Abstract
PI3K-δ inhibitors have shown impressive activity in lymphoid malignancies but have been hampered by autoimmune and infectious toxicities leading to market withdrawals. We previously demonstrated activity of the PI3K-δγ inhibitor duvelisib in T-cell lymphomas that was associated with inflammatory adverse events. We conducted a phase Ib/IIa study of duvelisib in combination with either romidepsin (n=66) or bortezomib (n=32) in patients with relapsed/refractory TCL and found that the addition of romidepsin, but not bortezomib, appeared to increase efficacy while attenuating PI3K inhibitor-driven toxicity. The primary endpoint of the study was to determine the safety and maximum tolerated dose of duvelisib, which was 75 mg twice daily when combined with romidepsin versus 25 mg twice daily when combined with bortezomib. The most common adverse events were neutropenia (42%, 25/59) and fatigue (37%, 22/59) in patients treated with duvelisib and romidepsin and diarrhea (48%, 11/23) and neutropenia (30%, 7/23) in patients treated with duvelisib and bortezomib. Duvelisib and romidepsin resulted in less grade 3/4 hepatotoxicity (14%, 8/59) compared with 40% (14/35) in our prior study with duvelisib monotherapy. This was associated with reductions in circulating inflammatory mediators and myeloid cell inflammatory gene expression. Secondary endpoints of overall and complete response rates were 55% (35/64) and 34% (22/64) for patients treated with duvelisib and romidepsin and 34% (11/32) and 13% (4/32) for patients treated with duvelisib and bortezomib. Among patients with peripheral T-cell lymphomas (PTCL), overall and complete response rates of duvelisib and romidepsin were 56% (27/48) and 44% (21/48) respectively, with exploratory analyses showing increased response rates in patients with a follicular helper T-cell subtype. These findings support further development of combined PI3K and HDAC inhibition in T-cell lymphomas and suggest a unique strategy to enable PI3K inhibitor-based combinations for additional patient populations. ClinicalTrials.gov Identifier: NCT02783625
INTRODUCTION
The phosphatidylinositol 3-kinase (PI3K) pathway has long been recognized for its crucial role in lymphoid malignancies. Constitutive activation of the PI3K/Akt/mTORC1 pathway is a common occurrence in both B- and T-cell lymphomas1–4. This activation can result from various factors, including known oncogenes like nucleophosmin/anaplastic lymphoma kinase5 or TEL-FGFR36, activating mutations within the PI3K pathway7–11, or the inactivation of phosphatase and tensin homolog (PTEN), a negative regulator of PI3K activity12–14. The δ and γ isoforms of PI3K have emerged as promising therapeutic targets in lymphoma due to their preferential expression in leukocytes and their non-redundant roles in T-cell development and function15,16.
Initial studies exploring PI3K-delta inhibitors (PI3Ki) like idelalisib and copanlisib, as well as the PI3K-delta/gamma inhibitor duvelisib, demonstrated impressive and durable responses, leading to their rapid approval for indolent B-cell lymphomas17–21. However, subsequent trials demonstrated late toxicities impacting overall survival (OS) 22–25 that were primarily autoimmune or infectious in nature. These toxicities correlated with the development of autoimmune reactions in p110δ knockout mice, likely arising from the inhibitory impact of this isoform on regulatory T-cells16,25,26. This culminated in the voluntary market withdrawal of all three agents for the treatment of follicular lymphoma in 2022. To date, combination strategies to overcome PI3Ki-driven toxicities have not been successfully developed.
T-cell lymphomas are rare, heterogeneous, and often aggressive non-Hodgkin’s lymphomas 27–30. They are typically classified into peripheral T-cell lymphomas (PTCL) and cutaneous T-cell lymphomas (CTCL) based upon clinical presentations and behavior. Treatment options for PTCL remain limited; most patients achieve either a suboptimal or limited remission following frontline therapy. Patients with relapsed and refractory disease have a median OS of six to twelve months31–33. Currently, consolidative allogeneic stem cell transplantation in the setting of clinical response or remission remains the sole curative option for relapsed T-cell lymphomas but is associated with frequent treatment-associated morbidity and mortality. Single agent overall response rates for relapsed or refractory T-cell lymphomas generally fall within the 20–35% range, with a median progression-free survival of less than four months and limited duration of response34–37. Thus, there is a pressing need for more effective therapies that can provide deep remissions to serve as a bridge to allogeneic stem cell transplantation or provide long-term disease control without cumulative toxicity for both PTCL and CTCL patients.
Duvelisib emerges as a promising solution - an orally bioavailable, selective, potent small-molecule inhibitor targeting the δ and γ isoforms of PI3K38,39. Preclinical studies indicated that duvelisib is effective in T-cell lymphomas with constitutive PI3K activation, inducing cell death in 3 out of 4 lymphoma cell lines with constitutive phospho-AKT (pAKT) expression, as opposed to 0 out of 7 lines lacking pAKT28. In the same study, mice engrafted with a PTCL patient-derived xenograft and treated with duvelisib showed a shift among tumor-associated macrophages from an immunosuppressive to an inflammatory phenotype28, consistent with parallel studies showing that PI3Kγ limits anti-tumor myeloid cell activity40,41. We previously performed a phase I study of duvelisib in 35 patients with relapsed or refractory T-cell lymphoma with encouraging results. Overall, 3/16 patients with peripheral T-cell lymphoma achieved complete responses (CR) and 5/16 had partial responses (PR) with single agent duvelisib when administered as a single agent at the maximum-tolerated dose of 75 mg twice daily. However, this dose was complicated by a high rate of transaminase elevation (57%) that limited tolerability over time28. Duvelisib is 10-fold less potent against PI3Kγ compared to PI3Kδ and may require 75 mg twice daily for maximal effect against both isoforms42 and preliminary data from a dedicated phase 2 study in patients with PTCL suggested improved early disease control with 75 mg twice daily as compared to 25 mg twice daily43.
Among approved single-agents for T-cell lymphomas, romidepsin and bortezomib showed preclinical combinatorial efficacy with duvelisib and were therefore selected for a phase Ib/IIa study with the goal of enhancing efficacy. Early signal during the dose escalation phase suggested reduced toxicity with romidepsin and duvelisib, possibly through effects on nonmalignant lymphocytes44. Based on this signal, we expanded the study with the combined goals of improved efficacy and reduced toxicity. This approach set the stage for a new therapeutic paradigm—one that combines potent anti-tumor activity with ameliorative effects on the toxicity associated with combination partners. In the present study, we report the development, clinical outcomes and biological correlates of response to duvelisib-based combination therapies in relapsed or refractory T-cell lymphomas.
RESULTS
Maximum tolerated dose of duvelisib-based combinations
Both romidepsin and bortezomib have single-agent activity in patients with CTCL or PTCL, with romidepsin listed in the NCCN guidelines for relapsed/refractory disease. We initiated a multicenter, investigator-initiated phase Ib/IIa study of duvelisib in combination with either romidepsin or bortezomib in patients with relapsed or refractory PTCL or CTCL (NCT#02783625). The primary endpoints of the study were to define the maximum tolerated dose (MTD) and characterize the safety and toxicity profile of extended treatment with the combination of duvelisib with either romidepsin or bortezomib in peripheral T-cell lymphoma (PTCL) and cutaneous T-cell lymphoma (CTCL). While dose limiting toxicity was defined in cycle 1, toxicities during subsequent cycles were considered when selecting the optimal dose for expansion. The secondary objectives of this study were to determine, within PTCL and CTCL, respectively, the overall response rate (ORR), complete response (CR) and partial response (PR) rate, time to response (TTR), duration of response (DOR) and event free survival (EFS) of duvelisib in combination with either romidepsin or bortezomib. Post-hoc analyses included overall survival (OS) and progression-free survival (PFS) of patients treated with duvelisib in combination with either romidepsin or bortezomib. Exploratory endpoints included correlation of tumor and peripheral blood-derived biomarkers with response rates and/or adverse events.
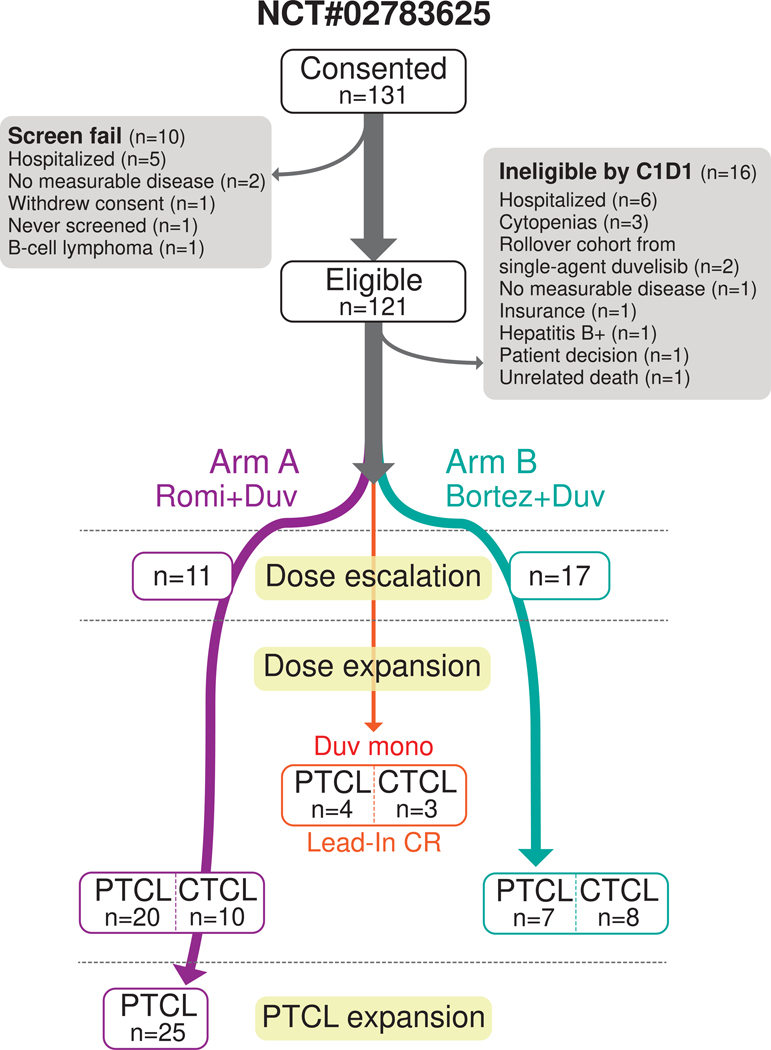
Key inclusion criteria included pathologically confirmed T-cell lymphoma that had relapsed or progressed after at least one systemic therapy with an ECOG performance status of ≤ 2. Key exclusion criteria included prior discontinuation of either duvelisib, romidepsin (Arm A), or bortezomib (Arm B) due to toxicity (see Methods for patient eligibility criteria). Prior monotherapy with duvelisib, bortezomib, or romidepsin was permitted if treatment was not discontinued due to toxicity. The study included a total of 105 patients, with 66 receiving duvelisib and romidepsin (Arm A), 32 receiving duvelisib and bortezomib (Arm B), and 7 patients who continued single agent duvelisib after achieving CR in the lead-in cycle (Fig. 1 and Extended Fig. 1). The rationale for the cohort size of the two respective arms can be found in the Methods. The first patient was enrolled on May 24, 2016, and the final patient was enrolled on October 14, 2020. Demographic characteristics revealed a median age of 62 years, predominantly male (62%), and white (76%) participants, with a majority having PTCL (71%) (Table 1). All subjects received prophylaxis against VZV and PJP, and antifungal prophylaxis with either nystatin or fluconazole was encouraged but not mandatory.
Figure 1. CONSORT diagram for NCT#02783625.
Study design and enrollment on NCT#02783625.
Table 1.
