Summary
Clinical application of autologous chimeric antigen receptor (CAR)-T cells is complicated by limited targeting of cancer types, as well as the time-consuming and costly manufacturing process. We develop CD70-targeted, induced pluripotent stem cell-derived CAR-natural killer (NK) (70CAR-iNK) cells as an approach for universal immune cell therapy. Besides the CD70-targeted CAR molecule, 70CAR-iNK cells are modified with CD70 gene knockout, a high-affinity non-cleavable CD16 (hnCD16), and an interleukin (IL)-15 receptor α/IL-15 fusion protein (IL15RF). Multi-gene-edited 70CAR-iNK cells exhibit robust cytotoxicity against a wide range of tumors. In vivo xenograft models further demonstrate their potency in effectively targeting lymphoma and renal cancers. Furthermore, we find that recipient alloreactive T cells express high levels of CD70 and can be eliminated by 70CAR-iNK cells, leading to improved survival and persistence of iNK cells. With the capability of tumor targeting and the potential to eliminate alloreactive T cells, 70CAR-iNK cells are potent candidates for next-generation universal immune cell therapy.
Keywords: CD70, induced pluripotent stem cell, multiplex gene editing, natural killer cell, chimeric antigen receptor, allogeneic rejection
Graphical abstract

Highlights
-
•
Human iPSC-derived, multi-gene-edited CD70 CAR-NK cells were successfully generated
-
•
70CAR-iNK cells exhibit potent cytotoxicity against multiple cancer types
-
•
70CAR-iNK cells can reduce the alloreactivity of alloreactive T cells
In this study, Wang et al. develop CD70-targeted, induced pluripotent stem cell-derived CAR-NK (70CAR-iNK) cell as an approach for universal immune cell therapy. Multi-gene-edited 70CAR-iNK cells exhibit robust cytotoxicity against a wide range of tumors and potently eliminate recipient alloreactive T cells, leading to their improved persistence.
Introduction
Adoptive cellular therapy has revolutionized the conventional concept of cancer therapy as a “living drug.”1 Notably, chimeric antigen receptor (CAR)-based modification has markedly enhanced the efficacy of immune therapy, offering a potentially curative approach for previously incurable hematological malignancies.2,3 However, despite the approval of seven CAR-T cell products by the US Food and Drug Administration, their range of indications remains limited, failing to adequately address the needs of a substantial population of patients with cancer. Compounded by the bespoke manufacturing process of autologous CAR-T cells, this therapeutic approach is plagued with high costs, inherent risk of manufacturing failure, and delays in accessibility.4,5,6 Therefore, we are aiming to develop the next generation of cellular products that exhibit broad applicability across a spectrum of tumor types, while also being amenable to industrial-scale production processes.
The immune checkpoint molecule CD70 has emerged as an important target and is extensively expressed in both hematological and solid tumors.7,8,9 CD70-targeted therapies, including antibody-drug conjugates, antibodies, and CAR-T cells, have shown promising efficacy in treating acute myeloid leukemia (AML), lymphoma, renal cell cancer (RCC), and others.10 For instance, cusatuzumab, an antibody developed against CD70, achieves an 83% complete remission (CR)/CR with incomplete blood count recovery (CRi) rate in treating AML/myelodysplastic syndrome (MDS) when combined with azacitidine. CTX130 (CD70 CAR-T) demonstrates a 70% overall response rate in cases of T cell lymphoma.11 ALLO-316 (CD70 CAR-T) shows an 89% disease control rate in treating advanced or metastatic RCCs.12 In addition, natural killer (NK) cells potently clear widespread tumors due to their intrinsic mechanisms.13,14 Clinical data have demonstrated the potential of adoptive NK cell infusion in treating AML/MDS, breast cancer, ovarian cancer, and others.14 However, the innate anti-tumor function of NK cells is usually impaired by the reduction in NCR/NKG2D ligands (NCR/NKG2DLs) on tumor cells, allowing cancer cells to escape. Therefore, a combination of CD70-targeted and NK cell-based cytotoxicity holds promise for broadening the coverage of cancer types to the greatest extent and could be effective in treating heterogeneous cancers.
As cellular therapy gains momentum, the development of universal cellular products has become a popular area of research in order to meet the needs of patients with reduced cost and immediate availability. However, unwanted graft-versus-host disease (GVHD) and host-versus-graft activities are significant obstacles in clinical applications, increasing the risk of toxicity and limiting their anti-tumor activity. To minimize GVHD, deletion of the αβ T cell receptor (TCR) is a common strategy for blocking T cell alloreactivity. In contrast, NK cells lacking TCR are considered innate universal cellular products.15 To eliminate host immune rejection, alemtuzumab (CD52 mAb) has primarily been used to nonspecifically clear host lymphocytes in participants of recent clinical trials, enabling the efficient proliferation of adoptive immune cells with CD52 knockout (KO).16,17 However, this approach often results in severe immunosuppression, greatly increasing the risk of fatal sepsis. Recently, an ingenious concept of targeting activated immune cells has emerged to suppress allogeneic rejection, with a lower risk of severe immunosuppression.18 Mo et al. engineered T cells with a chimeric 4-1BB-specific alloimmune defense receptor (ADR) that efficiently prevents the rejection of allogeneic T cells by specifically clearing activated T cells.19 Similarly, CD70 is a marker restricted to activated immune cells, involved in immune activation.9 Therefore, we propose that NK cells targeting CD70 may be a promising candidate for reducing allogeneic rejection before adopting universal cellular products.
Previously, we developed a platform for the robust differentiation and expansion of human induced pluripotent stem cell (iPSC)-derived NK (iNK) cells.20,21 Compared to NK cells isolated from peripheral blood or umbilical cord blood, multi-gene modification is more feasibly introduced to iPSCs, and more homogeneous NK cells can be derived from iPSCs on a clinical scale.22 Using this platform, we developed multi-gene-modified iNK cells, named 70CAR-iNK, for targeting multiple tumors. Besides, high-affinity and non-cleavable CD16 (hnCD16) was introduced into 70CAR-iNK cells to enhance antibody-dependent cellular cytotoxicity (ADCC). Moreover, an interleukin (IL)-15 receptor α/IL-15 fusion protein (IL15RF) was incorporated for superior persistence of iNK cells without the need for exogenous cytokine support, along with the KO of CD70 expression to prevent fratricide. Their anti-tumor efficacy was tested in vitro through co-culture with multiple tumor cell lines and further in vivo experiments using xenograft mice bearing human lymphomas and renal cancers. Additionally, we analyzed the dynamics of surface CD70 expression during the induction of alloreactive T (alloT) cells and validated the potency of 70CAR-iNK cells in suppressing allogeneic rejection activities.
