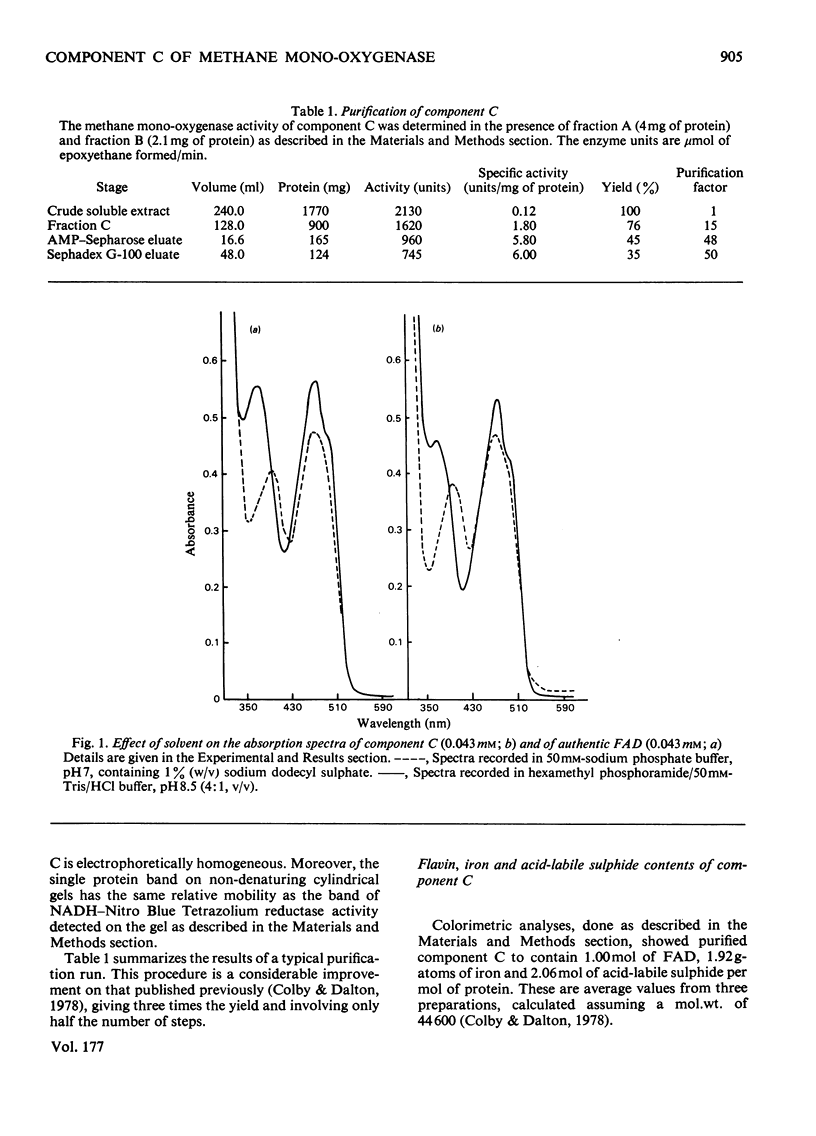
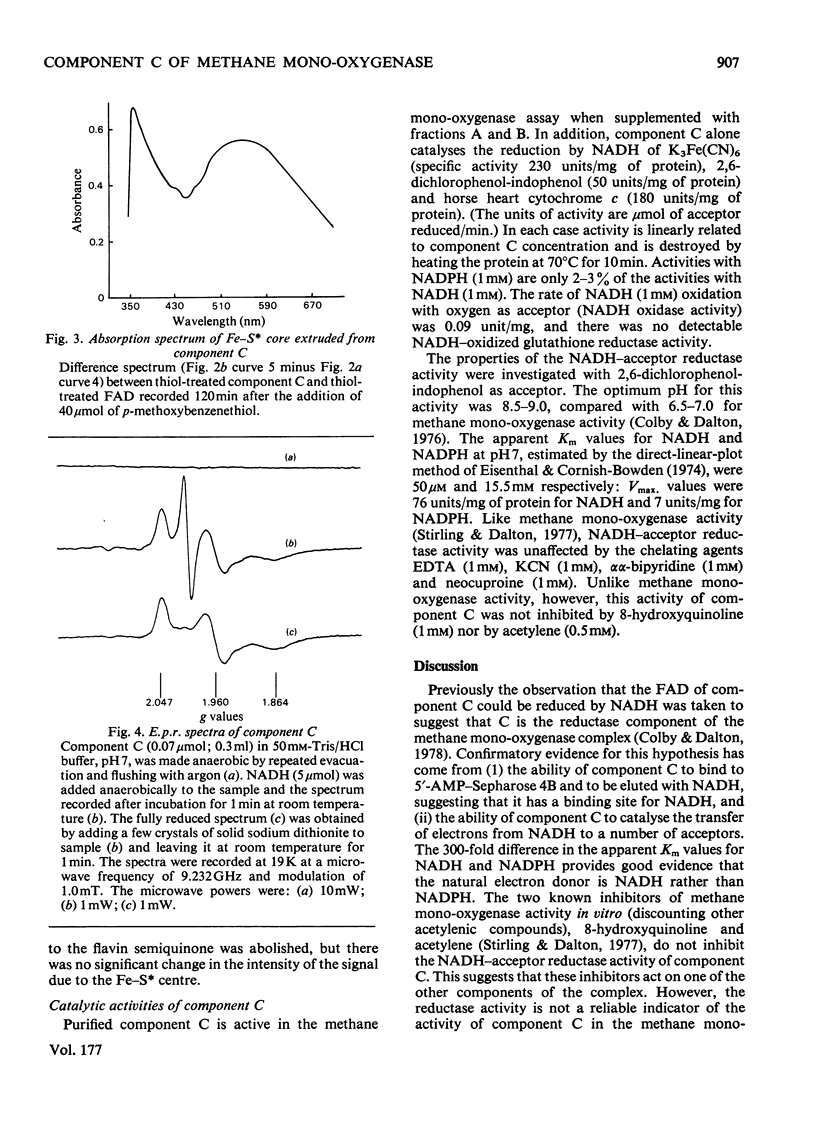
Abstract
1. A new two-step purification is described that routinely yields 100mg quantities of component C for biochemical studies. 2. Chemical analyses show component C purified by this procedure to contain 2 g-atoms of iron, 2 mol of acid-labile sulphide (S) and 1 mol of FAD per mol of protein. 3. The Fe-S core of component C was extruded by treating the protein with p-methoxybenzenethiol in hexamethyl phosphoramide/50mM-Tris/HCl buffer, pH 8.5 (4:1, v/v), under anaerobic conditions. The spectral properties of the extruded core suggest that component C contains 1 mol of [2Fe-2S(S-Cys)4] centre per mol of protein. 4. E.p.r. spectroscopy confirms the presence of a Fe-S centre in component C. 5. Component C catalyses the reduction by NADH of ferricyanide, 2,6-dichlorophenol-indophenol or horse heart cytochrome c, with specific activities of 50--230 units/mg of protein. 6. The optimum pH for the NADH-acceptor reductase activity is 8.5--9.0, and the apparent Km values for NADH and NADPH are 0.05mM and 15.5mM respectively. 7. Unlike methane mono-oxygenase activity, NADH-acceptor reductase activity of component C is not inhibited by 8-hydroxyquinoline or by acetylene.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRUMBY P. E., MILLER R. W., MASSEY V. THE CONTENT AND POSSIBLE CATALYTIC SIGNIFICANCE OF LABILE SULFIDE IN SOME METALLOFLAVOPROTEINS. J Biol Chem. 1965 May;240:2222–2228. [PubMed] [Google Scholar]
