrichment of the ARS306-proximal barrier had increased to 142% of the value at 30 min. For Psf2/Cdc102, the enrichment of the ARS305-proximal barrier at 40 min decreased to 84% of the value at 30 min, whereas the enrichment of the ARS306-proximal barrier increased to 143% of the value at 30 min. This experiment shows that the behavior of Pri1 and Psf2/Cdc102 at the RFBs was extremely similar. Overall, therefore, these experiments lead us to conclude that the replisome does not disassemble when a eukaryotic DNA replication fork pauses upon encountering a protein–DNA barrier. Instead, the paused fork retains an intact replisome, analogous to the replisome that is maintained at HU-stalled forks.
The Rrm3 helicase is specifically recruited to paused eukaryotic replisomes
It is possible that some proteins that function at DNA replication forks do not normally form part of the active replisome but instead are recruited specifically to paused or stalled replisomes. The Rrm3 DNA helicase plays an important role in helping forks progress past protein–DNA barriers at many sites across the budding yeast genome (Ivessa et al. 2003), including the Fob1–RFB in the rDNA, but it is not clear if Rrm3 assembles as part of the replisome at origins during the initiation of chromosome replication or instead is specifically recruited to paused DNA replication forks. We therefore examined the Rrm3 helicase in a strain with RFBs on chromosome 3, and saw that the protein accumulates specifically at the sites of the paused forks, with similar kinetics to other components of the paused replisome (Fig. 3C). Importantly, however, we could not detect significant association of Rrm3 with the two origins ARS305 and ARS306 (Fig. 3C, panel i), or with other sites between ARS305 and the corresponding RFB (Fig. 3C, panel ii). This suggests that Rrm3 is not normally a stable component of DNA replication forks but instead is principally recruited to paused replisomes.
Mrc1 is not essential for the replisome to pause at the Fob1–RFB
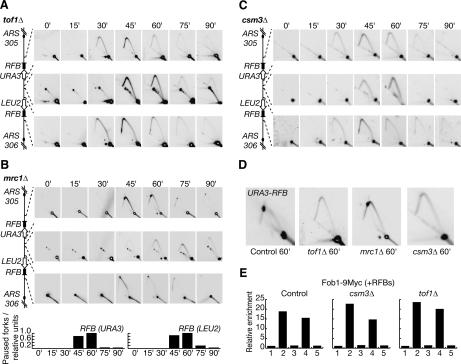
At HU-stalled forks, both Mrc1 and Tof1 play a similarly important role in restraining progression of the stalled replisome (Katou et al. 2003). We have shown that both proteins are also components of the paused replisome at the Fob1–RFB (Fig. 3), and it might be expected therefore that both Mrc1 and Tof1 are equally important for pausing, particularly as the fission yeast homolog of Tof1 has already been shown to be important for forks to pause within the mating-type locus and in the rDNA (Dalgaard and Klar 2000; Krings and Bastia 2004). We therefore used 2D DNA gels as above to examine the progression of forks from ARS305 and ARS306 in the absence of either Tof1 or Mrc1. As shown in Figure 4A, both forks pass the corresponding RFB sites with very little sign of pausing in a strain lacking the Tof1 protein. In striking contrast, forks from ARS305 and ARS306 are still able to pause at the RFBs in the absence of Mrc1 (Fig. 4B), thereby identifying an important difference in regulation between forks that pause at a protein–DNA barrier and HU-stalled forks.
Figure 4.
Mrc1 is not essential for DNA replication forks from ARS305 and ARS306 to pause at the RFBs on chromosome 3, but Tof1 and Csm3 are both important. Fork progression was examined as above in cells lacking Tof1 (A), Mrc1 (B), or Csm3 (C). (D) Larger versions of the data from the 60-min time point. (E) Fob1 still associates with the RFBs on chromosome 3 in the absence of Tof1 or Csm3. Cells were synchronized in G1 phase, and expression of Fob1-9Myc was then induced for 45 min.
Work with fission yeast has shown that another protein, Swi3, is also required in addition to the homolog of Tof1 for forks to pause in the mating locus and in the rDNA (Dalgaard and Klar 2000; Krings and Bastia 2004). The budding yeast protein Csm3 (Rabitsch et al. 2001) is similar in sequence to Swi3, and previous work has shown that Tof1 and Csm3 interact with each other (Mayer et al. 2004). We thus examined the progression of forks from ARS305 and ARS306 in cells lacking Csm3, and saw once again that pausing at the RFB sites was greatly reduced (Fig. 4C).
These experiments indicate that Tof1-Csm3 and Mrc1 are functionally distinct, at least in the context of a paused DNA replication fork (an enlarged version of the 60-min time point is shown in Figure 4D to illustrate the point more clearly). It was important to confirm that the barrier was still intact in the absence of Tof1 and Csm3, and we therefore used ChIP to show that Fob1 still associated with the two RFB sites on chromosome 3 in these strains, just as in the control (Fig. 4E).