Results
CD70 is expressed on a broad spectrum of cancer cells
To gain preliminary insights of CD70 expression pattern on different types of cancers, we conducted an analysis of the transcriptome data obtained from The Cancer Genome Atlas database. Among pan-tumors, a total of 14 different tumor types exhibited significantly higher levels of CD70 expression compared with their adjacent normal tissues (Figure S1A). Among the 33 different cancer types represented, CD70 demonstrates notably high expression levels in kidney renal clear cell carcinoma, diffuse large B cell lymphoma (DLBCL), and mesothelioma (Figure S1A).
Given the substantial progress of cellular therapy in hematological malignancies, we primarily focused on lymphoma and further dissected the preference for CD70 expression in different subtypes. Based on the transcriptome data from 55 lymphoma cell lines accessed from the Human Protein Atlas database, CD70 exhibits more extensive expression across multiple subtypes of lymphoma than conventional lymphoma targets, such as CD19, CD30, and ROR1 (Figure S1B). More specifically, CD70 is highly expressed in mantle cell lymphoma, followed by T cell non-Hodgkin lymphoma (Figure S1C). Interestingly, virus-infected lymphoma cells display significantly higher levels of CD70 expression (Figure S1D). High CD70 expression has been reported in relapsed/refractory DLBCL,23 which is associated with poor prognosis and can contribute to tumor immune suppression.24,25 Consistently, we observed that CD70 expression was associated with worse relapse-free survival in patients with DLBCL (although without statistical significance) and renal cell carcinoma (∗p = 0.011, Figures S1E and S1F). We further analyzed single-cell RNA sequencing databases of T cell lymphoma and renal cell carcinoma, revealing that tumor cells had higher CD70 expression compared to total, CD103+, and CD39+-infiltrating T cells (Figure S2). Overall, broad expression of CD70 on multiple cancer cell types renders a wide application range for CD70-targeted therapy.
CD70-KO has no impact on the function of NK cells or the differentiation process of iPSCs
Due to the activation-induced pattern of CD70 molecules on immune cells, fratricide is a concern in CD70-targeted immune cellular products without further modification, leading to limited persistence and efficacy.26 While CD70 deletion can prevent the fratricide of CD70-targeted CAR-T cells and reduce exhaustion levels,27 its impact on NK cells remains unknown. To explore the feasibility of introducing CD70-KO in iNK cells, we assessed its impact on iNK cell function and differentiation.
Using the CRISPR-Cas9 technology, we first knocked out CD70 on differentiated iNK and 19CAR-iNK cells (Figures S3A and S3B). When expanding on artificial antigen-presenting cells (aAPCs), CD70-KO iNK cells and 19CAR-iNK cells displayed comparable proliferation potential to their respective controls (Figure S3C). When challenged by tumor cell lines, the surface CD107a expression level of CD70-KO iNK cells reached the similar expression level as their unmodified controls (Figure S3D). Both the endogenous (against CD19− THP-1 cells) and CAR-mediated (against CD19+ Raji cells) cytotoxicity and IFN-γ production remained unaffected in CD70-KO iNK cells (Figures S3E and S3F). Furthermore, we performed tumor repetitive challenge assays, confirming that the long-term cytotoxicity of iNK cells was not affected by CD70-KO (Figure S3G).
Having excluded the adverse impact of CD70-KO on iNK function, we developed CD70-KO iPSCs to assess their feasibility of NK cell differentiation (named as CD70−/− iNK cells). The schematic is summarized in Figure 1A. Human iPSCs with confirmed CD70-KO maintained favorable stemness, with a 99.68% SSEA+ TRA-181+ population (Figures 1B and 1C). On day 8 of embryo body (EB) formation, the hemogenic endothelium displayed a normal ratio, with 29.22% ± 2.60% CD34bright proportion (Figure 1D). The proportion of CD56+ cells reached as high as 92.60% ± 1.85% on day 24 of NK cell differentiation (Figure 1E). After expansion on aAPCs, CD70− iNK cells exhibited overall similar profiles to unmodified controls, including the expression of maturation marker (CD57), activation markers (CD16, NKG2D, NKp30, NKp44, NKp46, DNAM-1, and FasL), inhibitory markers (TIGIT, KIR, NKG2A, NKG2C, and CD94), and exhaustion markers (PD1, TIM3, and LAG3) (Figures 1F and 1G). Furthermore, the innate cytotoxicity of CD70−/− iNKs was comparable to controls when challenged by THP-1 cells, Molm13 cells, or Caki-1 cells (Figure 1H). These results together suggested that CD70-KO can be successfully introduced to iNK cells without compromising the differentiation potential and cytotoxic function.
Figure 1.
Human iPSCs with CD70-KO can differentiate into functional NK cells
(A) Overview of procedures to generate gene-edited iPSCs and derive NK cells from iPSCs.
(B) Comparison of sequence of CD70-KO clone (obtained by Sanger sequencing) with CD70-WT sequence by Basic Local Alignment Search Tool showing frameshift mutations in both alleles.
(C) Flow cytometry analyses of stemness for CD70-KO iPSCs (marked with “SSEA4 and TRA-1-81”).
(D) Flow cytometry analyses of proportion of hemogenic endothelium (marked with “CD34”) at EB stage, and CD34bright proportions were summarized in bar plot (values are represented as means ± SEM, n = 3 batches of iPSC differentiation per group). ns, not significant using an unpaired t test.
(E) Flow cytometry analyses and bar plot of CD45+ and CD56+ proportion (values are represented as means ± SEM, n = 3 batches of iPSC differentiation per group). ns, not significant using an unpaired t test.
(F) Flow cytometry analyses of surface markers of activation, inhibition, and exhaustion of NK cells.
(G) Surface CD70 expression on mock iNK and CD70-KO (CD70−/−) iNK cells.
(H) Cytotoxicity of mock iNK and CD70−/− iNK cells co-cultured with THP-1, Molm13, and Caki-1 cells, respectively (values are represented as means ± SEM, n = 3 technical replicates per group).
Quadruple gene-edited hiPSCs can efficiently differentiate into NK cells with stable transgene expression and robust expansion
The limitations of cytotoxicity and persistence remain major bottlenecks in boosting the efficacy of NK cell therapy, which dictates the directions of developing multi-gene-engineered NK cells.18 To expand the application of NK cell cytotoxicity to a broader range of tumors, we adapted CD70-targeted CAR modalities to supplement innate NK anti-tumor function, along with CD70-KO editing to reduce fratricide. Additionally, we introduced hnCD16 to enhance the ADCC effect.28 Furthermore, to address the limited persistence of NK cells, we integrated IL15RF as a key element that is capable of supporting superior proliferation and survival by augmenting mTOR signaling and metabolism fitness.29,30 The quadruple gene-editing modalities are illustrated in Figure 2A.
