Abstract
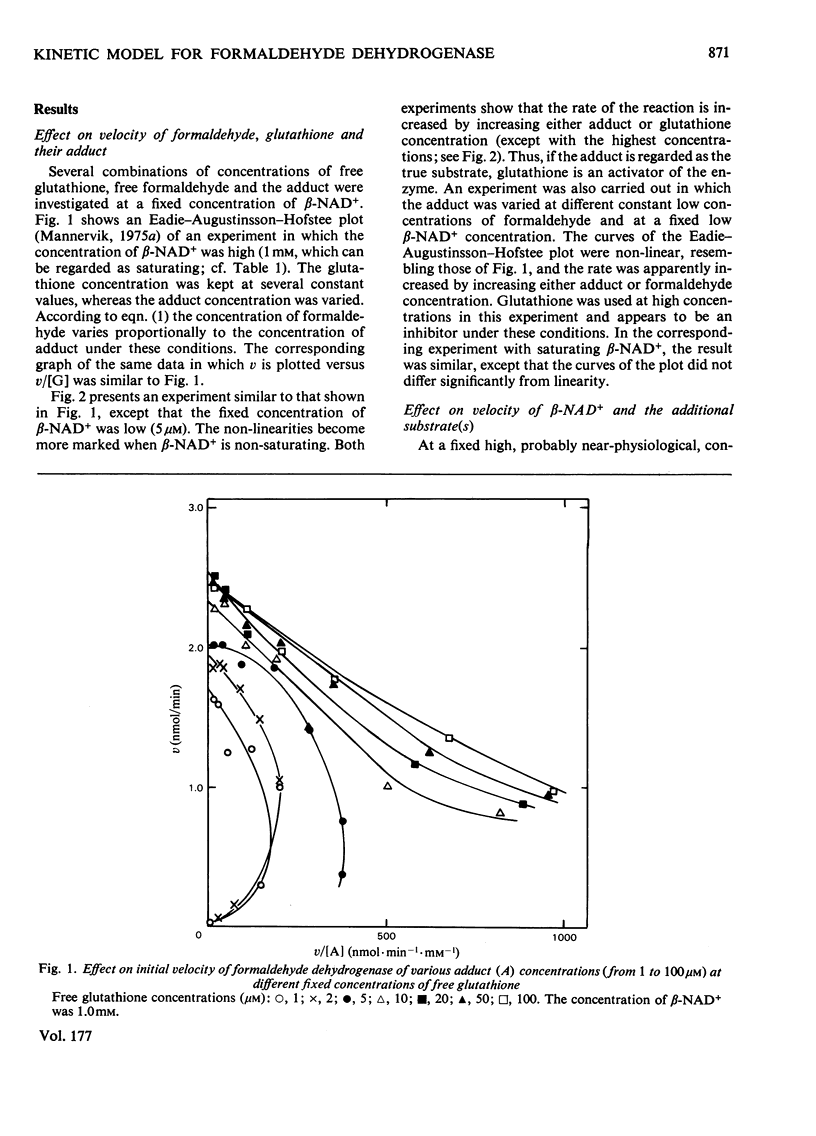
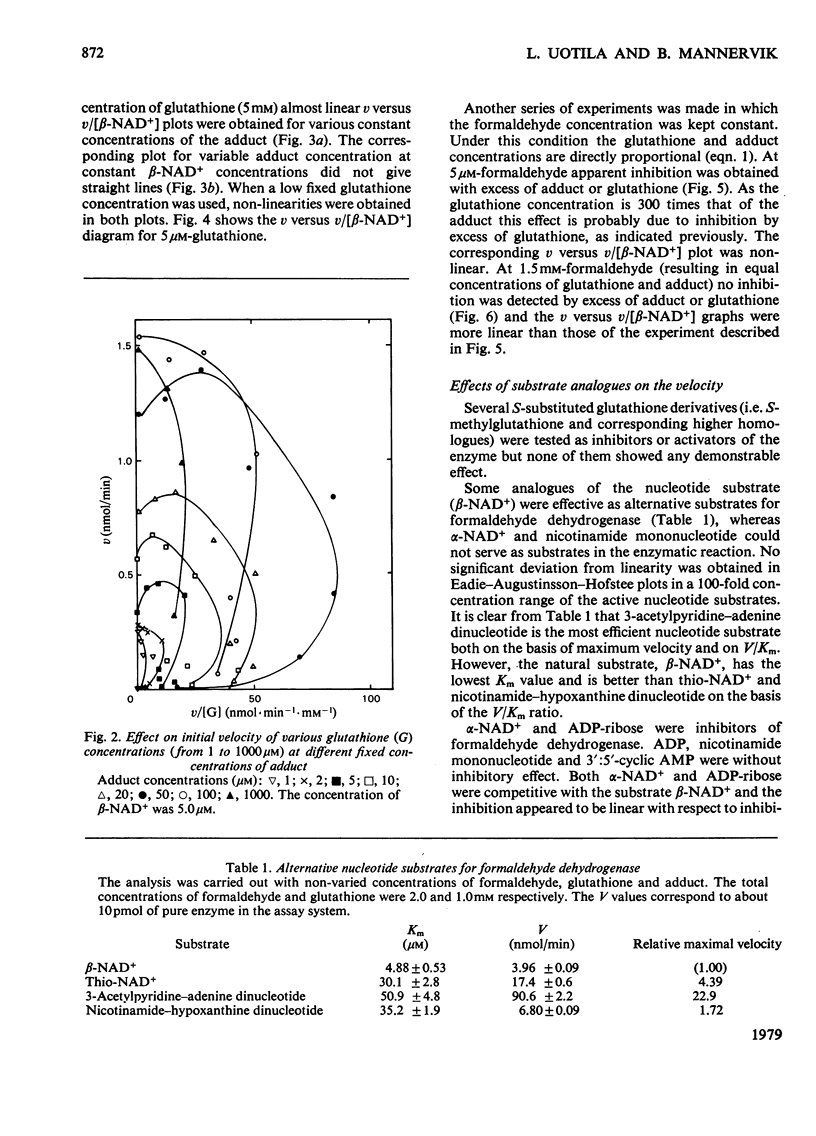
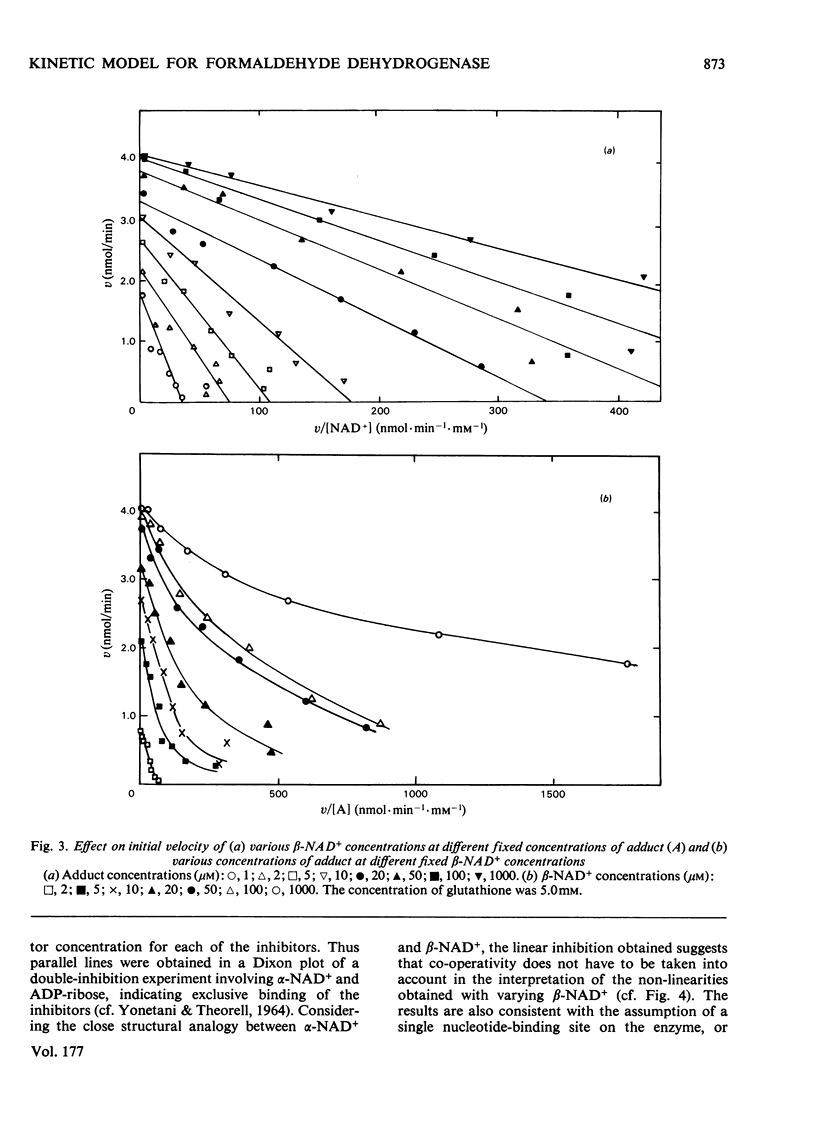
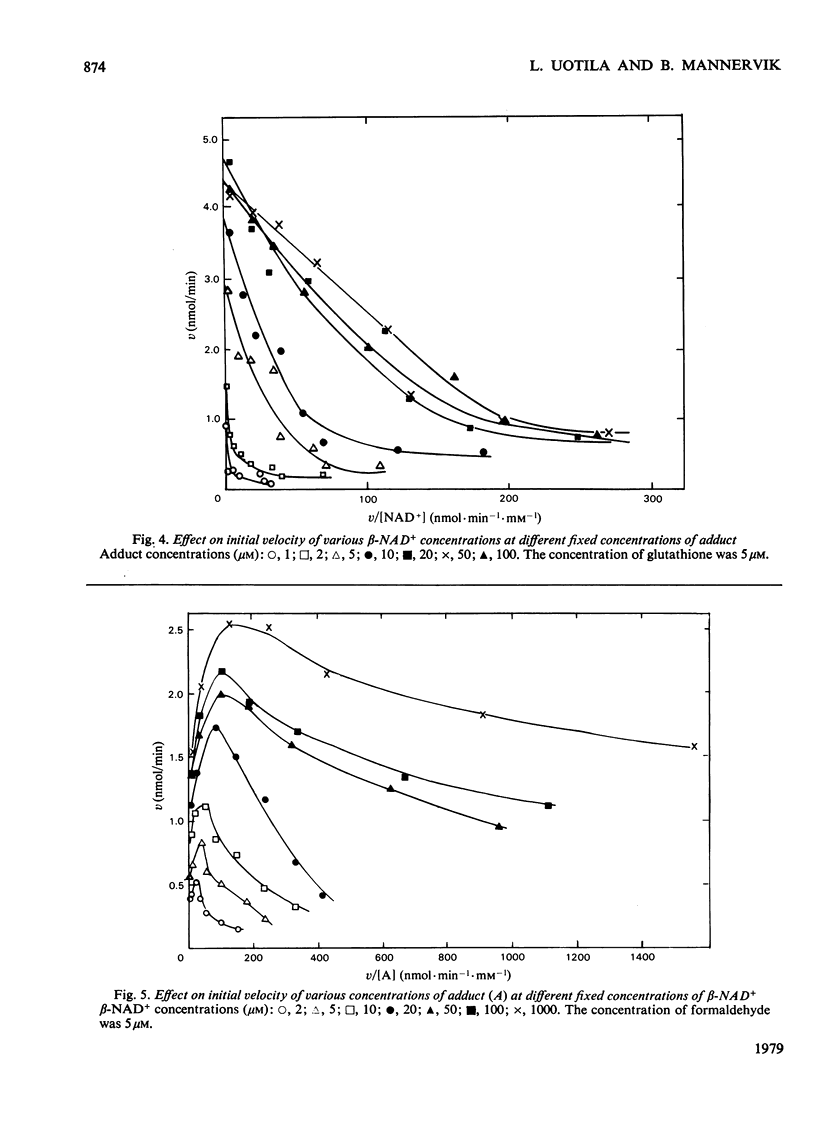
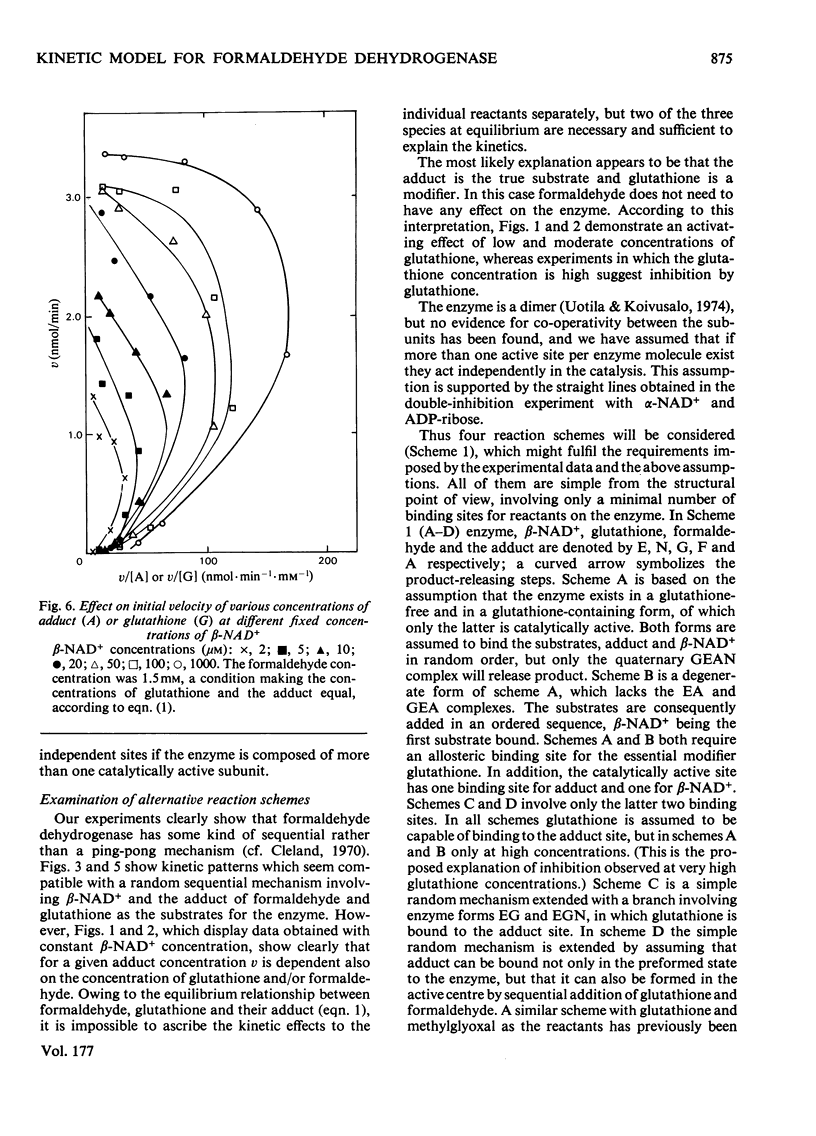
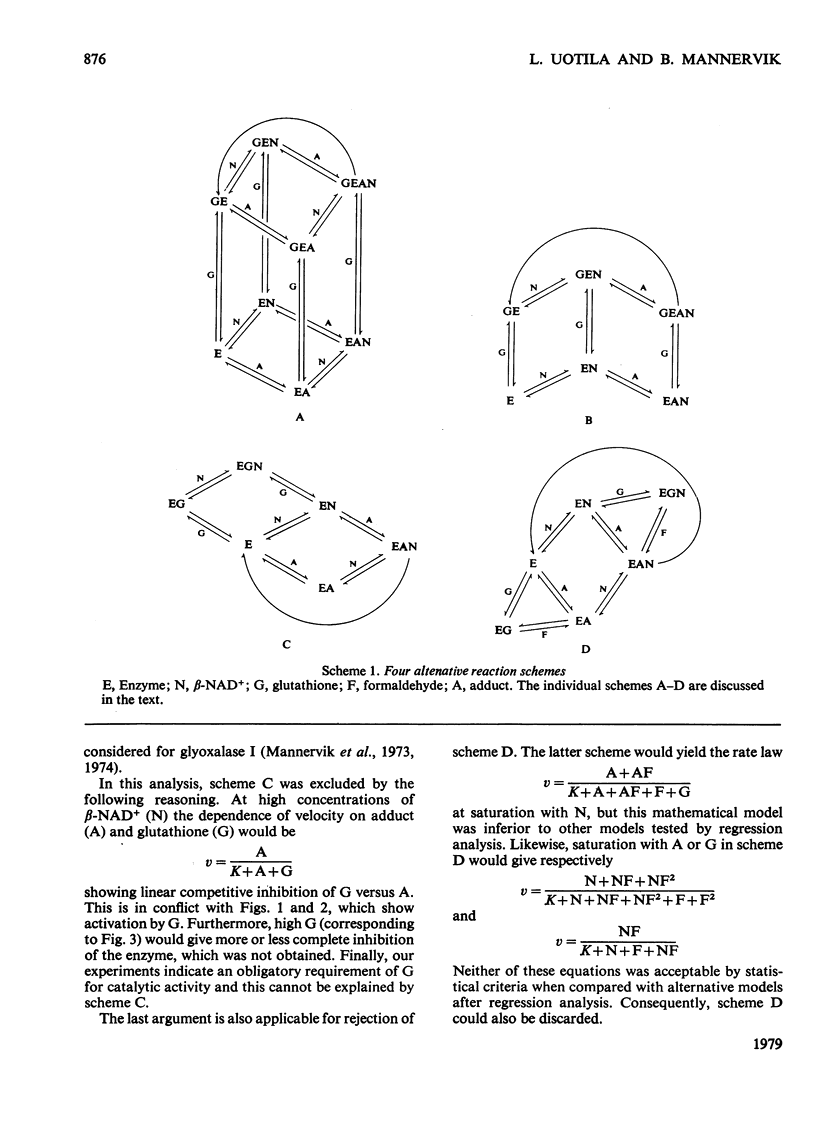
The steady-state kinetics of formaldehyde dehydrogenase from human liver have been explored. Non-linearities were obtained in v-versus-v[S] plots. It was necessary and sufficient to consider two reactants of the equilibrium mixture of formaldehyde, glutathione and their hemimercaptal adduct for a complete description of the kinetics. A random sequential reaction scheme is proposed in which adduct and beta-NAD+ are the substrates. In addition, glutathione can bind to an allosteric regulatory site and only the glutathione-containing enzyme is considered productive. Various alternative reaction models were examined but no simple alterative