Demographics of patients treated on NCT#02783625.
| Characteristics | Arm A Romidepsin + Duvelisib | Arm B Bortezomib + Duvelisib | Lead-in CRs Single Agent Duvelisib | Total |
|---|---|---|---|---|
| Sample size, n | 66 | 32 | 7 | 105 |
| Median Age (range) | 61.5 (20–81) | 66.5 (42–83) | 62 (46–78) | 62 (20–83) |
| Male, n (%) | 42 (64%) | 21 (66%) | 2 (29%) | 65 (62%) |
| Race, n (%) | ||||
| White | 52 (79%) | 22 (69%) | 6 (86%) | 80 (76%) |
| African American or Black | 8 (12%) | 5 (16%) | 1 (14%) | 14 (13%) |
| Asian | 4 (6%) | 2 (6%) | - | 6 (6%) |
| Unknown | 2 (3%) | 3 (9%) | - | 5 (5%) |
| Treatment History | ||||
| Prior # therapies, median (range)* | 3 (1–16) | 4 (1–14) | 4 (1–5) | 3 (1–16) |
| Prior romidepsin | 15 (23%) | 22 (69%) | 2 (29%) | 39 (37%) |
| Prior protease inhibitor | 1 (2%) | 3 (9%) | - | 4 (4%) |
| Prior Transplants, n (%) | ||||
| Auto | 10 (15%) | 5 (16%) | 2 (29%) | 17 (16%) |
| Allo | 3 (5%) | - | - | 3 (3%) |
| Disease Subtype, n (%) | ||||
| Peripheral T-cell Lymphoma | 55 (83%) | 16 (50%) | 4 (57%) | 75 (71%) |
| PTCL-NOS | 18 (27%) | 7 (22%) | - | 25 (24%) |
| AITL/PTCL-TFH | 21 (32%) | 4 (12%) | 4 (57%) | 29 (28%) |
| ALCL | 3 (5%) | 2 (6%) | - | 5 (5%) |
| pcGDTCL | 3 (5%) | - | - | 3 (3%) |
| HSTCL | 2 (3%) | - | - | 2 (2%) |
| MEITL | 2 (3%) | - | - | 2 (2%) |
| Epidermotropic CD8+ TCL | 2 (3%) | - | - | 2 (2%) |
| Other | 4 (6%) | 3 (9%) | - | 7 (7%) |
| T-PLL | 1 (2%) | 1 (3%) | - | 2 (2%) |
| ATLL | 1 (2%) | 1 (3%) | - | 2 (2%) |
| pcPTCL | 1 (2%) | - | - | 1 (1%) |
| PTLD | 1 (2%) | - | - | 1 (1%) |
| EN-NK/TCL | - | 1 (3%) | - | 1 (1%) |
| Cutaneous T-cell Lymphoma | 11 (17%) | 16 (50%) | 3 (43%) | 30 (29%) |
| MF | 7 (11%) | 9 (28%) | 3 (43%) | 19 (18%) |
| - Transformed | 3 (5%) | 4 (13%) | 1 (14%) | 8 (8%) |
| SS | 4 (6%) | 7 (22%) | - | 11 (10%) |
| - Transformed | 1 (2%) | 2 (6%) | - | 3 (3%) |
In Arm A, no dose-limiting toxicities (DLTs) were observed, establishing the highest dose level (DL3, romidepsin 10 mg/m2 intravenously and duvelisib 75 mg oral twice daily) as the maximum tolerated dose (Supplementary Table 1). In contrast, one patient in Arm B (duvelisib and bortezomib) experienced a DLT at the initial dose level (bortezomib 1 mg/m2 subcutaneously and duvelisib 25 mg twice daily), and subsequent adverse events, mainly grade 3 transaminase elevations, occurred at higher dose levels (Supplementary Table 2). As a result, the maximum tolerated dose for Arm B was established at dose level 1 (bortezomib 1 mg/m2 and duvelisib 25 mg twice daily).
Grade 3/4 adverse events (AEs) and AEs of any grade are listed in Table 2 and Supplementary Table 3, respectively. The most common adverse events (AEs) on study were nausea, dysgeusia, and fatigue, with a higher proportion noted in treatment Arm A. The most commonly occurring Grade 3/4 adverse event was neutropenia, reported in 21 (36%) patients in treatment Arm A and in 6 (26%) patients in treatment Arm B. Toxicity related dose-reductions and treatment discontinuations occurred at the MTD in 13 (22%) and 10 (17%) patients who received romidepsin and duvelisib, 5 (22%) and 4 (17%) patients who received bortezomib and duvelisib, and 1 (14%) and 1 (14%) patients who received duvelisib alone, respectively; these events are listed in Supplementary Table 4. The most common cause of dose reduction was neutropenia, and the most common cause of treatment discontinuation was diarrhea or colitis. Six patients passed away for reasons other than progression of disease (PD) while on study; their cases are listed in Supplementary Table 5. One patient died from Stevens-Johnson Syndrome, three patients died of infectious causes, and two died from toxicity associated with allogeneic transplant (pulmonary fibrosis and graft-versus-host disease).
Table 2.
Grade 3/4 Adverse Events in Patients at Maximum Tolerated Dose.
| Total Related Toxicities (inclusive of SAEs) | Arm A | Arm B | Lead-in CR | Lead-in CR | ||||
|---|---|---|---|---|---|---|---|---|
| Romidepsin + Duvelisib | Bortezomib + Duvelisib | Duvelisib (75 mg) | Duvelisib (25 mg) | |||||
| Grade 3 | Grade 4 | Grade 3 | Grade 4 | Grade 3 | Grade 4 | Grade 3 | Grade 4 | |
| % (n) | % (n) | % (n) | % (n) | % (n) | % (n) | % (n) | % (n) | |
| Sample size, n | 59 | 23 | 4 | 3 | ||||
| Hematological Toxicities | ||||||||
| Neutropenia | 20 (12) | 15 (9) | 22 (5) | 4 (1) | - | 25 (1) | 33 (1) | - |
| Thrombocytopenia | 8 (5) | 2 (1) | 4 (1) | - | - | - | - | - |
| Anemia | 5 (3) | - | 9 (2) | - | - | - | - | - |
| Lymphopenia | - | 3 (2) | 4 (1) | - | - | - | - | - |
| Lymphocytosis | 2 (1) | - | - | - | - | - | - | - |
| Febrile neutropenia | 2 (1) | - | 4 (1) | - | - | - | - | - |
| Liver Function and Electrolyte Toxicities | ||||||||
| ALT increased | 12 (7) | - | 4 (1) | - | 25 (1) | - | - | - |
| AST increased | 5 (3) | - | 4 (1) | - | 25 (1) | - | - | - |
| Blood bilirubin increased | 2 (1) | - | - | - | - | - | - | - |
| Alkaline phosphatase increased | 2 (1) | - | - | - | - | - | - | - |
| Decreased electrolytes | 8 (5) | 2 (1) | 4 (1) | - | - | - | - | - |
| Infections | ||||||||
| Candida esophagitis, Thrush | 3 (2) | - | - | - | - | - | - | - |
| Lung infections, URIs, Pneumonia, Cough | 3 (2) | - | 4 (1) | - | - | - | - | - |
| Skin infections | 2 (1) | - | - | - | - | - | - | - |
| Other infections | 2 (1) | - | - | - | - | - | - | - |
| Rash | ||||||||
| Maculo-papular rash | 7 (4) | - | 4 (1) | - | - | - | - | - |
| Other rash | 3 (2) | - | - | - | 25 (1) | - | - | - |
| Erythroderma | 2 (1) | - | - | - | - | - | - | - |
| Other Toxicities | ||||||||
| Diarrhea | 15 (9) | - | 4 (1) | - | - | - | - | - |
| Fatigue | 5 (3) | - | 4 (1) | - | - | - | - | - |
| Anorexia | 5 (3) | - | - | - | - | - | - | - |
| Constipation | 2 (1) | - | - | - | - | - | - | - |
| Dyspnea | - | - | 4 (1) | - | - | - | - | - |
| Hypertension | 2 (1) | - | 4 (1) | - | - | - | - | - |
| Abdominal pain | - | - | 4 (1) | - | - | - | - | - |
| Colitis | 2 (1) | - | - | - | 25 (1) | - | - | - |
| Complete heart block | - | 2 (1) | - | - | - | - | - | - |
| Electrocardiogram QTc interval prolonged | - | 2 (1) | - | - | - | - | - | - |
| Enterocolitis | 2 (1) | - | - | - | - | - | - | - |
| Pain of skin | 2 (1) | - | - | - | - | - | - | - |
| Peripheral sensory/motor neuropathy | - | - | 4 (1) | - | - | - | - | - |
| Pruritis | 2 (1) | - | - | - | - | - | - | - |
High response rate in tumors with a T-follicular helper phenotype
Secondary endpoints, including treatment responses by subtype across both arms are shown in Table 3. Duvelisib plus romidepsin showed greater efficacy at the MTD across disease subtypes, with an overall response rate (ORR) of 54% in Arm A (37% CRR, 18% PRR) compared with 30% in Arm B (9% CRR, 22% PRR). Patients with PTCL exhibited an ORR of 56% with duvelisib plus romidepsin compared with 42% in Arm B (Fig. 2A and Extended Fig. 2A), with a significantly higher rate of CRs in a post-hoc analysis (44% versus 17% in Arm B, p = 0.0257 by Fisher’s Exact Test) (Fig. 2B and Table 3). Median event-free survival (EFS) by intention-to-treat in PTCL patients on Arm A was 3.5 months (95% CI: 2.4–5.9) (Extended Fig. 2B). The median time to first response for PTCL patients was 1.6 months on both Arm A (range 0–3.5) and Arm B (range 0–8.4), with median duration of response (DOR) for patients with PTCL of 12 months (95% CI 6- not reached) on Arm A and 14 months (95% CI 1.8 – not reached) on Arm B.
Table 3.
Efficacy by Disease Subtype.