Figure 2.
Quadruple-gene-edited 70CAR-iPSCs can efficiently differentiate into NK cells with stable transgene expression and robust expansion
(A) Schema of 70CAR-iNK cells.
(B) Flow cytometry analyses of the stemness and transgene expression of 70CAR-iPSCs.
(C) Sanger sequencing of CD70-KO iPSC clone was analyzed via Basic Local Alignment Search Tool.
(D) Flow cytometry analyses of hemogenic endothelium at day 8 of embryonic body formation (values are represented as means ± SEM, n = 4 batches of differentiation per group). ns, not significant using an unpaired t test.
(E) Flow cytometry analyses of NK cell proportion at day 24 of NK cell differentiation (values are represented as means ± SEM, n = 4 batches of NK cell differentiation for each group). ns, not significant using an unpaired t test.
(F) Flow cytometry analyses of genetic modifications. Mock iNK cells were modified with empty vector, and backbone iNK cells were engineered with IL15RF and hnCD16.
(G and H) Mock iNK, backbone iNK, and CD70-KO iNK cells were labeled with CFSE, and then incubated with 70CAR-iNK cells, respectively. After 4 h, the mixtures were stained with propidium iodide to differentiate fratricide-induced dead cells (n = 3 biological replicates). Values are represented as means ± SEM. ∗∗∗∗p < 0.0001 using an unpaired t test comparing “backbone iNK” versus “CD70−/− iNK” in both E:T ratios.
(I) Fold expansion of iNK cells on aAPCs comparing mock iNK monoculture (“iNK”) to co-culture of mock and 70CAR-iNK cells (“iNK + 70CAR-iNK”) (n = 3 biological replicates). ∗∗p < 0.01 using a paired t test.
Following the screening of clones with multi-gene editing, we obtained an iPSC clone with favorable stemness and stable expression of CD16, IL-15, and CD70-CAR molecules (Figure 2B). The successful deletion of CD70 was confirmed by Sanger sequencing (Figure 2C). The entire differentiation process incorporates EB formation, NK cell differentiation, and NK cell expansion. Morphologically, 70CAR-iPSCs closely resembled unmodified controls throughout the entire differentiation process (Figure S4A). On day 8 of EB formation, we found that approximately 48.07% ± 6.00% of the components were CD34bright (Figure 2D). On day 24 of NK cell differentiation, 85.88% ± 2.17% of cells exhibited an NK cell phenotype defined as CD45+CD56+ (Figure 2E). Compared with mock iNK cells, 70CAR-iNK cells demonstrated stable expression of CD70 CAR, CD16, and IL-15 but lacked surface CD70 expression (Figure 2F). Similar to peripheral blood NK (PBNK) cells, iNK and 70CAR-iNK cells consist of a homogeneous population of CD56+ NK cells that co-express typical NK cell surface markers, including NKp30, NKp44, NKp46, FASL, NKG2C, NKG2A, and TIM3 (Figure S4B). To investigate the function of each genetic modification, we also produced mock iNK cells (no genetic modifications) and backbone iNK cells (with hnCD16, IL15RF, and CD70-KO, no CD70-CAR) (Figure S4C). CD70-unedited 70CAR-iNK cells were unable to be produced, likely due to CD70 expression during the differentiation process that caused fratricide. Therefore, we exploited alternative approaches to confirm the fratricide potential during NK cell expansion. Mock iNK cells were co-cultured with 70CAR-iNK cells, showing a significant reduction of expansion and induction of apoptosis (Figures 2G–2I). These results indicated that CD70-CAR-mediated killing can happen between iNK cells and supported the necessity of including CD70-KO in 70CAR-iNK products. Indeed, the in vitro expansion of 70CAR-iNK cells (with CD70-KO) was as robust as that of mock iNK cells (Figure S4D). We also noted that the iNK cells had heterogeneous expression of NKG2A. Using HLA-E-overexpressing target cells and NKG2A-KO iNK cells, we confirmed that NKG2A is dispensable for the cytotoxic function of iNK cells generated by our platform (Figures S4E–S4H).
70CAR-iNK cells exhibit potent in vitro cytotoxicity against multiple cancer types
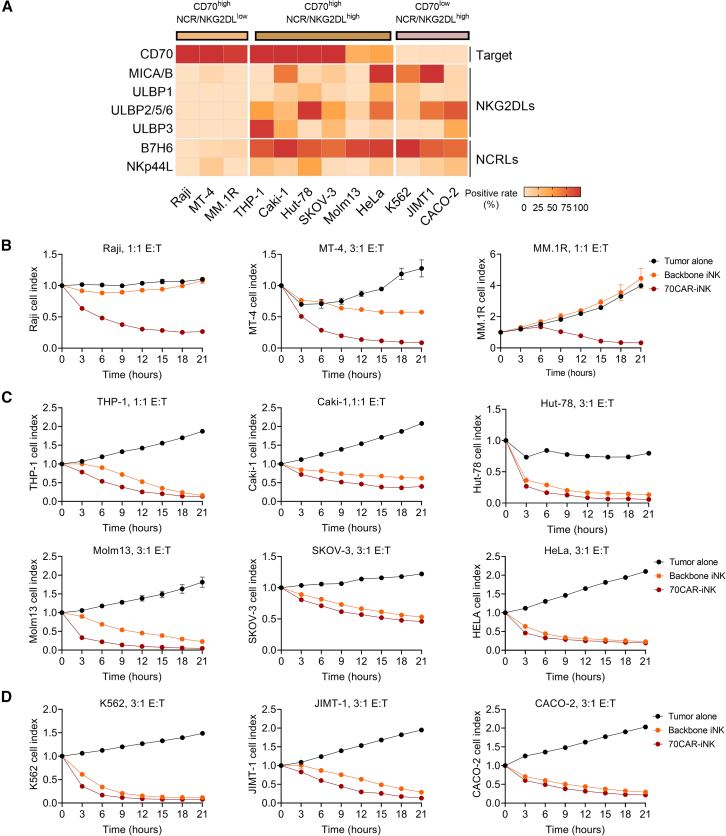
CAR-NK cells possess both innate and CAR-mediated anti-tumor functions. To assess the sensitivity of tumor cell lines to CAR-NK cells, we initially profiled the surface expression of NCR/NKG2DLs and CD70 on multiple tumor cell lines (Figure 3A). These cell lines represented a spectrum of cancer types, including lymphoma (Raji, MT-4, and Hut-78), myeloid leukemia (THP-1, Molm13, and K562), myeloma (MM.1R), breast cancer (JIMT-1), renal cancer (Caki-1), ovarian cancer (SKOV-3), colon cancer (CACO-2), and cervical cancer (HeLa). All of them could be categorized into three groups, including CD70highNCR/NKG2DLlow, CD70highNCR/NKG2DLhigh, and CD70lowNCR/NKG2DLhigh.