Previous work showed that progression of the replisome is uncoupled from DNA synthesis at HU-stalled forks in cells lacking Mrc1 or Tof1 (Katou et al. 2003). It was possible, therefore, that DNA synthesis might indeed be arrested at the RFB in cells lacking Mrc1, as indicated by the 2D gels, but the replisome (or components such as the MCM–Cdc45 helicase) may still progress beyond the RFB. In this case, both Mrc1 and Tof1 would be important to restrain progression of the replisome at a paused fork, just as at HU-stalled forks, but Tof1 (and Csm3) would have an additional role in restraining DNA synthesis, for example, by inhibiting progression of a DNA polymerase. To test this possibility, we used ChIP to determine the behavior of Cdc45 in cells lacking either Mrc1 or Tof1, and to aid the analysis we grew both strains in parallel in the same experiment (Fig. 5). Thirty minutes after release from G1 arrest, Cdc45 associated principally with ARS305 and ARS306 but also associated with the sites ∼10 kb from each origin, and the observed pattern of localization was very similar in cells lacking either Mrc1 or Tof1. Fifteen minutes later, however, the situation was very different in the two strains: In cells lacking Mrc1, the sites close to the two RFBs were strongly enriched in the immunoprecipitate of Cdc45 (Fig. 5, mrc1Δ, sites 2 and 4); in the absence of Tof1, however, Cdc45 did not accumulate at the RFB sites, and instead the site toward the middle of the region was strongly enriched in the Cdc45 immunoprecipitate (Fig. 5, tof1Δ, site 3). This experiment, together with the 2D data described above, shows that progression of the replisome is still coupled to DNA synthesis in the mrc1Δ strain. Mrc1 is not essential, therefore, for the replisome to pause at the Fob1–RFB, in striking contrast to the important role played by Tof1.
Figure 5.
Pausing of the replisome at the RFBs on chromosome 3 is independent of Mrc1 but requires Tof1. The association of Cdc45 with the region between ARS305 and ARS306 was examined by ChIP at 30 min or 45 min after release of cells from G1 arrest.
We wanted to test whether the observations made at the RFBs on chromosome 3 were also true for the endogenous RFB in the rDNA repeats on chromosome 12. We therefore examined replication of the rDNA repeats in the same experiments described above (the region of interest and the expected products in the 2D gels are shown in Fig. 1A, panels i,iii [right]; Supplementary Fig. 1C). As shown in Figure 6, a strong spot on the Y-arc is observed at the site of the RFB during replication of the rDNA repeats (Fig. 6A, panel i, Control), together with a “bubble arc” representing activation of the rDNA origin of replication. One or two vertical lines emerge from the RFB spot in the 2D gels; such structures probably represent reversed forks or “chicken feet” (Vengrova and Dalgaard 2004) at each of the two adjacent pause sites within the RFB (Brewer et al. 1992; Gruber et al. 2000). These structures are not detected at the RFB in vivo (Lucchini and Sogo 1994) and may thus form in vitro during the isolation of genomic DNA containing DNA replication forks. They form in the absence of recombination (see Fig. 9C, below), and the accumulation of forks at the unique RFB site within the multiple rDNA repeats may simply aid their detection.
Figure 6.
(A) Mrc1 is not essential for DNA replication forks to pause at the endogenous RFB within the rDNA, but Tof1 and Csm3 are both important for pausing. (B) Larger versions of the data from the 45-min time point. The arrows mark the site where forks normally pause at the RFB (1), as well as a region beyond this point where weak pausing can be seen in the absence of Tof1 or Csm3 (2).
Figure 9.
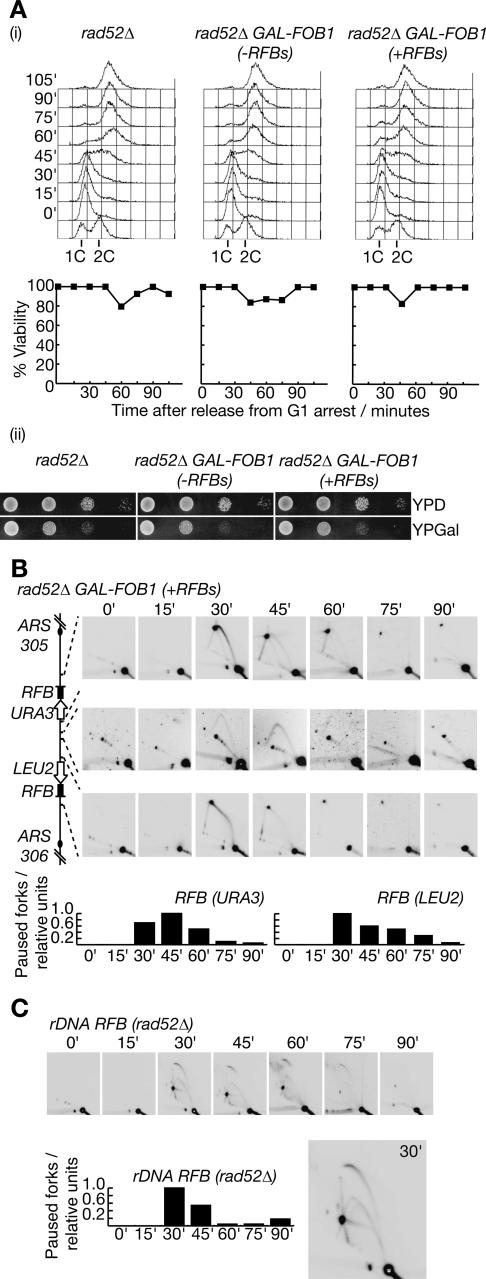
Recovery of DNA synthesis from forks paused at the Fob1–RFB does not require recombination. (A, panel i) Viability of the indicated strains was measured as cells were released from G1 phase in the presence of Fob1, as in previous experiments. DNA content was measured by flow cytometry. (Panel ii) Growth of cells lacking Rad52 is not affected by extended pausing of DNA replication forks at the RFBs on chromosome 3. Serial dilutions of cells were plated on YPD medium (inactive RFBs) and YPGal medium (active RFBs) and photographed after 48 h growth at 25°C. Note that yeast cells grow more slowly on YPGal medium. (B) Progression of DNA replication forks through the region between ARS305 and ARS306 was examined as before. (C) Replication of the rDNA repeats was examined in the same experiment.