- Bray R. C., Barber M. J., Lowe D. J. Electron-paramagnetic-resonance spectroscopy of complexes of xanthine oxidase with xanthine and uric acid. Biochem J. 1978 Jun 1;171(3):653–658. doi: 10.1042/bj1710653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Mortenson L. E. Inhibition of methylene blue formation during determination of the acid-labile sulfide of iron-sulfur protein samples containing dithionite. Anal Biochem. 1977 May 1;79(1-2):157–165. doi: 10.1016/0003-2697(77)90390-6. [DOI] [PubMed] [Google Scholar]
- Colby J., Dalton H. Resolution of the methane mono-oxygenase of Methylococcus capsulatus (Bath) into three components. Purification and properties of component C, a flavoprotein. Biochem J. 1978 May 1;171(2):461–468. doi: 10.1042/bj1710461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J., Dalton H. Some properties of a soluble methane mono-oxygenase from Methylococcus capsulatus strain Bath. Biochem J. 1976 Aug 1;157(2):495–497. doi: 10.1042/bj1570495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J., Stirling D. I., Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977 Aug 1;165(2):395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbes D. L., Burris R. H., Orme-Johnson W. H. On the iron-sulfur cluster in hydrogenase from Clostridium pasteurianum W5. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4795–4799. doi: 10.1073/pnas.72.12.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum W. O., Mortenson L. E., Chen J. S., Holm R. H. Quantitative extrusions of the Fe4S4 cores of the active sites of ferredoxins and the hydrogenase of Clostridium pasteurianum. J Am Chem Soc. 1977 Jan 19;99(2):584–595. doi: 10.1021/ja00444a044. [DOI] [PubMed] [Google Scholar]