Figure 3.
70CAR-iNK cells can potently eradicate a wide spectrum of hematological and solid tumor cell lines
(A) Heatmaps of the surface expression of CD70, NKG2D ligands, and NCR ligands on multiple tumor cell lines. The patterns of tumor cell lines were divided into three groups. CD70highNCR/NKG2DLlow cell lines include Raji (Burkitt lymphoma), MT-4 (T cell lymphoma), and MM.1R (multiple myeloma). CD70highNCR/NKG2DLhigh cell lines include THP-1 (acute monocytic leukemia), Caki-1 (renal cancer), Hut-78 (T cell lymphoma), SKOV-3 (ovarian cancer), Molm13 (acute myeloid leukemia), and HeLa (cervical adenocarcinoma). CD70lowNCR/NKG2DLhigh cell lines include K562 (chronic myeloid leukemia), JIMT-1 (breast ductal adenocarcinoma), and CACO-2 (colorectal adenocarcinoma).
(B–D) Backbone iNK and 70CAR-iNK cells were used as effectors at the indicated effector cell:target tumor cell (E:T) ratios in IncuCyte-based function assay with CD70highNCR/NKG2DLlow tumor cells (B), CD70highNCR/NKG2DLhigh tumor cells (C), and CD70lowNCR/NKG2DLhigh tumor cells (D). Values are represented as means ± SEM, n = 3–4 technical replicates per group.
70CAR-iNK cells exhibited a significant advantage of cytotoxicity compared with backbone iNK cells against CD70highNCR/NKG2DLlow cells (Raji, MT-4, and MM.1R, Figure 3B). When co-cultured with NCR/NKG2DLhigh tumor cells, 70CAR-iNK cells still demonstrated more potent cytolytic efficacy than backbone iNK cells to varying extents (Figures 3C and 3D). Given the substantial progress of cellular products in lymphoma therapy, we further evaluated the serial-killing and cytokine production potential of 70CAR-iNK cells against different lymphoma subtypes. Compared with controls, the remarkable cytotoxicity of 70CAR-iNK cells occurred in the first round of co-culture with the Raji and MT-4 cell lines (NCR/NKG2DLlow) and was further amplified after each round of rechallenges (Figures S5A and S5B). Moreover, although the Hut-78 cell line (NCR/NKG2DLhigh) was sensitive to backbone iNK and 70CAR-iNK cells, the advantages of 70CAR-iNK cells were amplified as the rechallenge round accumulated (Figure S5C). Notably, 70CAR-iNK cells demonstrated an overall higher production of cytotoxic cytokines during tumor-lytic activities than backbone iNK cells (Figures S5D and S5E). More specifically, when co-cultured with lymphoma cells and Caki-1 cells, 70CAR-iNK cells exhibited markedly higher levels of IL-2 and IFN-γ production than backbone iNK cells, while a higher level of TNF-α was only noted during the co-culture with Raji cells and Caki-1 cells (Figures S5D and S5E).
70CAR-iNK cells demonstrate enhanced ADCC function and superior in vitro persistence
To test the potential of CAR-independent, ADCC-mediated cytotoxicity of 70CAR-iNK cells, we first generated CD70-KO Raji cells, which are able to evade CAR-mediated killing and resemble antigen escape (Figure S6A). Given the low expression of endogenous CD16 (Figure 1F), we engineered CAR-iNK cells with hnCD16 to strengthen their ADCC function. Activation-induced cleavage of endogenous CD16 was reported on PBNK cells and was also observed in our unmodified iNK cells after challenging with tumor cells or in combination with antibodies (Figure S6B). In contrast, 70CAR-iNK cells equipped with hnCD16 maintained stable CD16 expression, irrespective of PMA/ionomycin-induced activation or tumor challenges (Figure S6B). When combined with the CD20-targeted antibody rituximab, 70CAR-iNK cells exhibited significantly higher levels of CD107a than mock iNK cells, suggesting an enhanced ADCC against CD70-KO Raji cells (Figure S6C). Consistently, using a tumor cell repetitive challenge assay, we confirmed that the addition of rituximab also increased the killing potency of 70CAR-iNK cells against CD70-KO Raji cells (Figures S6D and S6E). These results have together indicated that hnCD16 can enhance ADCC function as compared to the endogenous CD16 of iNK cells.
Limited persistence significantly hampers the anti-tumor function of iNK cells. In the absence of exogenous IL-2 support, unmodified iNK cells displayed minimal expansion capability, and their cell count decreased sharply within 3 days of in vitro culture (Figure S6F). Comparatively, 70CAR-iNK cells showed superior in vitro proliferation ability and prolonged survival times, even without exogenous IL-2 support (Figure S6F). Flow cytometry analysis revealed a stable expression of pSTAT3 and pSTAT5 in 70CAR-iNK cells, while the phosphorylation levels in mock iNK cells appeared to be IL-15 dependent (Figure S6G). IL15RF also improved the cytolytic activity of iNK cells (Figures S6H and S6I), consistent with previous reports.30,31
70CAR-iNK cells could potently control tumor progression in vivo
Since the potency of 70CAR-iNK cells against different types of tumor cells has been illustrated in vitro, we further evaluate their anti-tumor activity in lymphoma and renal cancer xenograft models. To build lymphoma xenograft models, we inoculated NOG mice with luciferase-expressing MT-4 cells (T cell lymphoma) or Raji cells (B cell lymphoma) through the tail vein, followed by treatment of mock-iNK, backbone iNK, or 70CAR-iNK cells (Figure 4). Then, tumor burden was monitored on a weekly basis. Here, we validated that 70CAR-iNK cells demonstrated anti-tumor potency in controlling the progression of both types of lymphoma (MT-4: p < 0.0001; Raji: p < 0.01 compared with backbone) and prolonged their survival time (MT-4: p < 0.01; Raji: p < 0.001 compared with backbone) (Figures 4B–4E and 4G–4I). Notably, these two tumor cell lines expressed minimal levels of NKG2DL (Figure 3A). Consequently, only 70CAR-iNK but not mock or backbone iNK cells were able to control the tumor growth and prolong the survival of tumor-bearing animals (Figure 4), indicating the critical function of the CD70-targeted CAR in iNK cells. All 3 batches of 70CAR-iNK cells can eradicate established tumors, but differences in expansion were observed between batches (Figures S7A–S7D), suggesting that a rigorous evaluation of cytotoxicity and expanding potency is required before selecting specific batches of cells for further expansion and clinical application.