In cells lacking Mrc1 or Tof1, replication of the rDNA occurs over a shorter period, perhaps reflecting changes in origin efficiency or copy number within the rDNA repeats; a similar though milder effect is observed in the absence of Csm3 (Fig. 6A). The behavior of DNA replication forks at the RFB is strikingly different, however, in the three strains. Forks still pause strongly at the RFB within the rDNA in the absence of Mrc1 (Fig. 6A, panel i), as shown above for chromosome 3. Pausing at the RFB is greatly reduced in the absence of Tof1 or Csm3 and is associated with weak pausing at a point beyond the normal RFB site (Fig. 6B). This latter pause site is independent of Fob1 (Supplementary Fig. 5), and may correspond to forks that clash with 3′ end of the 35S rRNA gene when pausing at the RFB is prevented, as reported previously (Takeuchi et al. 2003). The behavior of forks at the RFB within the rDNA repeats is thus strikingly different in cells lacking either Mrc1 or Tof1-Csm3, consistent with our observations of individual forks that pause at the RFBs on chromosome 3.
Pausing and recovery of eukaryotic forks without checkpoint kinases
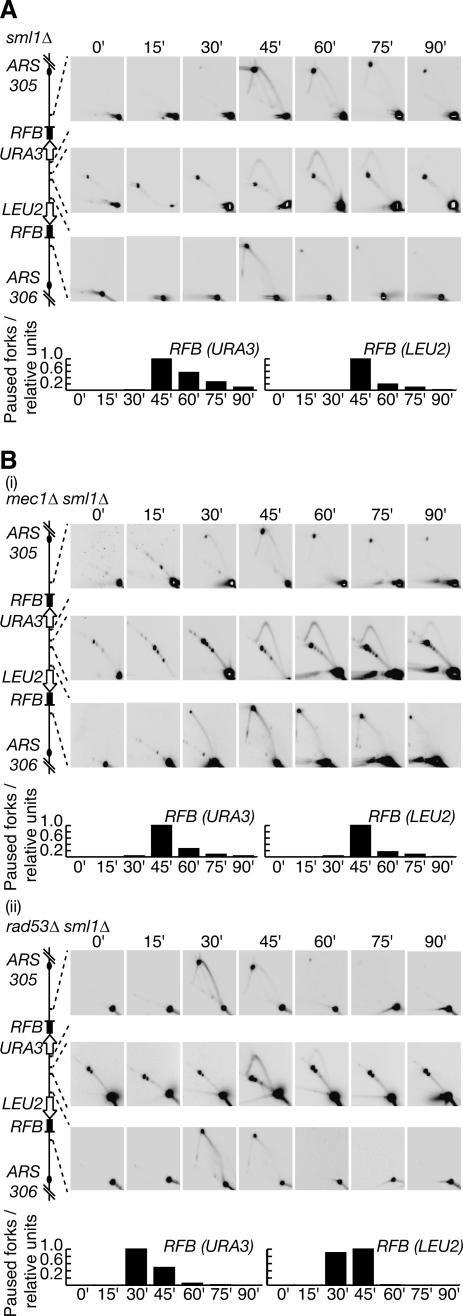
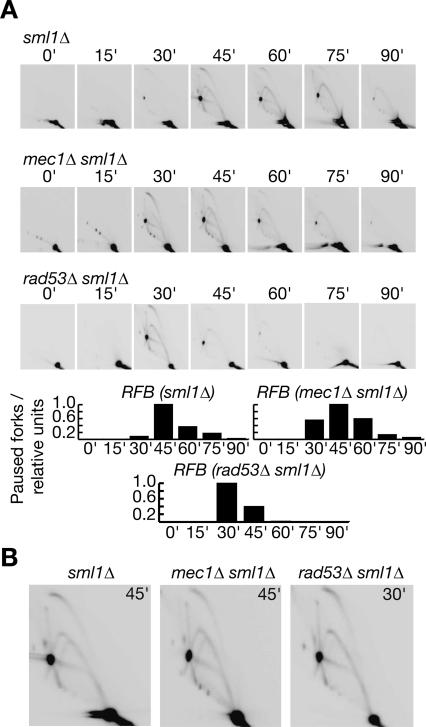
When DNA replication forks stall in response to the depletion of nucleotides, checkpoint kinases such as the budding yeast proteins Mec1 and Rad53 are essential to preserve the integrity of the stalled fork, and thus ensure that replication can resume subsequently (Lopes et al. 2001). As both Mec1 and Rad53 are also essential for cell viability even in the absence of replication stress, we examined cells lacking either kinase in addition to the Sml1 protein that inhibits ribonucleotide reductase (see Introduction). We first assayed the progression of forks from ARS305 and ARS306 in cells lacking Sml1. As shown in Figure 7A, both forks still paused at the corresponding RFB, just as in a wild-type strain. We then performed a similar experiment using cells lacking both Mec1 and Sml1. Strikingly, replication of the region between ARS305 and ARS306 was very similar to the sml1Δ control: The forks from each origin paused at the corresponding barrier as before, and DNA synthesis then resumed subsequently, so that replication of the region was completed (Fig. 7B, panel i). We also showed that the forks from ARS305 and ARS306 pause and recover in a similar manner in cells lacking both Rad53 and Sml1 (Fig. 7B, panel ii; note that the onset of S phase is slightly advanced in this strain); moreover, neither Mec1 nor Rad53 is essential for pausing or recovery of forks at the endogenous RFB within the rDNA (Fig. 8). We thus conclude that the integrity of forks that pause at the Fob1–RFB is independent of checkpoint kinases, contrasting once again with the regulation of HU-stalled forks.