- Hill C. L., Steenkamp D. J., Holm R. H., Singer T. P. Identification of the iron-sulfur center in trimethylamine dehydrogenase. Proc Natl Acad Sci U S A. 1977 Feb;74(2):547–551. doi: 10.1073/pnas.74.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. B., Lorsbach T., Que L. Iron-sulfur clusters and cysteine distribution in a ferredoxin from Azotobacter vinelandii. Biochem Biophys Res Commun. 1976 May 17;70(2):582–588. doi: 10.1016/0006-291x(76)91087-1. [DOI] [PubMed] [Google Scholar]
- Kennedy S. I., Fewson C. A. Enzymes of the mandelate pathway in Bacterium N.C.I.B. 8250. Biochem J. 1968 Apr;107(4):497–506. doi: 10.1042/bj1070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D. J., Lynden-Bell R. M., Bray R. C. Spin-spin interaction between molybdenum and one of the iron-sulphur systems of xanthine oxidase and its relevance to the enzymic mechanism. Biochem J. 1972 Nov;130(1):239–249. doi: 10.1042/bj1300239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme-Johnson W. H. Iron-sulfur proteins: structure and function. Annu Rev Biochem. 1973;42(0):159–204. doi: 10.1146/annurev.bi.42.070173.001111. [DOI] [PubMed] [Google Scholar]
- Que L., Jr, Holm R. H., Mortenson L. E. Letter: Extrusion of Fe2S2 and Fe4S4 cores from the active sites of ferredoxin proteins. J Am Chem Soc. 1975 Jan 22;97(2):463–464. doi: 10.1021/ja00835a064. [DOI] [PubMed] [Google Scholar]
- Stirling D. I., Colby J., Dalton H. A comparison of the substrate and electron-donor specificities of the methane mono-oxygenases from three strains of methane-oxidizing bacteria. Biochem J. 1979 Jan 1;177(1):361–364. doi: 10.1042/bj1770361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. I., Dalton H. Effect of metal-binding and other compounds on methane oxidation by two strains of Methylococcus capsulatus. Arch Microbiol. 1977 Jul 26;114(1):71–76. doi: 10.1007/BF00429633. [DOI] [PubMed] [Google Scholar]