Figure 4.
Adoptive transfer of 70CAR-iNK cells eradicated in vivo xenograft tumors
(A) Schematic diagram of in vivo anti-tumor functional evaluation of 70CAR-iNK cells using luciferase (Luc)-expressing MT-4 cells. NCG mice were inoculated intravenously with 1×106 Luc-expressing MT-4 cells. Tumor-bearing mice were left untreated, or treated with mock iNK, backbone iNK, or 70CAR-iNK cells (n = 5 biological replicates per group).
(B and C) Bioluminescent images and quantification of bioluminescent intensity (BLI) through day 35. Values are represented as means ± SEM. ∗∗∗∗p < 0.0001 calculated by two-way ANOVA comparing 70CAR-iNK with the other 3 groups.
(D) Pharmacokinetics of 70CAR-iNK cells in MT-4 murine model. Values are represented as means ± SEM.
(E) Kaplan-Meier curve representing the percent survival of the experimental groups. Significance of difference was determined by two-tailed log rank test comparing 70CAR-iNK with the other 3 groups. ∗p < 0.05, ∗∗p < 0.01.
(F) Schematic diagram of in vivo anti-tumor functional evaluation of 70CAR-iNK cells using Luc-expressing Raji cells. NCG mice were inoculated intraperitoneally with 2×105 Luc-expressing Raji cells. Tumor-bearing mice were left untreated, or treated with mock iNK, backbone iNK, or 70CAR-iNK cells (n = 6 biological replicates in each group).
(G and H) Bioluminescent images and quantification of BLI data through day 14. Values are represented as means ± SEM. Statistical significance was calculated on day 14 comparing 70CAR-iNK with the other 3 groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(I) Kaplan-Meier curve representing the percent survival of the experimental groups. Significance of difference was determined by log rank test (two-tailed) comparing 70CAR-iNK with the other 3 groups. ∗∗p < 0.01, ∗∗∗p < 0.001.
We also established a renal cancer xenograft model by intraperitoneally inoculating Caki-1 cells (Figure S7E), which were treated by mock-, backbone-, or 70CAR-iNK cells. All types of iNK cells were able to control tumor growth, consistent with the results of in vitro cytotoxicity (Figures S7F, S7G, and 3C). However, 70CAR-iNK cells still exhibited an enhanced in vivo persistence (Figure S7H), indicating that CAR-mediated activation might potentiate iNK cell survival on top of endogenous cytotoxicity. In another set of assays, we further confirmed that either intraperitoneal or intravenous injection of CD70-iNK cells can effectively target Caki-1 tumors (Figures S7I–S7L).
70CAR-iNK cells could reduce the alloreactivity of alloT cells
Allogeneic rejection is a major impediment that complicates universal NK cell therapy due to their shortened persistence in recipients. Given that CD70 expression is detected on activated immune cells,9 we aimed to investigate the potential of suppressing the function of alloT cells by treating them with 70CAR-iNK cells.
To delineate the dynamics of CD70 expression during the induction of alloT cells, we incubated iNK cells with allogeneic peripheral blood mononuclear cells isolated from healthy donors for approximately 2 weeks (Figure S8A). During this period, we monitored the expansion of T cells and their surface expression of CD70. A significant increase in the CD3+ proportion and number was observed between day 5 and day 8 (Figures S8B and S8C). Interestingly, on day 8, CD8+ T cells demonstrated a higher level of CD70 than CD8− T cells (Figure S8D). Moreover, high CD70 expression in T cells was associated with higher levels of other activation markers including CD69, CD107a, CD137, CD40 ligand, and CD25 (Figure S8E). Thereafter, the surface expression of CD70 continued to rise on alloT cells during the induction process (Figure S8F).
Having demonstrated that iNK cells can induce allogeneic T cell responses, we next evaluated the capability of 70CAR-iNK cells to suppress the function of alloT cells. We harvested iNK-induced allogeneic T cells and evaluated their alloreactivity against 70CAR or backbone iNK cells (Figure 5A). Notably, after 4 h of co-incubation, 70CAR-iNK cells can induce more potent cytotoxicity against alloT cells compared to backbone iNKs (Figure 5B). CD70+ T cells were completely depleted by 70CAR-iNK cells, together with a reduction of multiple surface activation markers on the T cell population, including CD137, CD69, CD25, CD40 ligand, and CD107a (Figure 5C). The alloreactivity was evaluated by the apoptosis status and quantification of co-cultured iNK cells, revealing that 70CAR-iNK cells can reduce allogeneic T cell function (Figure 5D). Next, to evaluate whether resistance to allogeneic T cell function can lead to increased in vivo persistence, we established an in vivo rejection model by injecting allogeneic T cells intravenously into NOG mice,19 followed by injection of backbone or 70CAR-iNK cells (Figures 5E and S8G). In this model, the allogeneic T cells eliminated backbone iNK cells, while 70CAR-iNK cells showed persistence (Figures 5F and 5G), suggesting that the CD70-CAR is critical for the resistance of iNK cells against allogeneic T cell rejection.
Figure 5.
70CAR-iNK cells effectively target CD70+ allogeneic T cells
(A) Experimental schema to test the function of alloT cells. Allogeneic potential was evaluated by alloT cell viability, activation, and alloreactivity (indicated by iNK viability). AlloT cells were used as target cells and co-cultured with backbone iNK or 70CAR-iNK at an E:T ratio of 1:1 for 24 h.
(B) Left: fluorescence-activated cell sorting (FACS) histogram plot of PI staining gated on CD3+ alloT cells after 6-h co-culture; right: viable T cells at the end of co-culture (values are represented as means ± SEM, n = 3 biological replicates per group). Error bars represent ±SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 calculated by one-way ANOVA.
(C) Surface activation markers and CD70 expression on alloT cells after 6-h incubation with backbone or 70CAR-iNK cells.
(D) Left: FACS histogram plot of PI staining gated on CD56+ iNK cells after 6-h co-culture; right: viable iNK cells at the end of co-culture (values are represented as means ± SEM, n = 3 technical replicates per group from donor 1 and donor 2, respectively). Error bars represent ±SEM. ∗p < 0.05, ∗∗p < 0.01 using an unpaired t test.
(E) Schematic diagram of the in vivo allogeneic rejection model.
(F and G) Representative FACS plot and in vivo persistence of backbone and 70CAR-iNK cells (values are represented as means ± SEM, n = 6 biological replicates per group). Error bars represent ±SEM. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 using an unpaired t test for each time point.