Figure 7.
Stability of forks that pause at the RFBs on chromosome 3 does not require checkpoint kinases. Progression of forks was examined as before in sml1Δ (A), mec1Δ sml1Δ (B, panel i) or rad53Δ sml1Δ (B, panel ii) strains.
Figure 8.
Stability of paused forks at the RFB in the rDNA is independent of checkpoint kinases. (A) Progression of forks was examined as before in sml1Δ, mec1Δ sml1Δ, or rad53Δ sml1Δ strains. (B) Larger versions of individual time points are shown for the three strains.
Recovery of paused forks at the Fob1–RFB without recombination
It has often been suggested that the pausing of DNA replication forks may lead to breakage that is then repaired by homologous recombination (Keil and McWilliams 1993; Ivessa et al. 2000, 2002, 2003; Weitao et al. 2003; Ahn et al. 2005; Lambert et al. 2005; Prado and Aguilera 2005), but previous studies have not established whether such events represent an important mechanism by which DNA synthesis normally resumes at paused forks in eukaryotic cells, or else result from rare failures to achieve recovery by other means. By studying the kinetics of replication of individual replicons that contain specific pause sites, we have shown that forks from ARS305 and ARS306 pause for an extended period with an intact replisome at the RFB sites between the two origins, before DNA synthesis resumes at each fork and replication of the region is rapidly completed. Our data suggest, therefore, that DNA synthesis normally resumes without breakage or recombination at the RFBs, and we have not observed recombination intermediates at the paused forks on chromosome 3 (X-shaped molecules with double the DNA content of the unreplicated restriction fragment, which would have been seen in the 2D gels as a vertical line emerging from the bottom-left corner of the Y-arc).
The Rad52 protein is essential for practically all forms of homologous recombination in eukaryotic cells, including recombination between the rDNA repeats (Gangloff et al. 1996; Park et al. 1999; Ivessa et al. 2000). Budding yeast cells lacking Rad52 frequently become arrested at the G2/M phase of the cell cycle and are highly sensitive to the drug HU that causes DNA replication forks to stall (Bennett et al. 2001; Chang et al. 2002; Shor et al. 2002), probably reflecting a failure to repair double-strand breaks in DNA that occur during chromosome replication. To examine the importance of recombination in the resumption of DNA synthesis after extended pausing of DNA replication forks at the RFBs on chromosome 3, we measured the viability of cells lacking Rad52 as they entered the cell cycle synchronously and replicated their chromosomes, either in the presence or absence of extended pausing of forks at the RFBs on chromosome 3.
As shown in Figure 9A, panel i, we only detected minor changes in the viability of rad52Δ cells throughout the experiment, regardless of the presence or absence of RFBs on chromosome 3. Moreover, cells lacking Rad52 were able to form colonies just as well in the presence of active barriers on chromosome 3 as in their absence (Fig. 9A, panel ii). We used 2D gels to confirm that the barriers were indeed active in the absence of Rad52, and as before, we saw that the forks from ARS305 and ARS306 paused for an extended period at the corresponding RFBs, before the resumption of DNA synthesis allowed replication of the intervening region to be completed (Fig. 9B). We also examined the endogenous RFB in the rDNA in the same experiment, and found that the paused forks behaved as in previous experiments (Fig. 9C; note the presence of the “pointers” emerging from the RFB, which are thus formed independently of recombination). We therefore conclude from these experiments that DNA synthesis at the paused forks can normally resume without recombination, consistent with our observation of a stable replisome at the RFBs.
Discussion
By introducing RFBs on chromosome 3 of budding yeast, we have generated a system that is ideal for studying the composition and regulation of the replisome at individual paused eukaryotic DNA replication forks. The forks from ARS305 and ARS306 arrive at the corresponding RFBs with similar kinetics in each cell cycle and then pause for a greatly extended period, relative to most other pause sites in the genome. Eventually, DNA synthesis resumes from each of the two paused forks, allowing us to study both the stability and recovery of paused replisomes, without the arrival of a neighboring fork that would induce termination. Within the endogenous rDNA, however, origins tend to be activated in small clusters of adjacent repeats, so that most forks that pause at the RFBs are resolved after a relatively short period by forks from neighboring repeats. The RFB system on chromosome 3 is therefore ideally suited to the study of individual paused replisomes, in contrast to the highly repetitive rDNA, where changes in the copy number or frequency of origin activation can also produce further complications.