| Histology | Arm A | Arm B | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Romidepsin + Duvelisib | Bortezomib + Duvelisib | |||||||||||||||
| All Patients | at MTD only | All Patients | at MTD only | |||||||||||||
| Evaluable | ORR | CRR | PRR | Evaluable | ORR | CRR | PRR | Evaluable | ORR | CRR | PRR | Evaluable | ORR | CRR | PRR | |
| n | % (n) | % (n) | % (n) | n | % (n) | % (n) | % (n) | n | % (n) | % (n) | % (n) | n | % (n) | % (n) | % (n) | |
| Peripheral T-cell Lymphoma | 53 | 58 (31) | 42 (22) | 17 (9) | 48 | 56 (27) | 44 (21) | 13 (6) | 16 | 44 (7) | 25 (4) | 19 (3) | 12 | 42 (5) | 17 (2) | 25 (3) |
| PTCL-NOS | 18 | 50 (9) | 28 (5) | 22 (4) | 17 | 47 (8) | 29 (5) | 18 (3) | 7 | 57 (4) | 29 (2) | 29 (2) | 6 | 50 (3) | 17 (1) | 33 (2) |
| AITL/PTCL-TFH | 20 | 70 (14) | 60 (12) | 10 (2) | 17 | 71 (12) | 65 (11) | 6 (1) | 4 | 75 (3) | 50 (2) | 25 (1) | 2 | 100 (2) | 50 (1) | 50 (1) |
| ALCL | 3 | 100 (3) | 67 (2) | 33 (1) | 3 | 100 (3) | 67 (2) | 33 (1) | 2 | 0 | - | - | 1 | 0 | - | - |
| pcGDTCL | 3 | 33 (1) | 33 (1) | - | 3 | 33 (1) | 33 (1) | - | - | - | - | - | - | - | - | |
| HSTCL | 2 | 50 (1) | - | 50 (1) | 1 | 0 | - | - | - | - | - | - | - | - | - | |
| MEITL | 1 | 0 | - | - | 1 | 0 | - | - | - | - | - | - | - | - | - | |
| Aggressive Epidermotropic CD8+ TCL | 2 | 50 (1) | 50 (1) | - | 2 | 50 (1) | 50 (1) | - | - | - | - | - | - | - | - | |
| Other | 4 | 50 (2) | 25 (1) | 25 (1) | 4 | 50 (2) | 25 (1) | 25 (1) | 3 | 0 | - | - | 3 | 0 | - | - |
| T-PLL | 1 | 0 | - | - | 1 | 0 | - | - | 1 | 0 | - | - | 1 | 0 | - | - |
| ATLL | 1 | 0 | - | - | 1 | 0 | - | - | 1 | 0 | - | - | 1 | 0 | - | - |
| pcPTCL | 1 | 100 (1) | - | 100 (1) | 1 | 100 (1) | - | 100 (1) | - | - | - | - | - | - | - | - |
| PTLD | 1 | 100 (1) | 100 (1) | - | 1 | 100 (1) | 100 (1) | - | - | - | - | - | - | - | - | - |
| ENNK/TCL | 0 | - | - | - | 0 | - | - | - | 1 | 0 | - | - | 1 | 0 | - | - |
| Cutaneous T-cell Lymphoma | 11 | 36 (4) | 0 | 36 (4) | 9 | 44 (4) | 0 | 44 (4) | 16 | 25 (4) | 0 | 25 (4) | 11 | 18 (2) | 0 | 18 (2) |
| MF | 7 | 29 (2) | - | 29 (2) | 5 | 40 (2) | - | 40 (2) | 9 | 11 (1) | - | 11 (1) | 7 | 14 (1) | - | 14 (1) |
| - Transformed | 3 | 0 | - | - | 1 | 0 | - | - | 4 | 0 | - | 2 | 0 | - | - | |
| SS | 4 | 50 (2) | - | 50 (2) | 4 | 50 (2) | - | 50 (2) | 7 | 43 (3) | - | 43 (3) | 4 | 25 (1) | - | 25 (1) |
| - Transformed | 1 | 0 | - | 1 | 0 | - | - | 2 | 0 | - | 2 | 0 | - | - | ||
| TOTAL | 64 | 55 (35) | 34 (22) | 20 (13) | 57 | 54 (31) | 37 (21) | 18 (10) | 32 | 34 (11) | 13 (4) | 22 (7) | 23 | 30 (7) | 9 (2) | 22 (5) |
PTCL-NOS, peripheral T-cell lymphoma not otherwise specified; AITL, angioimmunoblastic T-cell lymphoma, Tfh, T-follicular helper; ALCL, anaplastic large cell lymphoma; pcGDTCL, primary cutaneous gamma-delta T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; MEITL, monomorphic epitheliotrophic intestinal T-cell lymphoma; T-PLL, T-prolymphocytic leukemia; ATLL, acute T-cell leukemia/lymphoma; PTLD, peripheral T-cell lymphoproliferative disorder; EN-NK/TCL, extranodal natural killer/T-cell lymphoma; MF, mycosis fungoides; SS, Sezary syndrome.
Figure 2. Romidepsin and Duvelisib is a highly active combination treatment strategy in patients with peripheral T-cell lymphoma.
(a) Waterfall plot depicting maximal change in index lesions compared to baseline in patients with PTCL receiving romidepsin + duvelisib (Arm A). (b) Swimmer’s plot depicting responses to romidepsin + duvelisib (Arm A) or bortezomib + duvelisib (Arm B) as indicated. (c) Event-free (above) and overall (below) survival in patients with PTCL receiving romidepsin + duvelisib (Arm A), stratified by best response to therapy. (d) Overall survival of patients in PTCL treated on Arm A who underwent consolidated allogeneic stem cell transplant. P values in (c) conducted by log-rank test.
In a post-hoc analysis, median overall survival (OS) was 12 months (95% CI: 8.6-not reached). Achieving a CR significantly extended both EFS and OS compared to patients who achieved less than a CR (Fig. 2C). In an additional post-hoc analysis of 21 patients with PTCL who achieved a CR with romidepsin and duvelisib, the median time to complete response was 1.7 months (range 0.9–13.2), and 14 patients proceeded to allogeneic stem cell transplantation with curative intent. Among these patients, the median EFS was not reached (95% CI: 9.7 months-not reached); 6 patients remain disease free with 1 year of follow-up, 2 patients remain disease free with less than 6 months of follow-up, 4 patients experienced disease progression, and 2 patients passed away due to complications related to their transplant (Fig. 2D). In contrast to those with PTCL, clinical responses were observed in CTCL patients treated in both arms at the MTD (Arm A: 44%, Arm B: 18%), but none achieved a CR, and median EFS was relatively short in both arms (4.0 and 2.9 months on Arms A and B respectively) (Extended Fig. 2C-D).
Post-hoc analysis did not reveal any differences in outcomes in either arms based on evaluated clinical factors, including gender, age, race, or prior therapy. In treatment Arm A, the objective response rate among patients with either angioimmunoblastic T-cell lymphoma (AITL) or PTCL with a T-follicular helper phenotype (PTCL-Tfh) was 71% and CR rate was 65%, both of which compared favorably with the 25% ORR and 15% CR rate for single-agent romidepsin from the initial registration study in PTCL34 (Extended Fig. 3A and Table 3). To understand pre-treatment tumor features associated with response to therapy, we analyzed both the somatic mutational landscape and gene expression profiles of accessible pre-treatment tumor biopsies. Whole exome sequencing of available pre-treatment samples from patients with PTCL (n=25) revealed mutations in TET2, RHOA, and VAV1 exclusively in responders (Extended Fig. 3B-C). These mutations are highly enriched in PTCL tumors in which the cell of origin is a malignant T-follicular helper (Tfh) cells, which includes both AITL and PTCL-NOS with TFH features44,45. Notably, the majority of mutations in TET2 were frameshift loss-of-function (LOF) mutations, consistent with its role as a tumor suppressor in PTCL44,46, whereas mutations in RHOA and VAV1 were missense point mutations known to be gain-of-function. RHOA G17V mutations can promote Tfh differentiation47,48, whereas both mutations in the C-terminal SH3 domain and N-terminal CH domain potentiate Vav1 signaling and lymphomagenesis by disrupting feedback autoinhibition49,50, consistent with these being driver mutations in PTCL lymphomagenesis.
Cancer cell fraction is typically low in AITL and PTCL-Tfh phenotype, in part due to oligoclonal B-cell expansion51. We performed bulk RNA-sequencing of tumor samples with sufficient tissue for RNA extraction following processing for whole exome sequencing. Analysis of pre-treatment bulk RNA-sequencing of 13 PTCL tumors revealed enrichment of B-cell genes, including PAX5, CD180, CD19 and JCHAIN1 in responders52 (Extended Fig. 3D). Similarly, RNA deconvolution analysis of pre-treatment samples showed a strong enrichment of predicted B-cell tumor composition in patients who achieved a PR or CR compared with patients who did not (p = 0.0008, Extended Fig. 3E). As expected, based on the cancer cell fraction, we did not observe universal enrichment of Tfh genes in pre-treatment samples from responding patients. However, we did observe enrichment of IL21 transcript, which is produced primarily by Tfh cells and is required for germinal center formation, among responders (p=0.034) 54–56 (Extended Fig. 3F).
Analysis of enriched gene sets from the molecular signatures database (MSigDB) showed that the majority of genesets enriched in pre-treatment samples from responders were related to either B-cell function, complement pathway activation or phagocytosis, consistent with prior data from our group and others suggesting that PI3K-γ inhibition enables macrophage-mediated tumor clearance (Extended Fig. 4A) 40,53. In contrast to patients with PTCL, loss of both RET and NFKB2, either via copy number alterations or truncating mutations, were enriched in CTCL patients (n=18) who achieved a clinical response, but no significant changes in gene expression were observed in responding patients (Extended Fig. 4B-E).
Next, to understand mechanisms of response to romidepsin and duvelisib, we performed paired transcriptome sequencing of pre-treatment and on-treatment samples obtained from primary patient tumors as well as from patient-derived xenografts (PDXs) that responded to romidepsin plus duvelisib (Extended Fig. 5A). In both patient samples from the trial and PDXs, on-treatment specimens of responsive tumors exhibited a significant reduction in expression of genes related to mitotic progression, including mitotic spindle genes (NES = −2.11 and −1.67 for primary tumors and PDX tumors, respectively), E2F target genes (NES = −2.17 and −2.63), and G2M checkpoint genes (NES = −2.262 and −2.51) (Extended Fig. 5B), consistent with a reported role for HDACs in mitotic spindle assembly57 and G2M progression58.
Signal transduction pathway activity in patients with resistance to therapy
We next asked whether pre-existing or acquired tumor phenotypes were associated with either intrinsic or adaptive treatment resistance. Interestingly, activating mutations in JAK1 and JAK3 as well as copy number gains of STAT1, STAT3, or STAT5A were present in 58% of non-responders compared to only 15% of responders from whom pre-treatment whole exome analysis was available (Extended Fig. 3B and Extended Fig. 6A-C, p = 0.0414 by Fisher’s exact test). Activation of JAK/STAT signaling as a mechanism of resistance to PI3 kinase inhibition has previously been reported in breast cancer59. To confirm that cell autonomous activation of JAK/STAT signaling was associated resistance to combined therapy with romidepsin and duvelisib, we performed combined single-cell surface epitope identification, transcriptome sequencing, and VDJ sequencing using the 10x Genomics Single Cell Immune Profiling Platform on paired viable CD45+ peripheral blood mononuclear cells (PBMCs) from the two patients with PTCL-NOS who had sufficient circulating disease in the peripheral blood (>5% of CD45+ cells) (Extended Fig. 6D). UMAP projection of PBMCs from these patients showed that the putative malignant cells from Patient Y (non-responder) and Patient Z (responder) clustered separately. Malignant cells from Patient Y were phenotypically more de-differentiated with reduced expression of several T-cell lineage markers including CD2, CD3, CD4 and CD5; tumor cells from both patients were negative for CD7 and CD8 and expressed PD-1 (Extended Fig. 6E). Analysis of HALLMARK genesets revealed that pre-treatment malignant cells from patient Y (non-responder) were enriched in genes associated with JAK/STAT signaling prior to therapy, and these signatures increased following treatment with romidepsin and duvelisib. Furthermore, malignant cells from Patient Y, but not Patient Z significantly upregulated transcription factors known to be downstream of JAK signaling including STAT1 and IRF1 as well as known STAT1 and IRF1 gene targets such as CXCR3 and PRF1 (Extended Fig. 6F-G). Finally, bulk RNA-sequencing of AITL xenograft tumors treated with duvelisib at the time of disease progression demonstrated an enrichment of genes associated with JAK/STAT signaling as compared to pre-implantation samples (NES 1.53, p = 0.0145) (Extended Fig. 6H). These results are consistent with parallel JAK/STAT activation as a mechanism of intrinsic resistance to romidepsin and duvelisib. We previously reported that higher phosphorylation of S6 in tumor cells (consistent with activation of PI3K signaling) is associated with resistance to ruxolitinib in patients with PTCL, also consistent with a reciprocal relationship between dependence on PI3K or JAK/STAT in PTCL60.
Next, we asked whether adaptive resistance emerged during treatment with romidepsin and duvelisib by analyzing paired whole exome sequencing of biopsies obtained pre-treatment and following disease progression. This revealed acquired subclonal mutations in several genes related to growth factor-mediated signal transduction, including STAT3 in one patient and TSC2 and PTEN in another patient (Extended Fig. 7A). Similarly, our single cell analysis of circulating tumor cells showed transcription evidence of increased PI3K/Akt/MTOR signaling in tumor cells patient Y (non-responder) that increased following treatment with romidepsin and duvelisib (Extended Fig. 7B). We therefore asked whether either PTEN loss or TSC2 loss could enable resistance to treatment. We transduced a Cas9-expressing T-cell lymphoma cell line, OCI-ly13.2, with FgH1tUTG vectors that express guide RNAs targeting TSC2 or PTEN as indicated only in response to doxycycline (Extended Fig. 7C). In this line, TSC2 loss, but not PTEN loss, conferred resistance to treatment with duvelisib in vitro, consistent with downstream reactivation of Akt/MTOR signaling but not activation of PI3K upstream of duvelisib activity promoting resistance to duvelisib-based combination therapy (Extended Fig. 7D).