Discussion
Universal cellular therapy has attracted significant attention due to its broader accessibility by patients. Among immune cells, NK cells are one of the most promising candidates for universal products, given their favorable safety profiles and innate anti-tumor functions.32 Furthermore, human iPSCs provide a promising platform for multiplex gene editing and clone screening, serving as an unlimited source for generating functional NK cells with robust in vitro expansion capacities. Overall, our studies provided evidence that quadruple gene-edited, CD70-targeted, iPSC-derived NK cells can effectively target both tumor cells and allogeneic T cells, which simultaneously enable broad tumor coverage, superior proliferative abilities, enhanced ADCC functions, and resistance to allogeneic T cell rejection, with great potential to overcome current limitations in cellular therapy.15
We constructed 70CAR-iNK cells to enable both CAR-mediated and NK cell endogenous cytotoxicity and enhance tumor coverage. The feasibility of CD70-CAR is supported by favorable outcomes of CD70-targeted clinical trials, including antibody-drug conjugates, mono-antibodies, and CD70-targeted CAR-T cells.10 Besides, the contribution of CD70/CD27 axis in modulating the suppressive tumor microenvironment also supported the importance of CD70-targeted therapy.9 In our in vivo studies, 70CAR-iNK cells showed superior efficacy compared with backbone iNK cells against MT-4 and Raji (NKG2DLlow) tumor models. In the MT-4 model, 70CAR-iNK also outperformed backbone iNK cells in expansion. In the Caki model (NKG2DLhigh), the therapeutic benefit of 70CAR-iNK cells over backbone iNK is less significant, indicating that CAR-mediated cytotoxicity is more critical in eradicating tumors with low NKG2DL expression. More interestingly, the comparable anti-tumor function and pharmacokinetics between intraperitoneal and intravenous NK cell administrations suggested the ability of 70CAR-iNK cells to cross the peritoneal barriers, which would indicate the translational potential against other CD70-expressing intraperitoneal tumors such as cervical squamous cell carcinoma and mesothelioma (Figure S1A).
CD70 is upregulated on activated T cells and has been revealed for its controversial roles in T cell activation and exhaustion.9,33 As previously reported, CD70-KO can ameliorate the exhaustion patterns of CAR-T cells and contribute to enhanced proliferation and cytotoxicity.34,35 Regarding NK cells, Al Sayed et al. revealed that CD70 reverse signaling can stimulate the killing function of NK cells,36,37 but there has been little evidence about the direct impact of CD70-KO on NK cells. Our study reveals that CD70-KO has no adverse effects on iNK cell activation, proliferation, and cytotoxicity, providing evidence for an optimal choice to avoid fratricide in CD70-targeted NK cell therapy. Moreover, the successful derivation of NK cells from CD70-KO iPSCs provides valuable insights for optimizing modification strategies for CD70-targeted NK cell therapy. As CD70 expression is also associated with T cell activation, one potential concern of CD70-targeted therapy is the elimination of endogenous anti-tumor T cells. We analyzed three independent single-cell databases, revealing that tumor cells expressed higher levels of CD70 compared to the infiltrating T cells. Therefore, we believe that although the risk of compromising endogenous anti-tumor response cannot be fully excluded, the anti-tumor potency of CD70-targeted therapy might outweigh the effect of intratumoral T cell elimination.
Unwanted host-versus-graft reactions are the primary obstacles to the clinical application of universal cellular therapy, greatly limiting its anti-tumor efficacy.15 The limited persistence of therapeutic NK cells has been considered to be, at least partially, mediated by the rejection from recipient T cells.38 Alemtuzumab (CD52 mAb) has been used to nonspecifically eliminate host lymphocytes in clinical trials of allogeneic cell therapy. Nevertheless, severe immunosuppression exposes patients to a high risk of fatal sepsis.16,17 An ingenious approach is to target the activated allogeneic immune cells and suppress cellular rejection,18 such as ADR that recognized the activation marker 4-1BB.19 Here, using the same in vivo model, we validated that 70CAR-iNK cells had prolonged persistence in the presence of allogeneic T cells, indicating the potential of enhanced in vivo survival by suppressing the recipient T cell rejection.
Relapse is a major barrier against effective cellular therapy. On one hand, the challenge of antigen-negative relapse warrants multi-targeting strategy.39 Notably, CD16 can be exploited as a mediator of second target in the presence of antibodies. Considering the cleavable feature of CD16 during the activation of NK cells, we engineered iPSCs with hnCD16. These iNK cells demonstrated high CD16 stability as well as potent ADCC function when combined with antibodies, providing a feasible strategy of using the same 70CAR-iNK cells to target both CD70+ and CD70− tumors and limit antigen escape. Intriguingly, the co-activation of CAR and ADCC pathways did not significantly improve the cytotoxic activity of iNK cells, and the underlying mechanisms remain to be investigated in future studies. On the other hand, the clinical efficacy of adoptive NK cell therapy was also hindered by the finite potency of expansion and persistence of NK cells. The introduction of membrane-bound IL-15RF could render superior in vitro persistence to iNK cells, which will also be important for ameliorating anti-tumor efficacy.
Overall, this “modification-differentiation” paradigm of iPSCs is one of the most efficient strategies for developing multi-gene-engineered immune cells. The nature of iPSCs makes it feasible for the establishment of gene-engineered clone banks, and our platform of NK cell differentiation enables the stable generation of therapeutic NK cells harboring these genetic modifications. While we are aware that emerging clinical challenges of cellular therapy will guide further modifications of iNK cells, the 70CAR-iNK cells depicted in this study represent a successful combination of CD70 targeting (tumor and allogeneic T cells) with functional potentiation (IL15RF and hnCD16), incorporating into the translatable iNK platform, allowing for further clinical development.
Limitations of the study
A potential limitation of our study is that tumors used for xenograft models are cell line derived, while patient-derived xenograft model might be closer to the clinical scenarios. Another limitation is the lack of an orthotopic or subcutaneous solid tumor model due to inconsistent tumor engraftment of Caki-1 cells. Although our study has several limitations, it provides strong evidence for the potential of 70CAR-iNK cells as cellular products with broad applicability in tumor management, and preliminary evidence of CD70-targeted therapy in clearing activated alloreactive cells offers an approach for handling allorejection.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, He Huang (huanghe@zju.edu.cn).
Materials availability
This study did not generate new unique materials.
Data and code availability
-
•
This study analyzed publicly available datasets at Gene Expression Omnibus. GEO accession numbers are listed in the key resources table.
-
•
All the original codes have been deposited at Zenodo. Accession website is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
These studies received support from the National Natural Science Foundation of China (82341205, 82341206, 82271874, and 82270234), the Key Project of the Science and Technology Department of Zhejiang Province (2024SSYS0023), and Sanming Project of Medicine in Shenzhen (SZSM202111004). Some schematic diagrams and the graphical abstract were created in https://BioRender.com. The procedures of in vivo experiments in this study were evaluated and approved by the Yangtze Delta Region Institute of Tsinghua University, Zhejiang, Hangzhou.