Our study identifies both similarities and also important differences between the replisomes associated with paused and stalled eukaryotic DNA replication forks. Our data indicate that many components of the replisome remain associated with the paused fork, just as when forks stall in response to the depletion of nucleotides by HU treatment. We do not know whether the various components of the paused replisome associate with the fork in an uninterrupted fashion, or repeatedly disassociate and then rapidly reassociate, but it is clear that the replisome does not disassemble when a fork pauses at the Fob1–RFB. It is interesting that DNA polymerase α-primase remains associated with the paused fork, despite the fact that synthesis of the lagging-strand is likely to be completed rapidly upon pausing (Gruber et al. 2000). It is also notable that the paused replisome retains the putative MCM–Cdc45 helicase, as it has previously been shown that the removal of MCM2–7 proteins from HU-stalled DNA replication forks blocks irreversibly the subsequent resumption of DNA synthesis (Labib et al. 2000), suggesting that the MCM complex must be maintained continuously at forks from initiation to termination; our observations of paused replisomes are consistent with this idea. We note that our data also suggest that the MCM–Cdc45 complex does indeed travel with DNA replication forks as part of the replisome, rather than functioning at a significant distance away from forks as recently suggested (Laskey and Madine 2003).
Despite the association of both Mrc1 and Tof1 with paused DNA replication forks, our data show that Mrc1 is dispensable for pausing to occur, whereas Tof1 is crucially important for efficient pausing of the replisome at a protein–DNA barrier, as is the associated protein Csm3. Previous studies showed that both Mrc1 and Tof1 are equally important to restrain the progression of HU-stalled forks (Katou et al. 2003). The homolog of Mrc1 in vertebrate cells, Claspin, is required for checkpoint activation to occur in response to the stalling of DNA replication forks (Kumagai and Dunphy 2000), and in budding yeast, it appears that both Mrc1 and Tof1 are important for checkpoint activation in response to HU-stalled DNA replication forks (Alcasabas et al. 2001; Foss 2001; Katou et al. 2003). It therefore seems that Mrc1 and Tof1 play a similar role in the regulation of HU-stalled DNA replication forks. Our data identify an important functional distinction between these proteins when DNA replication forks pause at protein–DNA barriers. Although we cannot exclude that Mrc1 contributes to pausing in a manner that is redundant with Tof1-Csm3, the reverse is not true, and it is clear that Tof1 and Csm3 are both crucial for the efficient pausing of DNA replication forks at the Fob1–RFB, in contrast to Mrc1.
We have also shown that individual paused eukaryotic DNA replication forks can recruit a protein that is not normally part of the replisome. The Rrm3 helicase is important throughout the genome to help forks progress past protein–DNA barriers (Ivessa et al. 2003) and was shown previously by ChIP to associate with the rDNA in asynchronous cultures of budding yeast cells (Ivessa et al. 2000), though the timing and site of recruitment were not determined. By studying the kinetics of individual replicons containing RFBs, we have shown that Rrm3 is recruited specifically to the individual paused replisomes on chromosome 3, and does not associate significantly with newly established forks at ARS305 and ARS306. It is known that Rrm3 can interact with PCNA (Schmidt et al. 2002), which is loaded onto the lagging strand as a “sliding clamp” for DNA polymerase δ during the synthesis of each Okazaki fragment. When synthesis of the lagging-strand is completed soon after pausing of a DNA replication fork at a protein–DNA barrier, PCNA on the newly completed lagging strand may become available for interaction with other proteins such as Rrm3, which can thus gain access to the paused fork. It is interesting to note that the Rrm3 helicase moves along DNA with a 5′–3′ polarity (Ivessa et al. 2002), contrasting with the 3′–5′ polarity proposed for the MCM helicase (Ishimi 1997; Kelman et al. 1999; Chong et al. 2000; Shechter et al. 2000; Lee and Hurwitz 2001; Kaplan et al. 2003). Although the mechanism of the putative MCM–Cdc45 helicase has yet to be resolved, one attractive possibility would be that the MCM–Cdc45 helicase moves 3′–5′ along the template of the leading strand and then pauses upon encountering the Fob1–RFB, until Rrm3 loads onto the template of the lagging strand and unwinds the RFB 5′–3′, thus displacing Fob1 transiently and allowing progression of MCM–Cdc45 to continue (Fig. 10).
Figure 10.
A model for the pausing and recovery of DNA replication forks at a protein–DNA barrier; see text for details.
It is also clear that HU-stalled forks recruit specific proteins that are not normally part of the replisome, analogous to our finding that Rrm3 is specifically recruited to paused forks at the Fob1–RFB. These include the checkpoint kinase complex Mec1–Ddc2 (Katou et al. 2003; Osborn and Elledge 2003; Lucca et al. 2004), which together with Rad53 is essential to maintain the integrity of the stalled fork and prevent disassembly of the stalled replisome (Lopes et al. 2001; Cobb et al. 2004). We have shown that these checkpoint kinases are not required to preserve the integrity of forks that pause for prolonged periods at the Fob1–RFB, further highlighting the differences in regulation between paused and stalled forks. Although the mechanism by which Mec1 and Rad53 stabilize stalled forks is not understood, it has been suggested that these kinases are required to prevent breakage occurring due to the exposure of long regions of single-strand DNA at DNA replication forks (Lopes et al. 2001; Sogo et al. 2002). As a paused fork will rapidly become double stranded except at the junction with the parental duplex, the fork and its associated replisome may simply be inherently stable and thus would not require checkpoint kinases to preserve their integrity. Alternatively, the stability of paused forks may be maintained by another mechanism that remains to be identified.