Finally, we asked whether treatment-induced changes in the immune microenvironment predicted resistance to duvelisib and romidepsin. Data from both our group and others suggests that PI3Kγ inhibition can promote macrophage-driven cancer cell phagocytosis and tumor control40,53 Analysis of paired pre-treatment and on-treatment transcriptomes from PTCL patients who did not respond to romidepsin and duvelisib revealed that on-treatment tumors had substantially reduced expression of multiple genesets related to complement activation and phagocytosis (Extended Fig. 7E). In contrast, macrophage abundance as determined by RNA deconvolution did not differ significantly in responders versus non-responders or with treatment (Extended Fig. 7F).
Romidepsin attenuates duvelisib-driven hepatotoxicity.
Notably, the overall rate of grade 3/4 AEs and specifically the rate of treatment-induced grade 3/4 transaminase elevation was significantly lower in patients treated with romidepsin and duvelisib at 75 mg twice daily (8/59, 13.6%) compared to patients with duvelisib alone at 75 mg twice daily (14/35, 40%) in our previous phase I study (Fig. 3A) 53. This rate of grade 3/4 transaminase elevation was even lower when considering patients who initiated combination therapy from the onset of treatment (4/49, 8%), while patients who initiated single agent duvelisib and then escalated to dual therapy had a similar rate of grade 3/4 transaminase elevation (4/10, 40%). Together, these data suggest that romidepsin reduces duvelisib-associated hepatotoxicity; this effect appears specific to romidepsin as DLTs including transaminase elevations limited duvelisib to 25 mg twice daily in combination with bortezomib.
Figure 3. Adverse events in patients treated with romidepsin and duvelisib are associated with myeloid inflammation.
(a) Proportion of patients who developed grade 3/4 hepatotoxicity following treatment with romidepsin + duvelisib or duvelisib alone as part of the phase I PRIMO study. (b) Changes in plasma analytes between C1D8 and C1D1 samples in patients treated with duvelisib or duvelisib + romidepsin as indicated. (c) UMAP of myeloid cells isolated from pre- and on-treatment peripheral blood samples obtained from patients receiving romidepsin + duvelisib. (d) Dot plot depicting enrichment of MSigDB genesets in pre-treatment peripheral blood CD14+ monocytes from patient experiencing either clinically significant or non-significant hepatotoxicity, as indicated. (e) Differentially expressed genes in CD14+ monocytes between C1D8 and C1D1 in patients not experiencing clinically significant hepatotoxicity. P values calculated using Chi Squared Test (a), paired t-test (b) and two-sided Wilcoxon Rank-Sum Test with Bonferroni correction for multiple comparisons (e).
To determine the mechanism by which the addition of romidepsin attenuates duvelisib-driven hepatotoxicity, we performed O-link proteomic analysis on paired pre-treatment and on-treatment plasma samples from 12 patients who received duvelisib monotherapy as part of the PRIMO study and compared them with 9 patients who received duvelisib and romidepsin on the present study. On-treatment samples from duvelisib monotherapy patients showed a statistically significant increase in several circulating inflammatory mediators, such as IL-18, IL-18R1, CD40, and CX3CL1, all of which have been linked to liver inflammation (Fig. 3B and Extended Fig. 8A) 61–65. Additionally, these patients had statistically significant increases in serum hepatocyte growth factor (HGF) and Sprouty homolog 2 (Spry2), both of which are known to promote liver regeneration following injury66. Other than KRT19 (cytokeratin-19), which is neither expressed predominantly in the liver nor in immune cells, no circulating markers that were significantly elevated in duvelisib on-treatment samples were found to be elevated in patients who received combination therapy, suggesting that romidepsin attenuates duvelisib-induced elaboration of inflammatory mediators.
Finally, to identify the cell types that are responsible for inflammation-driven adverse events in response to duvelisib and romidepsin, we performed combined single-cell surface epitope identification, transcriptome sequencing, and VDJ sequencing on paired viable CD45+ peripheral blood mononuclear cells (PBMCs) from ten patients on C1D1 and C1D15 of treatment with romidepsin and duvelisib based on whether patients did or did not develop clinically meaningful (grade 2+) hepatotoxicity as compared with those who did not (grade 0–1). Excluding the two patients with >5% circulating tumor cells, Uniform Manifold and Projection of PBMCs revealed distinct clusters of B-cells, T-cells, and myeloid cells, along with small clusters of erythrocytes and platelets (Extended Fig. 8B-C). We did not observe any significant differences in the T-cell or B-cell compartments when stratified by development of hepatotoxicity. However, analysis of the circulating myeloid populations (Fig. 3C) revealed that CD14+ monocytes from patients with grade 2+ hepatotoxicity expressed genes related to inflammation and metabolism (Fig. 3D), including genes associated with glucose uptake (SLC2A3), lipid droplet formation (PLIN2, HILPDA), proliferation (G0S2), and self-renewal (SELL) (Extended Fig. 8D). In patients who did not develop toxicity, treatment with romidepsin and duvelisib was associated with a reduction in CD14+ monocyte counts (1076 cells vs. 1714 cells) and significant reduction in both transcriptional regulators of myeloid inflammation (MAFB, EGR1) as well as several members of the S100A protein family, which has established roles in amplifying inflammatory responses (S100A8, S100A9, S100A12) (Fig. 3E).
DISCUSSION
This phase Ib/IIa study demonstrated safety, tolerability and high overall and complete response rates with the combination of romidepsin and duvelisib, offering a promising combination strategy for treatment of relapsed or refractory T-cell lymphomas, a historically challenging group of malignancies. Responses were particularly robust in patients with AITL and PTCL-TFH subtypes. Our results offer an additional therapy that provides hope and improved prospects for patients across the diverse spectrum of PTCL subtypes. Romidepsin is still NCCN compendium listed for relapsed and refractory T-cell lymphomas despite being voluntarily withdrawn from consideration for FDA approval. Given the rarity of T-cell lymphomas, the results of this phase Ib/IIa study support the use of this agent in combination with duvelisib for patients with relapsed or refractory disease and support the development of either a dedicated phase II study in patients with AITL and PTCL-TFH subtypes or a Phase III study. Furthermore, the high rate of complete responses with romidepsin and duvelisib support the development of an upfront study assessing romidepsin + duvelisib in elderly or unfit patients with PTCL, most of whom experience substantial toxicity and poor responses to frontline CHOP-based therapy67,68.
One of the most notable aspects of this combination therapy is its tolerability. Importantly, the addition of romidepsin was able to significantly reduce duvelisib-induced hepatotoxicity, a common adverse event observed in our phase 1 study of duvelisib at a similar 75 mg twice daily dose28. Additionally, the romidepsin-duvelisib combination showed low rates of infections and an absence of colitis, although infectious prophylaxis was routinely administered to patients on study. Our mechanistic analyses point to potential myeloid-driven mechanisms of both efficacy and toxicity, suggesting that this cell type may play a key role in both the therapeutic benefit or toxicity of this combination. In contrast, the addition of bortezomib was not able to attenuate duvelisib-driven hepatotoxicity at 50 or 75 mg daily doses. Furthermore, this study introduces the concept of “therasoteric” agents, exemplified by romidepsin, which not only possess single-agent activity but also mitigate the toxicity of other combination partners. This approach could have broader applications in situations where the on-target effects of one agent interfere with the safety profile of another. Here, we highlight the particular value of this strategy to restore the utility of PI3Kδ inhibitors, whose development in B-cell lymphomas has been halted by toxicity concerns. As much needed studies centered around the potential for therasoteric combinations with PI3Kδ inhibitors are conducted, longitudinal sampling of both peripheral blood and tumor tissue will be essential to be able to identify and selectively engage anti-tumor myeloid cell efficacy while avoiding inflammatory myeloid toxicity. We also note that therasoteric strategies should not be viewed through the prism of limiting inflammatory toxicity alone; rather, we envision this strategy as a broader paradigm to maximize tumor-specific activity while mitigating toxicity. For example, the addition of MEK inhibition substantially reduced the development of cutaneous squamous cell carcinomas and keratoacanthomas caused by BRAF inhibitors69.
Finally, our data suggests that both intrinsic and adaptive resistance to romidepsin and duvelisib are associated with activation of either parallel or downstream signal transduction pathways, including JAK/STAT signaling and mTORC1 reactivation. We previously reported the single-agent efficacy of Ruxolitinib in peripheral T-cell lymphomas60 and are currently enrolling patients on a multi-agent, single-arm phase 1 study of Ruxolitinib and Duvelisib in patients with relapsed and refractory lymphomas (NCT#05010005). For patients with evidence of distal PI3K/mTOR activation, multiple agents are available, including everolimus, which has shown clinical activity in T-cell lymphomas, both alone and in combination with HDAC inhibition70,71.
There are limitations to this study. The lack of a duvelisib monotherapy arm limits the ability to directly compare the efficacy and toxicity of combination therapy with duvelisib alone. Furthermore, despite the high overall response rates, the durability of response was modest, particularly in patients not achieving a CR. Finally, combination treatment showed limited efficacy in patients with CTCL. Nevertheless, the combination of romidepsin and duvelisib shows substantial promise for the treatment of T-cell lymphomas. The data from this study establishing the feasibility of leveraging HDAC inhibition to enhance efficacy and attenuate toxicity of PI3K inhibitors in patients with lymphoma offers a unique strategy to reintroduce PI3 kinase inhibitor-based combination strategies across malignancy subtypes.
METHODS
Study Design
The full protocol can be found in the Supplementary Information. This study is a multicenter, open label, parallel, phase Ib/IIa trial of duvelisib in combination with either romidepsin or bortezomib with expansion cohorts in patients with relapsed or refractory peripheral T-cell lymphoma and cutaneous T-cell lymphoma. The study design is depicted in Fig. 1A and Supplementary Fig. 1A. Eligible patients were first assigned to the dose escalation phase of the study to receive duvelisib with either romidepsin or bortezomib to determine the maximum tolerated dose. The enrollment schema is summarized in Extended Fig. 1A. Two parallel 3+3 dose escalation schemes were used to determine the maximum tolerated dose (MTD) of duvelisib + romidepsin and duvelisib+ bortezomib, respectively. Three dose levels plus a “−1” dose level were planned for this study with a maximum possible sample size of 24 patients per arm as shown in Supplementary Table 6.
Each cycle was 28-days in duration. Duvelisib was administered orally on days 1–28 of the 28-day cycle. Patients received either romidepsin or bortezomib, based on which arm of the study they were enrolled in. In the duvelisib plus romidepsin arm, romidepsin was administered intravenously on days 1, 8, and 15 and in the duvelisib plus bortezomib arm, bortezomib was administered subcutaneously on days 1, 4, 8, and 11. If none of the initial cohort of 3 at dose level 1 has a DLT, the dose level will be escalated. If one has a DLT that dose level will be expanded with 3 more patients. Dose escalation will stop if 2 or more DLTs are seen at a dose level. The MTD is defined as the highest dose level at which at most 1 of the 6 patients treated at that level has a DLT.
The probabilities of dose escalation given the true DLT rates shown in Supplementary Table 7. A total of 28 patients were required to determine the MTD of Dose 3 for Arm A (n=11) and Dose 1 for Arm B (n=17). (Late toxicities in Arm B resulted in eventual MTD of Dose 1.) Following determination of MTD of the phase I portion of the study, enrollment of up to 20 patients with measurable disease was planned for the lead in phase of dose expansion (up to 10 CTCL and up to 10 PTCL patients in each Arm). During this phase, patients were treated with duvelisib alone at MTD for one cycle, and then proceed to duvelisib + either romidepsin or bortezomib determined by the MSK PI. The purpose of the lead in phase was to collect tissue for translational biological studies of evaluating mechanisms of response or resistance therapy and optimal combinations for mechanism-based therapy for T-cell lymphomas (see exploratory objectives). After this cycle patients in the lead in phase who did not achieve a complete remission were treated with romidepsin + duvelisib and romidepsin + bortezomib and thus were counted towards the expansion cohorts. Ultimately, 27 patients were treated in the lead-in phase. 14 patients were treated on Arm A, 4 of whom achieved a CR to duvelisib monotherapy and 10 of whom proceeded to combination therapy with romidepsin and duvelisib (2 CR, 4 PR), while 13 patients were treated on Arm B, 3 of whom achieved a CR to duvelisib monotherapy and 10 of whom proceeded to combination therapy with romidepsin and bortezomib (1 CR, 2 PR).