Author contributions
L.W., Y.W., X.H., and Z.M. designed the project, developed the methodology, conducted the experiments, validated assays, analyzed the data, and contributed to manuscript writing. Y.Y. and G.M. contributed to the design and development of methodology and provided technical resources and advice. K.H., K.W., Y.Z., W.Z., and P.Z. conducted plasmid construction and function validation. L.S., X.S., and X.J. participated in the stem cell differentiation process. Z.M., M.Z., X.L., G.M., N.C., Y.S., and M.X. conducted animal experiments. Z.M., R.H., L.Z., and Y.F. participated in data mining and analysis of the online database. H.H., L.Y., Y.H., and D.W. jointly supervised this work. All authors discussed and interpreted the results, and they approved the final version of the manuscript.
Declaration of interests
X.H., Y.Y., G.M., Y.Z., N.C., L.S., X.S., W.Z., X.J., Y.S., P.Z., M.X., and L.Y. are employed by Qihan Biotech Inc., and L.Y. is the chief executive officer.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC-Labeled Human CD70 Protein | ACRObiosystems | Cat#CDD-HF2D4, RRID:AB_3662727 |
| Anti-human CD70-PE | BD Biosciences | Cat#555835,RRID:AB_396158 |
| Anti-human CD70-FITC | BD Biosciences | Cat#555834, RRID:AB_396157 |
| Anti-human IL-15-APC | Thermo Fisher Scientific | Cat#MA5-23627, RRID:AB_2608838 |
| Anti-human CD16-PE | BD Biosciences | Cat#555407, RRID:AB_395807 |
| Anti-human TRA-1-81-AF647 | BD Biosciences | Cat#560793, RRID:AB_10550550 |
| Anti-human SSEA-4-PE | BD Biosciences | Cat#560128, RRID:AB_1645533 |
| Anti-human CD34-APC | BD Biosciences | Cat#555824, RRID:AB_398614 |
| Anti-human CD43-FITC | BD Biosciences | Cat#555475, RRID:AB_395867 |
| Anti-human CD45-FITC | BD Biosciences | Cat#555482, RRID:AB_395874 |
| Anti-human CD56-APC | BD Biosciences | Cat#341026, RRID:AB_400559 |
| Anti-human CD57-PE | BD Biosciences | Cat#560844, RRID:AB_2033965 |
| Anti-human CD3-PE | BD Biosciences | Cat# 552127, RRID:AB_394342 |
| Anti-human CD8-PE | BD Biosciences | Cat#557086, RRID:AB_396581 |
| Anti-human HLA-E-PE | BioLegend | Cat#342604, RRID:AB_1659249 |
| Anti-human CD69-PE | BD Biosciences | Cat#560968, RRID:AB_10565972 |
| Anti-human CD137-PE | BioLegend | Cat# 309804, RRID:AB_314783 |
| Anti-human CD40 ligand-FITC | Thermo Fisher Scientific | Cat#11-1548-41, RRID:AB_10667882 |
| Anti-human CD25-PE | BioLegend | Cat#356104, RRID:AB_2561861 |
| Anti-human NKG2A-PE | BioLegend | Cat#375104, RRID:AB_2888861 |
| Anti-human NKG2C-PE | R&D Systems | Cat#FAB138P, RRID:AB_2132983 |
| Anti-human NKG2D-PE | BD Biosciences | Cat#557940, RRID:AB_396951 |
| Anti-human NKp30-PE | BioLegend | Cat#325208, RRID:AB_756112 |
| Anti-human NKp44-PE | BD Biosciences | Cat#558563, RRID:AB_647239 |
| Anti-human NKp46-PE | BD Biosciences | Cat#557991, RRID:AB_396974 |
| Anti-human DNAM-1-PE | R&D Systems | Cat#FAB666P, RRID:AB_2072625 |
| Anti-human FasL-PE | BD Biosciences | Cat#564261, RRID:AB_2738713 |
| Anti-human CD158a-PE | Beckman Coulter | Cat#IM2278U, RRID:AB_2728104 |
| Anti-human CD158b-APC | Beckman Coulter | Cat#A22333, RRID:AB_3662729 |
| Anti-human CD158e-PE | Beckman Coulter | Cat#IM3292, RRID:AB_131339 |
| Anti-human TIGIT-PE | BioLegend | Cat#372704, RRID:AB_2632730 |
| Anti-human PD1-PE | BioLegend | Cat#329906, RRID:AB_940483 |
| Anti-human TIM3-PE | BioLegend | Cat#345006, RRID:AB_2116576 |
| Anti-human LAG3-PE | BioLegend | Cat#369206, RRID:AB_2632656 |
| Anti-human CD94-PE | Thermo Fisher Scientific | Cat#12-0949-42, RRID:AB_10854417 |
| Anti-human MICA/B | Thermo Fisher Scientific | Cat#12-5788-42, RRID:AB_10854117 |
| Anti-human ULBP1 | R&D Systems | Cat#FAB1380P, RRID:AB_2687471 |
| Anti-human ULBP2/5/6 | R&D Systems | Cat#FAB1298P, RRID:AB_2214693 |
| Anti-human ULBP3 | R&D Systems | Cat#FAB1517P, RRID:AB_10719122 |
| Anti-human B7H6 | Thermo Fisher Scientific | Cat#12-6526-42, RRID:AB_2784629 |
| Anti-human NKp44L | USBiological | Cat#212013, RRID:AB_3662728 |
| Anti-human CD107a-PE | BD Biosciences | Cat#555801, RRID:AB_396135 |
| PE Mouse IgG1 κ Isotype Control | BD Biosciences | Cat#555749, RRID:AB_396091 |
| FITC Mouse IgG1 κ Isotype Control | BD Biosciences | Cat#555748, RRID:AB_396090 |
| APC Mouse IgG1 κ Isotype Control | BD Biosciences | Cat#555751, RRID:AB_398613 |
| Stat3 (pY705) PE | BD Biosciences | Cat#612569, RRID:AB_399860 |
| Stat5 (pY694) Alexa 647 | BD Biosciences | Cat#562076, RRID:AB_11154412 |
| Chemicals, peptides, and recombinant proteins | ||
| Rituximab | Bio-Rad | Cat#MCA2260, RRID:AB_323787 |
| Trastuzumab (Anti-HER2) | Selleck Chemicals | CAS number: A2007 |
| Cas9 protein v2 | Thermo Fisher Scientific | Cat#A36498 |
| Recombinant human IL-2 | Beijing Four Rings Biopharmaceutical | Cat#20200714 |
| PMA | Sigma | Cat#P1585 |
| Ionomycin | TOCRIS | Cat#2092 |
| Propidium iodide | Thermo Fisher Scientific | Cat#P3566 |
| Bright-Glo Luciferase assay system | Promega | Cat#E2610 |
| Critical commercial assays | ||
| P3 Primary Cell 4D-Nucleofector X | Lonza | Cat#V4XP-3032 |