When forks pause for an extended period at the Fob1–RFB, we have shown that the resumption of DNA synthesis does not normally require recombination, indicating that breakage of such paused forks does not occur commonly. It has been shown previously, however, that recombination levels are increased in the absence of the Rrm3 helicase (Keil and McWilliams 1993; Ivessa et al. 2000, 2002, 2003), indicating that increased pausing of DNA replication forks may provide an important source of genomic instability. Recent studies have demonstrated that pausing of forks at specific loci does indeed promote an increased rate of Rad52-dependent recombination events (Ahn et al. 2005; Lambert et al. 2005; Prado and Aguilera 2005), again supporting the idea that the pausing of forks may contribute to breakage and recombination. Our data do not contradict this possibility but instead indicate that DNA synthesis normally resumes from paused forks without breakage and recombination, as the replisome at the Fob1–RFB is generally stable even after extended pausing, so that breakage events are likely to be relatively rare. The Fob1 protein can indeed promote recombination between sequences that are derived from the rDNA, but it is interesting to note that this requires RNA PolI transcription (Keil and Roeder 1984; Voelkel-Meiman et al. 1987; Stewart and Roeder 1989; Huang and Keil 1995). Stimulation of recombination under such conditions does not correlate with the pausing of DNA replication forks at the RFB (Ward et al. 2000), and it is not known whether clashes between RNA PolI transcription and DNA replication forks are the key to increased recombination in this case. It will be interesting to determine in the future whether the majority of paused eukaryotic replisomes resume synthesis subsequently without breakage of the fork and so without recombination, as suggested by our analysis of the Fob1–RFB, or whether certain kinds of barriers to the progression of DNA replication forks are more likely to cause breakage than others.
It appears that active pausing of DNA replication forks at protein–DNA barriers allows cells to couple DNA synthesis to other cellular processes. For example, mating type switching in fission yeast requires the homologs of Tof1 and Csm3, in order for a specific DNA replication fork to pause at a particular site and allow the establishment of a genomic imprint (Dalgaard and Klar 1999, 2000; Kaykov and Arcangioli 2004; Vengrova and Dalgaard 2004). It is interesting to note that the fission yeast mrc1 gene was not isolated in the original screen for mutations causing defects in mating type switching, despite the isolation of several alleles of swi1 and swi3 (Egel et al. 1984), consistent with our finding that Mrc1 is not essential for the pausing of DNA replication forks at a protein–DNA barrier in budding yeast. It will be interesting in the future to investigate the roles of homologs of Mrc1, Tof1, and Csm3 in the pausing of DNA replication forks at protein–DNA barriers in higher eukaryotes.
Materials and methods
Yeast growth
All our budding yeast strains are based on the “W303” genetic background. Cells were grown at 24°C in YP medium (1% yeast extract [Difco], 2% peptone [Oxoid]) supplemented with 2% glucose (YPD), 2% raffinose (YPRaff), or 2% galactose (YPGal). To synchronize cells in the G1 phase of the cell cycle, α-factor mating pheromone was added to a final concentration of 7.5 μg/mL for one generation time. To induce expression from the GAL1,10 promoter, cells were grown in YPRaff medium and then centrifuged before resuspending in YPGal medium for 45 min.
Construction of a yeast strain with RFBs on chromosome 3
We used PCR to amplify from yeast genomic DNA a 450-bp fragment containing the RFB from the rDNA repeats, corresponding to nucleotides 460470–460919 of chromosome 12, preceded by a BamHI site and followed by a SmaI site. We then generated the plasmid pBH3 by inserting the SmaI–BamHI RFB fragment into the plasmid pRS306 (Sikorski and Hieter 1989), adjacent to an XhoI–SmaI fragment containing the budding yeast LEU2 gene. We made the plasmid pBH7 in an analogous fashion by inserting the SmaI–BamHI RFB fragment into the plasmid pRS305 (Sikorski and Hieter 1989), adjacent to an XhoI–SmaI fragment containing the budding yeast URA3 gene. We used pBH7 and pBH3 to replace nucleotides 48540–48608 of chromosome 3 with RFB-URA3 and nucleotides 64547–64642 with LEU2-RFB, as described in the legend for Supplementary Figure 1. We used PCR and Southern blotting to confirm that the correct integrations had indeed occurred in the resultant yeast strain, YBH17.
Two-dimensional DNA gels
DNA samples for 2D neutral-neutral gel electrophoresis were prepared and analyzed as described previously (Friedman and Brewer 1995; Lopes et al. 2001); DNA was digested with the restriction enzymes BclI and SalI and detected using the probes indicated in Supplementary Figure 1B. Gels for the first dimension had an agarose concentration of 0.4% and were run for 38 h at 0.7 V/cm; gels for the second dimension had an agarose concentration of 1% and were run for 8 h at 5 V/cm. To quantify the pausing of forks at the RFBs in our experiments, we measured the signal corresponding to the “RFB spot” and the “linear spot” (see Fig. 1) using a Storm 860 PhosphorImager (Molecular Dynamics) and ImageQuant 5.1 software; the RFB signal was then expressed as a proportion of the linear signal, and the ratio denoted “paused forks/relative units”.
Tagging yeast proteins with multiple copies of the c-myc epitope
We used a “one-step PCR” approach (Knop et al. 1999) to introduce multiple copies of the c-myc or Flag epitopes at the C terminus of yeast proteins in the diploid strain W303-1. The correct integrations were confirmed by PCR and by immunoblotting with the anti-myc antibody 9E11 (Neomarkers “c-myc Ab-1”) or the anti-Flag antibody M2 (Sigma), and diploid colonies were then sporulated and tetrad analysis performed in order to isolate the corresponding haploids.