was superior to the model chosen. The discrimination was largely based on results of non-linear regression analysis. Several S-substituted glutathione derivatives were tested as activators or inhibitors of the enzyme, but all were without effect. Thio-NAD+, nicotinamide--hypoxanthine dinucleotide and 3-acetylpyridine-adenine dinucleotide could substitute for beta-NAD+ as the nucleotide substrate. alpha-NAD+ and ADP-ribose were competitive inhibitors with respect to beta-NAD+ and non-competitive with glutathione and the adduct. When used simultaneously, the inhibitors were linear competitive versus each other, indicating a single nucleotide-binding site or, if more than one, non-co-operative binding sites.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Askelöf P., Korsfeldt M., Mannervik B. Error structure of enzyme kinetic experiments. Implications for weighting in regression analysis of experimental data. Eur J Biochem. 1976 Oct 1;69(1):61–67. doi: 10.1111/j.1432-1033.1976.tb10858.x. [DOI] [PubMed] [Google Scholar]
- Bardsley W. G., Childs R. E. Sigmoid curves, non-linear double-reciprocal plots and allosterism. Biochem J. 1975 Aug;149(2):313–328. doi: 10.1042/bj1490313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártfai T., Mannervik B. A procedure based on statistical criteria for discrimination between steady state kinetic models. FEBS Lett. 1972 Oct 1;26(1):252–256. doi: 10.1016/0014-5793(72)80585-4. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Hurst R., Pincock A., Broekhoven L. H. Model discrimination and nonlinear parameter estimation in the analysis of the mechanism of action of beta-hydroxybutyrate dehydrogenase from Rhodopseudomonas spheroides. Biochim Biophys Acta. 1973 Sep 15;321(1):1–26. doi: 