Additional patients were subsequently enrolled on an expansion cohort of each arm with upfront combination (no lead-in component), with the primary goal of increasing the number of patients treated at the MTD to obtain better estimates for efficacy. The CTCL cohort for Arm A enrolled an additional 5 patients. Similarly, the PTCL cohort for Arm A enrolled an additional 5 patients. Another expansion of 9 additional patients were subsequently added to the PTCL Arm A cohort to enroll a total of 14 patients. In this second expansion, 10 patients total were enrolled as 1 of these patients was enrolled to replace 1 of the 2 inevaluable patients. A total of 15 patients were ultimately enrolled in the expansion of upfront combination. Following an amendment, an additional 25 patients were added in a second expansion to this cohort to reach a final total of 44 patients with measurable treated with combination therapy at the MTD. The justification for this expansion is below. Arm B enrolled a total of 20 patients, including 5 patients treated at the MTD in the dose escalation phase and 10 patients treated with combination therapy during the lead-in phase and so enrolled an additional 5 patients in the expansion with upfront combination.
An early stopping rule was used during this expansion phase for safety. In the unlikely event that more than 6 DLTs (out of the 20 patients, per arm) were observed in the expansion cohorts, we planned to halt the enrollment for patients’ safety protection. All toxicities not clearly unrelated to study treatment were thoroughly reviewed by investigators. Guidelines for early stopping are provided in Supplementary Table 8.
For the PTCL Arm A patients, due to the moderate sample size of up to 26 (20 from expansion cohort and up to 6 from the phase I part) for each arm, we did not set up a formal hypothesis test but planned to preliminarily assess the expectation of a response rate greater than 30% (current response rate) by an absolute margin of 15–25%. The probability of observing >=12 responses among 26 patients (corresponding to a sample proportion of 46% responses) was estimated to be 6.0% if the true response rate was 30%, 16.2% if the true response rate was 35%, 32.6% if the true response rate was 40%, 52.9% if the true response rate was 45%, 72.1% if the true response rate was 50%, and 86.5% if the true response rate was 55%.
AMENDMENT for Justification of 25 patient expansion of PTCL Pts
An additional 25 patient expansion of the PTCL Cohort of Arm A (duvelisib + romidepsin) was added. The purpose of this expansion is to show an ORR between 50–60%, with increased statistical confidence that treatment with romidepsin + duvelisib was more efficacious than treatment with single agent romidepsin (a standard FDA approved option) in relapsed/refractory PTCL patients when compared to the PIVOTAL romidepsin study (ORR: 25% [33/130]; CI: 17.6%−32.4%). With the addition of these 25 patients there will be 44 PTCL patients with measurable disease treated with the Romidepsin + Duvelisib combination at the MTD (dose level 3: duvelisib 75mg + romidepsin 10 mg/m2), and 5 PTCL patients treated below the MTD, exclusive of patients that initiated therapy with single agent duvelisib as part of the lead in cohort. Based on an interim analysis of the current study we can estimate target ORR rates of 50% to within +/−15% with 40 patients. This calculation uses normal approximation. Confidence intervals would be 35% - 65% which do not overlap with the PIVOTAL Romidepsin study.
Institutional review boards at each of the study sites approved the study protocol. This study was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent prior to treatment. Data safety was monitored by a Data Safety Monitoring Board comprised of the Primary multicenter team, the Clinical Research Quality Assurance Team, and the MSK Data Safety Monitoring Committee.
Patient Population
Subject Inclusion Criteria
Pathologically confirmed T-cell lymphomas at the enrolling institution, including stage ≥ Ib CTCL, which has relapsed or progressed after at least one systemic therapy.
Age ≥ 18,
Previous systemic anti-cancer therapy for T-cell lymphoma must have been discontinued at least 3 weeks prior to treatment. For the dose expansion phase, in progressing subjects, a 2-week washout may be allowed after discussion with the MSK Principal Investigator.
-
Previous radiation and/or surgery must have been discontinued or completed at least 2 weeks prior to treatment in this study and adverse effects must have resolved to Grade 1 or baseline. Lymph node or other diagnostic biopsies within 2 weeks are not considered exclusionary.
Patients who have received localized RT as part of their immediate prior therapy may be allowed to enroll with shorter washout period after discussion with the MSK Principal Investigator.
ECOG ≤ 2
- Meet the following laboratory criteria without use of growth factor support or platelet transfusions for 1 week:
- Absolute neutrophil count ≥ 1.0 K/mcL
- Platelet count ≥ 80 K/μl (in the expansion cohorts, if thrombocytopenia is due to bone marrow involvement platelet count must be ≥ 50 K/μL),
- Patients enrolled in the dose escalation phase who are not enrolled on the expansion cohorts must have calculated creatinine clearance ≥ 50ml/min by Cockcroft-Gault formula. Patients enrolled in the Dose Expansion phase must have calculated creatinine clearance ≥ 40ml/min by Cockcroft-Gault formula.
- Total bilirubin ≤ 1.5 × upper limit of normal (ULN) or ≤ 3 × ULN if documented hepatic involvement with lymphoma, or ≤ 5 × ULN if history of Gilbert’s syndrome; AST and ALT ≤ 3 × ULN; ≤ 5 × ULN if due to lymphoma involvement.
- Measurable disease for dose expansion and lead in phases only. Measurable disease defined by:
- Revised International Working Group (Cheson, 2007) Classification for systemic lymphoma or
- Atypical and or malignant lymphocytes quantifiable by flow cytometry or morphology in blood or bone marrow
- mSWAT > 0 or Sezary count ≥ 1000 cells/μL
- Short course systemic corticosteroids for disease control, improvement of performance status or non-cancer indication (< 7 days) must have been discontinued at least 6 days prior to study treatment. Stable ongoing corticosteroid use (≥ 30 days) up to an equivalent dose of 20 mg of prednisone is permissible.
- CTCL: Topical steroids that have been used for > 3 weeks may be continued
- All other histologies (not CTCL): Topical steroids use is permissible without restriction.
Women of reproductive potential† must have a negative serum or urine β human chorionic gonadotropin (βhCG) pregnancy test. All women of reproductive potential, all sexually active male patients, and all partners of patients must agree to use adequate methods of birth control (e.g. latex condoms) throughout the study and for 30 days after the last dose of study drug.
Subject Exclusion Criteria
Any serious medical condition, laboratory abnormality, or psychiatric illness that would prevent the subject from signing the informed consent form.
Pregnant females. (Lactating females must agree not to breast feed while taking the study medications).
Prior use of duvelisib if discontinued due to toxicity.
For the romidepsin arm of the study, prior therapy with romidepsin if discontinued due to toxicity.
For the bortezomib arm of the study, prior therapy with a proteasome inhibitor if discontinued due to toxicity.
For the bortezomib arm of the study, patients with grade ≥2 peripheral neuropathy.
History of chronic liver disease, veno-occlusive disease, or current alcohol abuse.
Administration of a live vaccine within 6 weeks of first dose of study drug.
Prior surgery or gastrointestinal dysfunction that may affect drug absorption (e.g., gastric bypass surgery, gastrectomy)
Known seropositive and requiring anti-viral therapy for human immunodeficiency virus (HIV), hepatitis B virus (HBV) or hepatitis C virus (HCV).
- Subjects with chronic hepatitis B or C as defined as test subjects with positive Hep B serology:
- Subjects with a negative HBsAg and a positive HBcAb require an undetectable/negative hepatitis B DNA test (e.g., polymerase chain reaction [PCR] test) to be enrolled and will require prophylactic antiviral treatment initiated prior to the first dose of study drug, and continued until approximately 6 to 12 months after completion of study drug(s).
Patients with positive hepatitis C virus
Subjects with active EBV unrelated to underlying lymphoma (positive serology for anti-EBV VCA IgM antibody and negative for anti-EBV EBNA IgG antibody, or clinical manifestations and positive EBV PCR consistent with active EBV infection.
Subjects with active CMV (positive serology for anti-CMV IgM antibody and negative for anti-CMV IgG antibody and positive CMV PCR with clinical manifestations consistent with active CMV infection) and requiring therapy will be excluded from participation in the study. Carriers will be monitored per institutional guidelines.
- Receiving systemic therapy for another primary malignancy (other than T-cell lymphoma)
- Patients with more than one type of lymphoma may be enrolled after discussion with the MSK Principal Investigator.
- Adjuvant or maintenance therapy to reduce the risk of recurrence of other malignancy (other than T-cell lymphoma) is permissible after discussion with the MSK Principal Investigator.
Known central nervous system or meningeal involvement (in the absence of symptoms, investigation into central nervous system involvement is not required).
Uncontrolled infection requiring systemic antimicrobials.
- The following known cardiac abnormalities:
- Congenital long QT syndrome.
- QTc/QTf interval ≥ 480 milliseconds; unless secondary to pacemaker or bundle branch block.
- Myocardial infarction within 6 months. (Subjects with a history of myocardial infarction within the last 6 to12 months who are asymptomatic and have had a negative cardiac risk assessment (treadmill stress test, nuclear medicine stress test, or stress echocardiogram) since the event may participate.)
- Other significant ECG abnormalities including 2nd degree atrio- ventricular (AV) block type II, 3rd degree AV block.
- Symptomatic coronary artery disease (CAD), e.g., angina Canadian Class IIIV (see Appendix B). In any patient in whom there is doubt, the patient should have a stress imaging study and, if abnormal, angiography to define whether CAD is present.
- An ECG recorded at screening showing evidence of cardiac ischemia (ST depression of ≥2 mm, measured from isoelectric line to the ST segment). If in any doubt, the patient should have a stress imaging study and, if abnormal, angiography to define whether CAD is present.
- Congestive heart failure (CHF) that meets New York Heart Association (NYHA) Class II to IV definitions (see Appendix C) and/or ejection fraction <45% by MUGA, echocardiogram, or cardiac MRI.
- A known history of sustained ventricular tachycardia (VT), ventricular fibrillation (VF), Torsade de Pointes, or cardiac arrest unless currently addressed with an automatic implantable cardioverter defibrillator (AICD).
- Hypertrophic cardiomegaly or restrictive cardiomyopathy from prior treatment or other causes.
- Uncontrolled hypertension, i.e., blood pressure (BP) of ≥170/95; patients who have a history of hypertension controlled by medication must be on a stable dose (for at least one month prior to study registration) and meet all other inclusion criteria.
- Any cardiac arrhythmia requiring an anti-arrhythmic medication (excluding stable doses of beta-blockers)
- For patients enrolling on the Romidepsin arm, taking drugs associated with significant QTc/QTf prolongation, unless able to be switched to non-QTc/QTf prolonging medication or on a stable dose without significant QT prolongation (>470 msec).
- Caution should be used when administering study drugs to patients taking medications significantly metabolized by these enzymes refer to (http://medicine.iupui.edu/clinpharm/ddis/clinical-table/) for clinically relevant medications. Particular attention should be paid to patients receiving warfarin. Patient should have coagulation parameters monitored regularly, and warfarin dose adjusted accordingly. If these drugs cannot be discontinued or replaced enrollment may be allowed after discussion with MSK PI.