ChIP
We performed ChIP experiments as described previously (Kamimura et al. 2001), except that the immunoprecipitated DNA was purified using the Qiagen PCR purification kit instead of Phenol/Chloroform/Iso-amyl alcohol. In all experiments except that described in Figure 3B, we performed two immunoprecipitations for each cell extract: one using the mouse monoclonal antibodies 9E11 or M2 that recognize the c-myc or Flag epitopes attached to the target protein, and a second using an equivalent mouse monoclonal antibody, 12CA5, which served as a negative control. For the experiment in Figure 3B, we used 9E11, M2, and 12CA5 to perform three immunoprecipitations. We used real-time PCR to quantify for each immunoprecipitate the amount of DNA corresponding to the specific genomic loci shown in Supplementary Figure 5. We set up 25 μL PCR reactions containing 1 μL purified DNA from a particular immunoprecipitate, 1.125 μL 10 μM 5′ oligonucleotide primer, 1.125 μL 10 μM 3′ oligonucleotide primer, 1 μL 5 μM FAM-TAMRA Taqman probe (synthesised by ABI), 12.5 μL 2× Taqman reaction mix (ABI), and 8 μL dH20. Reactions were analyzed using an ABI 7900 thermal cycler according to the manufacturer's instructions. We performed two independent duplicates of each PCR reaction and calculated the mean “threshold cycle number” (or Ct value). The specific enrichment of the target protein for a particular sample was calculated using the following formula: specific enrichment = 2(Ct12CA5 – Ct 9E11/M2), where Ct 12CA5 is the Ct value for the control immunoprecipitate, and Ct 9E11/M2 is the Ct value for the 9E11 or M2 immunoprecipitate. The specific enrichments for each time point were then normalized relative to the lowest background value observed in G1-arrested cells for nonorigin sequences, which was determined in each experiment (except those in Figs. 3B, 5, where the number of samples precluded analysis of G1 cells—we thus did not normalize these data). On each occasion, we also performed two control PCR reactions: a negative control without input DNA and a positive control using genomic DNA purified from the corresponding cell extract.
Acknowledgments
We thank members of our groups for helpful discussions, Marco Foiani for his 2D gel protocol, and Aloys Schepers for adviceconcerning real-time PCR. This work was funded by Cancer Research U.K., from whom K.L. receives a Senior Cancer Research Fellowship, and by the Instituto de Salud Carlos III (F.I.S. grant no. 03-1255), together with a PGC grant awarded to A.B. by the Spanish science ministry. K.L. is supported by the EMBO Young Investigator program, and A.C. received a short-term fellowship from EMBO and is supported by the Sistema Nacional de Salud (grant no. 02-3059). M.K. is funded by a JSPS Post-Doctoral Fellowship for Research Abroad.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.337205.
References
- Ahn J.S., Osman, F., and Whitby, M.C. 2005. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J. 24: 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Allen J.B., Zhou, Z., Siede, W., Friedberg, E.C., and Elledge, S.J. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes & Dev. 8: 2401–2415. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein, D.M., and Bell, S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM complexes and Cdc45p during S phase. Cell 91: 59–69. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout, A.M., and Bell, S.P. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. 96: 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.B., Lewis, L.K., Karthikeyan, G., Lobachev, K.S., Jin, Y.H., Sterling, J.F., Snipe, J.R., and Resnick, M.A. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29: 426–434. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471. [DOI] [PubMed] [Google Scholar]
- ____. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. [DOI] [PubMed] [Google Scholar]
- Brewer B.J., Lockshon, D., and Fangman, W.L. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–276. [DOI] [PubMed] [Google Scholar]
- Cha R.S. and Kleckner, N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606. [DOI] [PubMed] [Google Scholar]
- Chabes A., Domkin, V., and Thelander, L. 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274: 36679–36683. [DOI] [PubMed] [Google Scholar]
- Chang M., Bellaoui, M., Boone, C., and Brown, G.W. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. 99: 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.P., Hayashi, M.K., Simon, M.N., Xu, R.M., and Stillman, B. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. 97: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek, L., Shimada, K., Frei, C., and Gasser, S.M. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22: 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Shimada, K., and Gasser, S.M. 2004. Redundancy, insult-specific sensors and thresholds: Unlocking the S-phase checkpoint response. Curr. Opin. Genet. Dev. 14: 292–300. [DOI] [PubMed] [Google Scholar]
- Dalgaard J.Z. and Klar, A.J. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400: 181–184. [DOI] [PubMed] [Google Scholar]
- ____. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751. [DOI] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas, A.A., Bachant, J.B., and Elledge, S.J. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14: R778–R786. [DOI] [PubMed] [Google Scholar]
- Egel R., Beach, D.H., and Klar, A.J. 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. 81: 3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E.J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer, B.J. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262: 613–627. [DOI] [PubMed] [Google Scholar]
- Gangloff S., Zou, H., and Rothstein, R. 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15: 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S.A. and Newlon, C.S. 1992. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 12: 4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M., Wellinger, R.E., and Sogo, J.M. 2000. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol. 20: 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.S. and Keil, R.L. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. 1997. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272: 24508–24513. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.Q., and Zakian, V.A 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.Q., Schulz, V.P., Monson, E.K., and Zakian, V.A. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes & Dev. 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Lenzmeier, B.A., Bessler, J.B., Goudsouzian, L.K., Schnakenberg, S.L., and Zakian, VA. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol. Cell 12: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Tak, Y.S., Sugino, A., and Araki, H. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20: 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M., Sanchez-Diaz, A., Gambus, A., and Labib, K. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–725. [DOI] [PubMed] [Google Scholar]
- Kaplan D.L., Davey, M.J., and O'Donnell, M. 2003. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 278: 49171–49182. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kaykov A. and Arcangioli, B. 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr. Biol. 14: 1924–1928. [DOI] [PubMed] [Google Scholar]