10.1016/0005-2744(73)90055-7. [DOI] [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Bartfai T., Górna-Hall B. Random pathway mechanism involving parallel one- and two- substrate branches for glyoxalase I from yeast. J Biol Chem. 1974 Feb 10;249(3):901–903. [PubMed] [Google Scholar]
- Mannervik B., Bártfai T. Discrimination between mathematical models of biological systems exemplified by enzyme steady state kinetics. Acta Biol Med Ger. 1973;31(2):203–215. [PubMed] [Google Scholar]
- Mannervik B. Graphical analysis of steady-state kinetic data of multireactant enzymes. Anal Biochem. 1975 Jan;63(1):12–16. doi: 10.1016/0003-2697(75)90184-0. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Górna-Hall B., Bártfai T. The steady-state kinetics of glyoxalase I from porcine erythrocytes. Evidence for a random-pathway mechanism involving one- and two-substrate branches. Eur J Biochem. 1973 Aug 17;37(2):270–281. doi: 10.1111/j.1432-1033.1973.tb02985.x. [DOI] [PubMed] [Google Scholar]
- Mannervik B. Nonlinear regression methods in design of experiments and mathematical modelling. Applications to the analysis of the steady-state kinetics of glutathione reductase. Biosystems. 1975 Jul;7(1):101–119. doi: 10.1016/0303-2647(75)90048-9. [DOI] [PubMed] [Google Scholar]
- Pfleiderer G., Woenckhaus C., Nelböck-Hochstetter M. Die intramolekulare Wechselwirkung zwischen Heterocyclen in Coenzymen. II. Alpha-Nicotinamid-adenin-dinucleotid. Justus Liebigs Ann Chem. 1965 Dec;690:170–176. doi: 10.1002/jlac.19656900116. [DOI] [PubMed] [Google Scholar]
- ROSE Z. B., RACKER E. Formaldehyde dehydrogenase from bakers' yeast. J Biol Chem. 1962 Oct;237:3279–3281. [PubMed] [Google Scholar]
- STRITTMATTER P., BALL E. G. Formaldehyde dehydrogenase, a glutathionedependent enzyme system. J Biol Chem. 1955 Mar;213(1):445–461. [PubMed] [Google Scholar]
- Uotila L., Koivusalo M. Formaldehyde dehydrogenase from human liver. Purification, properties, and evidence for the formation of glutathione thiol esters by the enzyme. J Biol Chem. 1974 Dec 10;249(23):7653–7663. [PubMed] [Google Scholar]
- Uotila L. Preparation and assay of glutathione thiol esters. Survey of human liver glutathione thiol esterases. Biochemistry. 1973 Sep 25;12(20):3938–3943. doi: 10.1021/bi00744a024. [DOI] [PubMed] [Google Scholar]
- WONG J. T., HANES C. S. Kinetic formulations for enzymic reactions involving two substrates. Can J Biochem Physiol. 1962 Jun;40:763–804. [PubMed] [Google Scholar]
- YONETANI T., THEORELL H. STUDIES ON LIVER ALCOHOL HYDROGENASE COMPLEXES. 3. MULTIPLE INHIBITION KINETICS IN THE PRESENCE OF TWO COMPETITIVE INHIBITORS. Arch Biochem Biophys. 1964 Jul 20;106:243–251. doi: 10.1016/0003-9861(64)90184-5. [DOI] [PubMed] [Google Scholar]