Study Outcomes
The primary endpoint of the study was to determine the maximum tolerated dose of treatment with the combination of either duvelisib plus bortezomib or duvelisib plus romidepsin in patients with relapsed or refractory T-cell lymphomas. Additional efficacy endpoints for duvelisib plus bortezomib and duvelisib plus romidepsin collectively as well as disease subtypes during the dose expansion phase and safety were assessed as secondary endpoints. The additional efficacy endpoints included overall response rate, complete response rate, partial response rate, time to response, event-free survival and duration of response. Time to response is measured from treatment start date to date of first response, and duration of response is measured from date of first response to date of progression or last follow up. Patients who had other treatment-ending events were censored at the event dates. Event-free survival time is measured from treatment start date to progression, or other treatment-ending event (e.g. toxicity, clinical decision to end treatment). Patients who went on to SCT or withdrew consent were censored at that event date, In addition, tissue samples were collected from the enrolled patients, which was utilized for exploratory translational biological studies aimed at determining mechanisms of response or resistance to PI3K-δ and PI3K-γ inhibition.
Statistical Analysis
This study estimated a sample size of 49 patients in the dose escalation phase. The expansion cohort size was planned to be increased to 74 patients, to increase the number of patients at each maximum tolerated dose to obtain better estimates of efficacy. Patients with evaluable disease treated at the maximum tolerated dose in the dose escalation phase were combined with the dose expansion cohorts for estimating efficacy. Due to the moderate sample size in each treatment arm, no formal hypothesis test was set, but the preliminarily goal was to assess the expectation of a response rate greater than 30% (current response rate) by an absolute margin of 15–25%. For the efficacy endpoints (response rates), sample proportions and confidence intervals have been provided for each treatment arm. Similar estimates were calculated for each disease subtype. Time to response, event-free survival, and duration of response were assessed using Kaplan-Meier methods. EFS and OS were compared between PTCL patients on arm A who achieved complete response versus those who achieved only partial response or stable disease. In these analyses, EFS and OS were started at best response date to avoid guarantee time bias. The exploratory outcomes were evaluated using translational biological studies of response or resistance therapy and optimal combinations for mechanism-based therapy for T-cell lymphomas. Results have been reported qualitatively and no statistical analysis is required.
Key Trial Amendments.
Amendment 4 (Approved 12/13/2016) – A rollover cohort was added to account for patients receiving long-term continued treatment on a previous duvelisib study closed by the sponsor. This is Cohort 3 and two patients were enrolled and are not evaluable for the purposes of this study.
Amendment 7 (Approved 1/9/2018) – Addition of 9 patients to the Arm A PTCL cohort. Change of pharmaceutical company from Infinity Pharmaceuticals to Verastem. Additional inclusion/exclusion criteria clarified for washout periods, and to allow long term maintenance therapy for previous malignancy.
Amendment 8 (Approved 5/23/2018) – Change in Subject Eligibility to allow all patients, not just CTCL patients who have received localized RT as part of their immediate prior therapy to be considered for shorter washout period after discussion with PI Amendment 10 (Approved 6/12/2019) – Addition of 25 patients to the Arm A PTCL Cohort. Additional clarifications throughout the entire protocol.
Amendment 11 (Approved 8/28/2019) – Exclusion criteria updated to include language on uncontrolled infection requiring systemic antimicrobial medications)
Amendment 14 (Approved 4/27/2021) – SecuraBio acquired global rights for Duvelisib from Verastem. Updates were made in this document in all sections to reflect that Secura Bio is now the funder for this study.
Amendment 15 (Approved 6/22/2021) – Protocol updated to allow patients who came off for reasons other than disease progression to be followed for disease and survival status until progression or until the study ends.
In vivo patient-derived xenograft studies
Xenograft studies were performed using NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice (Stock #005557) purchased from Jackson Laboratories. All animal experiments were performed with approval of the Dana-Farber Cancer Institute (DFCI) Institutional Animal Care and Use Committee approved protocol #13–034. Mice were housed and allowed to acclimate to the housing facility for at least 1 week. All mice had free access to food and water and were randomly assigned to experimental groups.
A full description of each PDX model is available online at the Public Repository of Xenografts (www.PRoXe.org). For AITL PDX model (DFTL-47880), viably frozen PDX cells were thawed and washed in 1 × PBS before tail-vein injection at 1 × 106 cells per mouse. Tumor burden was monitored periodically by flow cytometry of peripheral blood. Blood was processed with Red Blood Cell Lysis Buffer (Qiagen) before staining with antibodies against human CD45 (BD Biosciences #566026) and human CD2 (Invitrogen #17–9459) in 1 × PBS with 1 mM EDTA plus 1% fetal bovine serum. Data were acquired on a BD LSRFortessa flow cytometer and analyzed with FlowJo Software. Upon engraftment with (>1% for survival, >10% for pharmacodynamic [PD] study) hCD45+/hCD2+ cells in peripheral blood, mice were randomized to vehicle or treatment arms. Mice were treated with either single or combined with duvelisib (50 mg per kg, b.i.d., p.o.) and romidepsin (1 mg per kg, weekly, i.p.). The vehicle was given orally by gavage on the same schedule as duvelisib. Mice were monitored daily for clinical signs of disease and humanely euthanized when they reached a clinical end point or PD time point (7 days after treatment). Cells from spleen and bone marrow were collected for flow cytometry analysis. Mouse cells were then depleted by using EasySep Mouse/Human Chimera Isolation Kit (Stem Cell Technologies, #19849). Purified human tumor cells were then analyzed by whole exome and transcriptome sequencing.
Patient tumor whole exome sequencing
Whole exome sequencing of patient tumors was performed by BostonGene as previously described:
Preprocessing:
Whole exome sequencing (WES) was performed on 75 tumor samples, of which 49 also had paired WES of normal tissue. Baseline WES samples were available for 43 patients (25 PTCL, 18 CTCL). Additionally, we performed dynamic WES analysis of samples on treatment or at the end of treatment or both for 12 patients. Exome capture was performed using the SureSelect XT HS2 RNA system (Agilent) according to the manufacturer’s protocol. In this study two different probe sets were utilized for the hybridization procedures, SureSelect Human All Exon V7 (Agilent, Cat#5191–4029) and a custom probe set, V7+ UTR, that was designed to cover all the coding exons and 5’ and 3’ utr regions. Sequencing libraries were sequenced on HiSeq2500 as paired end 100bp with average 78M reads per sample and median exon coverage of 78X.
NGS data quality control:
FastQC v0.11.5, FastQ Screen v0.11.1, RSeQC v3.0.0 and MultiQC v1.6 were used to perform quality control (QC) of all NGS samples. Sample correspondence was checked by HLA comparison from RNA-Seq using OptiType72.
Alignment:
low quality reads were filtered using FilterByTile/BBMap v37.90 and aligned to human reference genome GRCh38 (GRCh38.d1.vd1 assembly) using BWA v0.7.17. Duplicate reads were removed using Picard’s v2.6.0 MarkDuplicates, indels were realigned by IndelRealigner and recalibrated by BaseRecalibrator and ApplyBQSR (last three tools from GATK v3.8.1).
Variant calling:
Both germline and somatic single nucleotide variations (sSNVs), small insertions and deletions were all detected using Strelka v2.9. All variants, insertions and deletions were annotated using Variant Effect Predictor v92.1. Copy number alterations were evaluated with a customized version of Sequenza v2.1.2.
Tumor whole transcriptome sequencing
RNA extraction:
Flash Frozen tissue specimens were processed using Maxwell RSC SimplyCells RNA extraction kit. FFPE specimens were cut into 10-micron thick sections and utilized in either Qiagen AllPrep DNA/RNA extraction kits, or Maxwell RSC FFPE RNA extraction kits. All extractions were performed as per manufacturer’s instructions.
Library Preparation and Sequencing:
PolyA libraries were generated using the Illumina Truseq Stranded mRNA kit (Illumina, Cat#20040532). Cleanup procedures were utilized by Agencourt AMPure XP beads (Beckman Coulter, Cat A63881). These libraries were generated as described by the manufacturer’s instructions. All RNA libraries were sequenced on a NovaSeq 6000. Samples were sequenced with a target of 50M reads, with 151bp paired end sequencing.
Analysis:
All RNA libraries were analyzed using 130 nt + 21 nt paired-end sequencing on Illumina sequencers. Adapter sequences were removed from read 1 (130 nt), and UMI sequences were extracted from read 2 (21 nt). Differential gene-expression testing was performed using DESEQ2 v1.34.0. Gene-set enrichment against hallmark, gene ontology, and canonical pathway gene sets was computed using the pre-ranked module in gseapy v1.0.6.
Deconvolution of bulk RNA-seq
The Kassandra machine learning algorithm was used to predict cell percentages from bulk RNA-seq73. The model consisted of a two-level hierarchical ensemble that used LightGBM as building blocks. The model was trained on artificial RNA-seq mixtures of different cell types (T cells, B cells, NK, macrophages, cancer-associated fibroblasts, and endothelial cells) obtained from multiple datasets of sorted cells. All datasets were isolated from poly-A or total RNA-seq profiled human tissues with read lengths higher than 31 bp and at least 4 million coding read counts. These datasets passed quality control by FASTQC with minimal contamination (< 2%). The model was trained to predict the percentage of RNA belonging to specific cell types. Predicted percentages of RNA were later converted into percentages of cells using the methodology described previously74.
Single-cell transcriptomic profiling of peripheral blood
Single cell RNA FASTQ data was processed using Cellranger v6.0.2 count workflow to generate gene expression count matrices. TCR data was processed using Cellranger’s vdj pipeline to generate cell-clonotype annotations. Processed RNA and ADT matrices were combined into a single Seurat v4.0.0 object (using the CreateSeuratObject function), into which TCR clonotype information was subsequently incorporated using scRepertoire v1.7.2 combineExpression function. Filtering was applied upon the merged data to only retain cells with (1) greater than 500 and less than 3000 detected unique RNA features, (2) less than 10000 total RNA molecules (3) less than 5 percent mitochondrial RNA reads and (4) less than 2500 total ADT molecules. RNA and ADT data were normalized using Seurat’s NormalizeData function with ‘LogNormalize’ and ‘CLR’ methods specified for each, respectively.
PCA was performed (using Seurat’s RunPCA function) and to mitigate the effect of lane-specific and sample-specific covariates/batch effects, Harmony v1.0.0 RunHarmony function was used to correct embeddings. Elbow plots and jack straw plots were manually inspected to determine the number of principal components to use downstream and a global UMAP was constructed using Seurat’s weighted nearest-neighbor workflow upon corrected RNA and ADT data (n=16969). Using clustree v.0.5.0 produced tree diagrams for various resolution input values, cluster stability was evaluated to avoid over-clustering and optimize the number of communities selected. Subsequently, separate Seurat object for Myeloid (monocytes and dendritic cells), B and T-lymphocytes were produced by evaluation of and isolation by canonical marker expression. T-lymphocytes were further separated into CD8 and CD4 compartments by gating on each of those markers using the ADT assay data. The previously described UMAP construction and clustering steps were performed for each cell-subtype object. Specialized cell-types were labeled by manually evaluating differentially expressed genes and surface protein markers across clusters. Treatment-induced transcriptional and proteomic changes were characterized using Seurat’s implementation of the Wilcoxan-Rank Sum Test. Fgsea v.1.24.0 and escape v1.8.0 were used to evaluate-functional module level expression changes of various gene sets taken from the Molecular Signatures Database (MSigDB).
Western Blot analysis
Sample preparation of whole cell lysates, SDS-PAGE, membrane transfer and blotting were performed according to standard protocols. Briefly, cells were lysed in Cell Lysis Buffer (Cell Signaling Technologies, #9803) supplemented with anti-protease and anti-phosphatase cocktails (Thermo Fisher, #78442). Protein concentration was determined using BCA Protein Assay Kit (Pierce, #23225). 5~15 μg of protein was resolved by SDS-PAGE and transferred onto NC or PVDF membranes. Membranes were blocked and incubated overnight at 4°C with gentle agitation with primary antibodies. Then primary antibodies were conjugated to secondary HRP-conjugated antibodies (Bio-Rad #170–6515, Invitrogen #62–6520) and the signal was detected using ECL Kit (Sigma, GERPN2232) and acquired on Chemidoc MP System (Bio-Rad). Antibodies to TSC2 (#4308, D93F12) and PTEN (#9559, 138G6) were purchased from Cell Signaling Technologies. Antibodies to β-actin (#A5441) was purchased from Sigma Aldrich. Antibodies were used at 1:1000 dilution, or at 1:5000 dilution (for β-actin).