- Kearsey S.E. and Cotterill, S. 2003. Enigmatic variations: Divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12: 1067–1075. [DOI] [PubMed] [Google Scholar]
- Keil R.L. and McWilliams, A.D. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R.L. and Roeder, G.S. 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386. [DOI] [PubMed] [Google Scholar]
- Kelman Z., Lee, J.K., and Hurwitz, J. 1999. The single minichromosome maintenance protein of methanobacterium thermoautotrophicum ΔH contains DNA helicase activity. Proc. Natl. Acad. Sci. 96: 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23: 9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. and Horiuchi, T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–474. [DOI] [PubMed] [Google Scholar]
- Krings G. and Bastia, D. 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 101: 14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Takase, Y., Komori, Y., Hashimoto, Y., Arata, T., Kamimura, Y., Araki, H., and Takisawa, H. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes & Dev. 17: 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy, W.G. 2000. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6: 839–849. [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero, J.A., and Diffley, J.F.X. 2000. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647. [DOI] [PubMed] [Google Scholar]
- Lambert S., Watson, A., Sheedy, D.M., Martin, B., and Carr, A.M. 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702. [DOI] [PubMed] [Google Scholar]
- Laskey R.A. and Madine, M.A. 2003. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 4: 26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K. and Hurwitz, J. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. 98: 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M.H. and Huberman, J.A. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Pellicioli, A., Cotta-Ramusino, C., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C., and Foiani, M. 2001. The checkpoint response stabilizes stalled DNA replication forks. Nature 412: 599–602. [DOI] [PubMed] [Google Scholar]
- Lucca C., Vanoli, F., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Haber, J., and Foiani, M. 2004. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23: 1206–1213. [DOI] [PubMed] [Google Scholar]
- Lucchini R. and Sogo, J.M. 1994. Chromatin structure and transcriptional activity around the replication forks arrested at the 3′ end of the yeast ribosomal-RNA genes. Mol. Cell. Biol. 14: 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S., Herskowitz, I., and Blackburn, E.H. 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 24: 4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Sugino, A., and Araki, H. 2000. Dpb11 controls the association between DNA polymerases α and ε and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20: 2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L., Pot, I., Chang, M., Xu, H., Aneliunas, V., Kwok, T., Newitt, R., Aebersold, R., Boone, C., Brown, G.W., et al. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15: 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K. and Bastia, D. 2004. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 279: 1932–1941. [DOI] [PubMed] [Google Scholar]
- Nedelcheva M.N., Roguev, A., Dolapchiev, L.B., Shevchenko, A., Taskov, H.B., Stewart, A.F., and Stoynov, S.S. 2005. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J. Mol. Biol. 347: 509–521. [DOI] [PubMed] [Google Scholar]
- Osborn A.J. and Elledge, S.J. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes & Dev. 17: 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M. and Walter, J.C. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23: 3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P.U., Defossez, P.A., and Guarente, L. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F. and Aguilera, A. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24: 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K.P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A.M., Moreno-Borchart, A.C., Primig, M., et al. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler, E.A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D.L., Davis, R.W., Brewer, B.J., and Fangman, W.L. 2001. Replication dynamics of the yeast genome. Science 294: 115–121. [DOI] [PubMed] [Google Scholar]
- Reynolds A.E., McCarroll, R.M., Newlon, C.S., and Fangman, W.L. 1989. Time of replication of ARS elements along yeast chromosome III. Mol. Cell. Biol. 9: 4488–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.H., Derry, K.L., and Kolodner, R.D. 2002. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 277: 45331–45337. [DOI] [PubMed] [Google Scholar]
- Shechter D.F., Ying, C.Y., and Gautier, J. 2000. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum ΔH minichromosome maintenance protein. J. Biol. Chem. 275: 15049–15059. [DOI] [PubMed] [Google Scholar]
- ____. 2004. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J. Biol. Chem. 279: 45586–45593. [DOI] [PubMed] [Google Scholar]
- Shor E., Gangloff, S., Wagner, M., Weinstein, J., Price, G., and Rothstein, R. 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes, M., and Foiani, M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. [DOI] [PubMed] [Google Scholar]
- Stewart S.E. and Roeder, G.S. 1989. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 3464–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. 2005. Origin recognition and the chromosome cycle. FEBS Lett. 579: 877–884. [DOI] [PubMed] [Google Scholar]
- Takayama Y., Kamimura, Y., Okawa, M., Muramatsu, S., Sugino, A., and Araki, H. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & Dev. 17: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y., Horiuchi, T., and Kobayashi, T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes & Dev. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. and Nasmyth, K. 1998. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17: 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A. and Diffley, J.F. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- Vengrova S. and Dalgaard, J.Z. 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes & Dev. 18: 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil, R.L., and Roeder, G.S. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Wang Y., Vujcic, M., and Kowalski, D. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21: 4938–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T.R., Hoang, M.L., Prusty, R., Lau, C.K., Keil, R.L., Fangman, W.L., and Brewer, B.J. 2000. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: Shared sequences but independent activities. Mol. Cell. Biol. 20: 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T., Budd, M., Hoopes, L.L., and Campbell, J.L. 2003. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 278: 22513–22522. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller, E.G., and Rothstein, R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]
- Zhao X., Chabes, A., Domkin, V., Thelander, L., and Rothstein, R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]