TSC2 and PTEN dependence in transformed cell lines
OCI-ly13.2 cells are a transformed human T-cell non-Hodgkin lymphoma cell line most consistent with an anaplastic large cell lymphoma and expressing the pan-T-cell-specific markers CD2, CD3, and CD775. For inducible sgRNA constructs, the FgH1tUTG (Addgene #70183) plasmid was used. SgRNA sequences included a 4-bp overhang for the forward (TCCC) and complementary reverse (AAAC) oligos to enable cloning into the BsmBI sites of the lentiviral construct. The sequences of sgRNA are listed in Supplementary Table 9. For CRISPR knockdown, OCI-Ly13.2 cells with stable Cas9 expression were transduced with the inducible sgRNA vector containing the designed sequences by lentivirus. For treatment of cell lines to induce expression of the sgRNA, doxycycline hyclate (Sigma-Aldrich #D9891) was added to tissue culture media to a final concentration of 1 μg/ml. To assess the impact of TSC2 and/or PTEN CRISPR editing on cell proliferation, we used the CellTiter-Glo Luminescent Cell Viability reagent (Promega) in accordance with the manufacturer’s protocol on T-cells cultured in the presence or absence of duvelisib ranging from a final concentration of 1 nM to 10 μM as indicated. Assays were conducted in triplicate, and statistical significance was assessed by two-tailed Student’s t-test assuming unequal variance.
O-link proteomic analysis of plasma
O-link analysis of circulating plasma protein concentrations was performed using a proximity extension assay (PEA) (Olink Proteomics, Uppsala, Sweden) which allows for the simultaneous analysis of 92 protein biomarkers on each panel, as previously described76. Briefly, one microliter of plasma from freshly thawed samples was incubated overnight and allowed to bind with oligonucleotide-labeled antibody pairs to form specific DNA duplexes. This template was then extended and pre-amplified, and the individual protein markers were measured using high-throughput microfluidic real-time PCR (Fluidigm, South San Francisco, CA). The resulting Ct values were normalized and adjusted with a correction factor according to the manufacturer’s instructions to calculate a normalized protein expression value (NPX) in log2 scale. Samples were processed in batches with pooled quality control samples included in each batch. For the present study, plasma concentrations of proteins were measured using the ProSeek Inflammation and Immune Response panels.
Extended Data
Extended Figure 1. Expanded enrollment diagram for NCT#02783625.
Expanded diagram showing enrollment on NCT#02783625.
Extended Figure 2. Study design and outcomes of NCT#02783625 in patients with PTCL and CTCL.
(a) Swimmer’s plot depicting responses of patients with PTCL to bortezomib + duvelisib (Arm B). (b) Kaplan-Meier curve showing event-free survival of all PTCL patients stratified by treatment arm. (c) Swimmer’s plot depicting responses of patients with CTCL to romidepsin + duvelisib (Arm a) or bortezomib + duvelisib (Arm B) as indicated. (d) Kaplan-Meier curve showing event-free survival of patients with CTCL, stratified by treatment arm.
Extended Figure 3. A tumor-specific T follicular-helper phenotype is associated with response to romidepsin and duvelisib.
(a) Best response of patients with PTCL receiving romidepsin + duvelisib in Arm A compared with NCT00426764, stratified by whether PTCL were of follicular helper subtype. (b) Oncoprint of somatic mutations observed in pre-treatment biopsy samples from patients with PTCL, organized by treatment response and frequency. Stars indicate genes in which mutations are enriched in either responders (R) or non-responders (NR). (c) Lollipop plot of individual mutations in genes commonly mutated in PTCL with Tfh features. (d) Genes enriched in responders versus non-responders in pre-treatment samples obtained from patients with PTCL. (e) B-cell proportions estimated by CIBERSORT deconvolution in pre-treatment samples from responders (N=13) versus non-responders (N=10). Data are presented as box plots with center depicting median, bounds of box depicting interquartile range, and whiskers depicting minima and maxima. (f) Enrichment of IL21 in pre-treatment bulk RNA-seq samples from PTCL responders (N=5) versus non-responders (N=7) to romidepsin and duvelisib. Data are presented as box plots with center depicting median, bounds of box depicting interquartile range, and whiskers depicting minima and maxima. P values calculated by Chi squared test (a), Wald Test corrected for False Discovery Rate using the Benjamini-Hochberg procedure (d), student’s t-test (e), and two-sided Wilcoxon Rank-Sum Test (f).
Extended Figure 4. Pre-treatment predictors of response to romidepsin and duvelisib in patients with PTCL and CTCL.
(a) Dot plot depicting enrichment of MSigDB genesets in pre-treatment bulk RNA-seq from responding vs. non-responding patients with PTCL. (b) Oncoprint of somatic mutations observed in pre-treatment biopsy samples from patients with CTCL, organized by treatment response and frequency. (c) Lollipop plot of individual mutations in NFKB2 in patients with CTCL. (d) Copy number plot depicting alterations by chromosome in patients with CTCL. RET and NFKB2 genes loci are indicated. (e) Genes enriched in responders versus non-responders in pre-treatment samples obtained from patients with CTCL. P values determined by Wald Test corrected for False Discovery Rate using the Benjamini-Hochberg procedure (e).
Extended Figure 5. Mechanisms of response to romidepsin and duvelisib in PTCL xenografts and patient samples.
(a) Experimental design and survival of mice xenografted with an AITL xenograft and treated with either vehicle, romidepsin, duvelisib, or the combination. (b) Decrease in mitosis-associated MSigDB genesets in on-treatment versus pre-treatment samples from PTCL patients who responded to therapy (left) and AITL xenografts (right). Normalized enrichment score (NES), false discovery rate (FDR) and proportion of genes expressed in each gene set are as described. P values calculated using log-rank test (a) and estimated as the number of random gene set enrichment scores with the same or more extreme values divided by the total number of randomly generated gene sets, corrected for False Discovery Rate using the Benjamini-Hochberg procedure (b).
Extended Figure 6. JAK/STAT-driven resistance to romidepsin and duvelisib.
(a) Proportion of patients with activation mutations or amplifications of JAK/STAT genes, stratified by response. (b) Copy number plot depicting alterations by chromosome in patients with PTCL. STAT1 locus on chromosome 2 is highlighted. (c) Lollipop plot of pre-treatment mutations in JAK1 and JAK3 in non-responders to romidepsin + duvelisib. (d) UMAP of CD45+ cells isolated from pre- and on-treatment peripheral blood samples obtained from patients with high-burden circulating disease receiving romidepsin + duvelisib. (e) Expression of T-cell markers on malignant cells from Patients Y and Z visualized in 2-dimensional UMAP space. (f) Differently expressed genes in malignant cells isolated from the peripheral blood of a patient who did not respond to romidepsin + duvelisib. (g) Expression of IL6_JAK_STAT3 genes in pre-treatment and on-treatment circulating malignant T-cells from patients with (Z) or without (Y) response to romidepsin + duvelisib. (h) Gene set enrichment analysis of HALLMARK_IL6_JAK_STAT3_SIGNALING in PDX tumors refractory to duvelisib monotherapy. P values determined by Fisher’s exact test (a), two-sided Wilcoxon Rank-Sum Test with Bonferroni correction for multiple comparisons (f), student’s t-test (g), and estimated as the number of random gene set enrichment scores with the same or more extreme values divided by the total number of randomly generated gene sets, corrected for False Discovery Rate using the Benjamini-Hochberg procedure (h).
Extended Figure 7. PI3K/mTORC1 reactivation in tumors resistant to romidepsin and duvelisib.
(a) Emergence of clones with indicated mutations in patients treated with romidepsin + duvelisib who ultimately experienced disease progression. (b) Expression of PI3K_AKT_MTOR genes in pre-treatment and on-treatment circulating malignant T-cells from patients with (Z) or without (Y) response to romidepsin + duvelisib. (c) Efficient CRISPR editing of TSC2 and PTEN expression by 2 independent guide RNAs as shown by western blot. (d) Sensitivity of OCI-ly13.2 cells expressing Cas9 and doxycycline-inducible sgRNA targeting TSC2 (above) or PTEN (below) to duvelisib in vitro, with (N=3) or without (N=3) the addition of doxycycline to induce gene editing. Data presented as mean values +/− SD and are representative of two independent experiments. (e) Decrease in phagocytosis-associated MSigDB genesets in on-treatment versus pre-treatment samples from PTCL patients who did not respond to therapy. (f) Estimated intratumoral macrophage abundance, based on CIBERSORT deconvolution, in pre-treatment and on-treatment biopsies of patients undergoing treatment with romidepsin + duvelisib, stratified by response (non-responder pre-treatment N=11, non-responder on-treatment N=10, responder pre-treatment N=12, responder on-treatment N=10). Data are presented as box plots with center depicting median, bounds of box depicting interquartile range, and whiskers depicting minima and maxima. P values determined using student’s t-test (b) and estimated as the number of random gene set enrichment scores with the same or more extreme values divided by the total number of randomly generated gene sets, corrected for False Discovery Rate using the Benjamini-Hochberg procedure (e).
Extended Figure 8. Persistent NF-kB signaling and proliferation in patients with clinically significant adverse events following romidepsin and duvelisib.
(a) Changes in plasma analytes between C1D8 and C1D1 samples in patients treated with duvelisib or duvelisib + romidepsin as indicated. (b) UMAP of CD45+ cells isolated from pre- and on-treatment peripheral blood samples obtained from patients without circulating disease receiving romidepsin + duvelisib. (c) Dot plot depicting average expression and frequency of expression of genes defining CD4+T, CD8+T, and myeloid cell subsets as indicated. (d) Differentially expressed genes in pre-treatment peripheral blood CD14+ monocytes from patient experiencing clinically significant (Grade 2–3) versus non-significant (Grade 0–1) hepatotoxicity. P values calculated using two-sided Wilcoxon Rank-Sum Test with Bonferroni correction for multiple comparisons (d).
Extended Figure 9. Uncropped Western Blots.
Uncropped Western blots from CRISPR-edited OCI-ly13.2 Cas9-expressing cells transduced with either control guide RNAs, TSC2-targeting guide RNAs, or PTEN-targeting guide RNAs.
Supplementary Material
ACKNOWLEDGMENTS
This was an investigator-initiated study funded initially by Infinity Pharmaceuticals and subsequently by Verastem and Secura Bio®, Inc. as well as LLS SCOR 7011–16, LLS SCOR 7026–21 and NCI R35 CA231958. Medical writing support provided by Shilpa Lalchandani, PhD. S.M.H. was supported by Nonna’s Garden Foundation. A.J.N. was supported by NCI R00 CA256497. S.A.V. was supported by a Steven A. Greenberg Lymphoma Research Grant. We would like to thank our patients and their families, and the faculty and research staff of our various institutions.
COMPETING INTERESTS STATEMENT
D.M.W. is an employee of Merck and Co., owns equity in Merck and Co., Bantam, Ajax, and Travera, received consulting fees from Astra Zeneca, Secura, Novartis, and Roche/Genentech, and received research support from Daiichi Sankyo, Astra Zeneca, Verastem, Abbvie, Novartis, Abcura, and Surface Oncology. S.A.V. has served as an advisor for Immunai and has received consulting fees from ADC Therapeutics and Koch Disruptive Technologies. JJB received consulting fees from Sobi, Omeros, Bluebird Bio, Sanofi, Immusoft, SmartImmune, Bluerock and Advanced Clinical. NMS has received institutional clinical trial funding from AstraZeneca, Bristol Myers-Squibb, Celgene, C4 Therapeutics, Corvus Pharmaceuticals, Daiichi Sankyo, Genentech/Roche, Innate Pharmaceuticals, Secura Bio/Verastem, Yingli Pharmaceuticals and Dizal pharmaceuticals. She has received compensation for service as a consultant for Astrazeneca, Secura Bio/Verastem, Daiichi Sankyo, C4 Therapeutics, Genentech, Janssen Karyopharm Therapeutics, and Kyowa Hakko Kirin. NMS is funded as a Scholar in Clinical Research through the Leukemia Lymphoma Society and this study was reviewed as part of the ASCO/AACR Workshop. S.M.H. has received research support from ADC Therapeutics, Affimed, C4, Celgene, Crispr Therapeutics, Daiichi Sankyo, Dren, Kyowa Hakko Kirin, Millennium /Takeda, Seattle Genetics, and SecuraBio. He has received consulting fees from Affimed, Abcuro Inc, Corvus, Daiichi Sankyo, Kyowa Hakko Kirin, ONO Pharmaceuticals, SeaGen, SecuraBio, Takeda and Yingli Pharmaceuticals. A.J.M. has received research support from Beigene, Seattle Genetics, Merck, Bristol-Myers Squibb, Incyte, Affimed and Astra-Zeneca. She has received consulting fees from Affimed, Merck, Seattle Genetics, and Takeda.
Footnotes
The remaining authors declare no competing interests.
A female of reproductive potential is a sexually mature female who: has not undergone a hysterectomy or bilateral oophorectomy; or has not been naturally postmenopausal for at least 24 consecutive months (i.e. has had menses at any time in the preceding 24 consecutive months).
DATA AVAILABILITY
For original clinical data, plase contact horwitzs@mskcc.org. Deidentified participant data can be shared. External data requests will be answered within one month. The study protocols are included as a data supplement available with the online version of this article. Bulk RNA-sequencing data are available under the Sequence Read Archive (PRJNA1098020). Single-cell transcriptomic data are available under the Gene Expression Omnibus (GSE266760).
REFERENCES
- 1.Leseux L, et al. Syk-dependent mTOR activation in follicular lymphoma cells. Blood 108, 4156–4162 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Rudelius M, et al. Constitutive activation of Akt contributes to the pathogenesis and survival of mantle cell lymphoma. Blood 108, 1668–1676 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rassidakis GZ, et al. Inhibition of Akt increases p27Kip1 levels and induces cell cycle arrest in anaplastic large cell lymphoma. Blood 105, 827–829 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uddin S, et al. Role of phosphatidylinositol 3’-kinase/AKT pathway in diffuse large B-cell lymphoma survival. Blood 108, 4178–4186 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Slupianek A, et al. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res 61, 2194–2199 (2001). [PubMed] [Google Scholar]
- 6.Maeda T, Yagasaki F, Ishikawa M, Takahashi N & Bessho M Transforming property of TEL-FGFR3 mediated through PI3-K in a T-cell lymphoma that subsequently progressed to AML. Blood 105, 2115–2123 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Vallois D, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 128, 1490–1502 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Li Z, et al. Recurrent mutations in epigenetic modifiers and the PI3K/AKT/mTOR pathway in subcutaneous panniculitis-like T-cell lymphoma. Br J Haematol 181, 406–410 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Heavican TB, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood 133, 1664–1676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watatani Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia 33, 2867–2883 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Baohua Y, Xiaoyan Z, Tiecheng Z, Tao Q & Daren S Mutations of the PIK3CA gene in diffuse large B cell lymphoma. Diagn Mol Pathol 17, 159–165 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Pfeifer M, et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 110, 12420–12425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarisbrick JJ, Woolford AJ, Russell-Jones R & Whittaker SJ Loss of heterozygosity on 10q and microsatellite instability in advanced stages of primary cutaneous T-cell lymphoma and possible association with homozygous deletion of PTEN. Blood 95, 2937–2942 (2000). [PubMed] [Google Scholar]
- 14.Nakahata S, et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat Commun 5, 3393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki T, et al. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science 287, 1040–1046 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science 297, 1031–1034 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Flinn IW, et al. The phase 3 DUO trial: duvelisib vs ofatumumab in relapsed and refractory CLL/SLL. Blood 132, 2446–2455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flinn IW, et al. DYNAMO: A Phase II Study of Duvelisib (IPI-145) in Patients With Refractory Indolent Non-Hodgkin Lymphoma. J Clin Oncol 37, 912–922 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Dreyling M, et al. Phase II study of copanlisib, a PI3K inhibitor, in relapsed or refractory, indolent or aggressive lymphoma. Ann Oncol 28, 2169–2178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopal AK, et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med 370, 1008–1018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furman RR, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 370, 997–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson NC, Kasamon Y, Pazdur R & Gormley N The saga of PI3K inhibitors in haematological malignancies: survival is the ultimate safety endpoint. Lancet Oncol 23, 563–566 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Matasar MJ, et al. Copanlisib plus rituximab versus placebo plus rituximab in patients with relapsed indolent non-Hodgkin lymphoma (CHRONOS-3): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 22, 678–689 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Barr PM, et al. Phase 2 study of idelalisib and entospletinib: pneumonitis limits combination therapy in relapsed refractory CLL and NHL. Blood 127, 2411–2415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampson BL, et al. Idelalisib given front-line for treatment of chronic lymphocytic leukemia causes frequent immune-mediated hepatotoxicity. Blood 128, 195–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chellappa S, et al. The PI3K p110delta Isoform Inhibitor Idelalisib Preferentially Inhibits Human Regulatory T Cell Function. J Immunol 202, 1397–1405 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Campo E, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood 117, 5019–5032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz SM, et al. Activity of the PI3K-δ,γ inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood 131, 888–898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alaggio R, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 36, 1720–1748 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campo E, et al. The International Consensus Classification of Mature Lymphoid Neoplasms: a report from the Clinical Advisory Committee. Blood 140, 1229–1253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H & Abdul-Hay M T-cell lymphomas, a challenging disease: types, treatments, and future. Int J Clin Oncol 22, 18–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vose J, Armitage J & Weisenburger D International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 26, 4124–4130 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Bellei M, et al. The outcome of peripheral T-cell lymphoma patients failing first-line therapy: a report from the prospective, International T-Cell Project. Haematologica 103, 1191–1197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coiffier B, et al. Results from a pivotal, open-label, phase II study of romidepsin in relapsed or refractory peripheral T-cell lymphoma after prior systemic therapy. J Clin Oncol 30, 631–636 (2012). [DOI] [PubMed] [Google Scholar]
- 35.O’Connor OA, et al. Belinostat in Patients With Relapsed or Refractory Peripheral T-Cell Lymphoma: Results of the Pivotal Phase II BELIEF (CLN-19) Study. J Clin Oncol 33, 2492–2499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horwitz SM, et al. Objective responses in relapsed T-cell lymphomas with single-agent brentuximab vedotin. Blood 123, 3095–3100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connor OA, et al. Pralatrexate in patients with relapsed or refractory peripheral T-cell lymphoma: results from the pivotal PROPEL study. J Clin Oncol 29, 1182–1189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler DG, et al. PI3K-δ and PI3K-γ inhibition by IPI-145 abrogates immune responses and suppresses activity in autoimmune and inflammatory disease models. Chem Biol 20, 1364–1374 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Dong S, et al. IPI-145 antagonizes intrinsic and extrinsic survival signals in chronic lymphocytic leukemia cells. Blood 124, 3583–3586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneda MM, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Henau O, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539, 443–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkler DG, et al. PI3K-delta and PI3K-gamma inhibition by IPI-145 abrogates immune responses and suppresses activity in autoimmune and inflammatory disease models. Chem Biol 20, 1364–1374 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Horwitz SM, et al. Dose Optimization of Duvelisib in Patients with Relapsed or Refractory Peripheral T-Cell Lymphoma from the Phase 2 Primo Trial: Selection of Regimen for the Dose-Expansion Phase. Blood 134, 1567–1567 (2019). [Google Scholar]
- 44.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120, 1466–1469 (2012). [DOI] [PubMed] [Google Scholar]
- 45.de Leval L, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 109, 4952–4963 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Couronne L, Bastard C & Bernard OA TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 366, 95–96 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Cortes JR, et al. RHOA G17V Induces T Follicular Helper Cell Specification and Promotes Lymphomagenesis. Cancer Cell 33, 259–273 e257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng SY, et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood 132, 935–947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaefer A, Reinhard NR & Hordijk PL Toward understanding RhoGTPase specificity: structure, function and local activation. Small GTPases 5, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Y & Jaffe ES How I Diagnose Angioimmunoblastic T-Cell Lymphoma. Am J Clin Pathol 156, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakatsuki Y, Neurath MF, Max EE & Strober W The B cell-specific transcription factor BSAP regulates B cell proliferation. J Exp Med 179, 1099–1108 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horwitz SM, et al. Activity of the PI3K-delta,gamma inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood 131, 888–898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogelzang A, et al. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity 29, 127–137 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol 173, 68–78 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Ishii S, Kurasawa Y, Wong J & Yu-Lee LY Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc Natl Acad Sci U S A 105, 4179–4184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noh EJ & Lee JS Functional interplay between modulation of histone deacetylase activity and its regulatory role in G2-M transition. Biochem Biophys Res Commun 310, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Britschgi A, et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 22, 796–811 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Moskowitz AJ, et al. A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood 138, 2828–2837 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bourd-Boittin K, et al. CX3CL1/fractalkine shedding by human hepatic stellate cells: contribution to chronic inflammation in the liver. J Cell Mol Med 13, 1526–1535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen X, et al. CD4 T cells promote tissue inflammation via CD40 signaling without de novo activation in a murine model of liver ischemia/reperfusion injury. Hepatology 50, 1537–1546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimura K, Nagaki M, Takai S, Satake S & Moriwaki H Pivotal role of nuclear factor kappaB signaling in anti-CD40-induced liver injury in mice. Hepatology 40, 1180–1189 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Ikeda A, et al. Progression of autoimmune hepatitis is mediated by IL-18-producing dendritic cells and hepatic CXCL9 expression in mice. Hepatology 60, 224–236 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Tsutsui H, et al. IL-18 accounts for both TNF-alpha- and Fas ligand-mediated hepatotoxic pathways in endotoxin-induced liver injury in mice. J Immunol 159, 3961–3967 (1997). [PubMed] [Google Scholar]
- 66.Ho RT, Liew CT & Lai KN The expression of hepatocyte growth factor (HGF) and interleukin 6 (IL-6) in damaged human liver and kidney tissues. Hepatogastroenterology 46, 1904–1909 (1999). [PubMed] [Google Scholar]
- 67.Jo JC, et al. Peripheral T cell lymphomas in elderly patients: a retrospective analysis from the Hematology Association of South East Korea (HASEK). Ann Hematol 95, 619–624 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Mehta N, et al. A retrospective analysis of peripheral T-cell lymphoma treated with the intention to transplant in the first remission. Clin Lymphoma Myeloma Leuk 13, 664–670 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 372, 30–39 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Oki Y, et al. Phase I study of panobinostat plus everolimus in patients with relapsed or refractory lymphoma. Clin Cancer Res 19, 6882–6890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witzig TE, et al. The mTORC1 inhibitor everolimus has antitumor activity in vitro and produces tumor responses in patients with relapsed T-cell lymphoma. Blood 126, 328–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 72.Szolek A, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zaitsev A, et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell 40, 879–894 e816 (2022). [DOI] [PubMed] [Google Scholar]
- 74.Racle J, de Jonge K, Baumgaertner P, Speiser DE & Gfeller D Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng SY, et al. Targetable vulnerabilities in T- and NK-cell lymphomas identified through preclinical models. Nat Commun 9, 2024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su Y, et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 183, 1479–1495 e1420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For original clinical data, plase contact horwitzs@mskcc.org. Deidentified participant data can be shared. External data requests will be answered within one month. The study protocols are included as a data supplement available with the online version of this article. Bulk RNA-sequencing data are available under the Sequence Read Archive (PRJNA1098020). Single-cell transcriptomic data are available under the Gene Expression Omnibus (GSE266760).