| BD phosflow Fix buffer I | BD Biosciences | Cat#557870 |
| BD phosflow Perm buffer III | BD Biosciences | Cat#558050 |
| CD3/CD28 dynabeads | Thermo Fisher Scientific | Cat#11131D |
| TransDetect PCR Mycoplasma Detection Kit | Transgen Biotech | Cat#FM3-11-01 |
| Deposited data | ||
| Bulk RNA seq (pan-tumor) | The Cancer Genome Atlas | https://portal.gdc.cancer.gov |
| Bulk RNA seq (cancer cell lines) | Human Protein Atlas | https://www.proteinatlas.org |
| Bulk RNA seq | GEPIA database | http://gepia.cancer-pku.cn/ |
| scRNA-seq (cutaneous T cell lymphoma) | Gene Expression Omnibus | GEO: GSE182861, GSE122703 |
| scRNA-seq (clear cell renal cell carcinoma) | Gene Expression Omnibus | GEO: GSE222703 |
| Experimental models: Cell lines | ||
| Raji | ATCC | Cat#CCL-86 |
| THP-1 | ATCC | Cat#TIB-202 |
| HeLa | ATCC | Cat#CCL-2 |
| K562 | Mingzhoubio | Cat#MZ-0102 |
| MM.1R | Mingzhoubio | Cat#B240408 |
| JIMT-1 | Mingzhoubio | Cat#MZ-0578 |
| CACO-2 | Mingzhoubio | Cat#MZ-0039 |
| SKOV-3 | Mingzhoubio | Cat#MZ-0169 |
| Caki-1 | Mingzhoubio | Cat#B164110 |
| MT-4 | Procell | Cat#CL-0655 |
| Hut-78 | Procell | Cat#CL-0364 |
| Molm13 | Beyotime | Cat#C6600 |
| Human artificial antigen-presenting cells | Hangzhou Qihan Biotech | N/A |
| Human PBNK | Shanghai Saily Biotechnology | N/A |
| Human CBNK | Hangzhou Primitive Biotechnology | N/A |
| Human PBMC | Shanghai Saily Biotechnology & Oricellbio Biotechnology | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.Cg-PrkdcscidIl2rgtm1Sug/JicCrl (NOG) | Beijing Vital River Laboratory Animal Technology | N/A |
| Mouse: NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt (NCG) | GemPharmatech | N/A |
| Oligonucleotides | ||
| CD70 targeted gRNA-1: ‘5-GCCCGCAGGACGCACCCATA-3’ | GenScript Biotech Corporation | Custom order |
| CD70 targeted gRNA-2: ‘5-GGCCGGGTTCTAGAGTGCCA-3’ | GenScript Biotech Corporation | Custom order |
| CD70 forward primer: 5′-CGCTAGCGGAGGTGATCG-3′ | Beijing Tsingke Biotech | Custom order |
| CD70 reverse primer: 5′-AAGTGACTCGAGCGGCAG-3′ | Beijing Tsingke Biotech | Custom order |
| Recombinant DNA | ||
| HLA-E construct | This manuscript | N/A |
| Anti-CD70 scFv | This manuscript | N/A |
| Anti-CD19 scFv | This manuscript | N/A |
| Software and algorithms | ||
| Graphpad Prism version 9.0 | GraphPad software | https://www.graphpad.com |
| FlowJo 10.0 | FlowJo, LLC | https://www.flowjo.com/ |
| Incucyte Base Software | Sartorius | https://www.sartorius.com/en/products/live-cell-imaging-analysis/live-cell-analysis-software |
| Living Image software | PerkinElmer | N/A |
| Photon IMAGER™ OPTIMA | Biospace Lab | N/A |
| R version 4.2.1 | R Core Team 2017 | https://www.r-project.org |
| Seurat package | Rahul Satija Lab | https://github.com/satijalab/seurat |
| Copykat | Nicholas E Navin Lab | https://github.com/navinlabcode/copykat |
| scDblFinder | Mark D Robinson Lab | https://github.com/plger/scDblFinder |
| Publicly available gene expression datasets | Gene Expression Omnibus | GSE182861, GSE122703, and GSE222703 |
| Manuscript source code | Current publication | https://doi.org/10.5281/zenodo.14233570 |
Experimental model and study participant details
Cell lines and cell culture
Raji (ATCC, CCL-86), THP-1 (ATCC, TIB-202), and HeLa (ATCC, CCL-2) cells were obtained from the American Tissue Culture Collection. K562 (Mingzhoubio, MZ-0102), MM.1R (Mingzhoubio, B240408), JIMT-1 (Mingzhoubio, MZ-0578), CACO-2 (Mingzhoubio, MZ-0039), SKOV-3 (Mingzhoubio, MZ-0169), and Caki-1 (Mingzhoubio, B164110) cells were obtained from Mingzhoubio. MT-4 (Procell, CL-0655) and Hut-78 (Procell, CL-0364) cells were obtained from Procell. Molm13 (Beyotime, C6600) cells were purchased from Beyotime. Raji, THP-1, Molm13, K562, MM.1R, and MT-4 cells were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco). Additional 2-mercaptoethanol (Gibco) was added into THP-1 media at a final concentration of 0.05 mM. Furthermore, CACO-2 and HeLa cells were maintained in MEM (Gibco) supplemented with 20% and 10% FBS, respectively. SKOV-3 and Caki-1 cells were cultured in McCoy’s 5a (Gibco) with 10% FBS. JIMT-1 was cultured in DMEM (Gibco) with 10% FBS, and Hut-78 was cultured in IMDM (Gibco) with 20% FBS. All of these complete medium were supplemented with 1% penicillin/streptomycin (Gibco). They were maintained at 37°C and 5% CO2. All cells were tested monthly via TransDetect PCR Mycoplasma Detection Kit (Transgen Biotech) to ensure they were free of mycoplasma.
Mice
Female NOG mice (Vital River Laboratories, Beijing, China) or NCG mice (GemPharmatech, Jiangsu, China) at the age of 6–8 weeks were used for each batch of in vivo studies. The husbandry and experimental procedures of mice were conducted strictly according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All experiments were reviewed and approved by the Institutional Animal Care and Use Committee of QR Bio Technology Co., Ltd. (reference no. QDAE20220321001).
Method details
Analysis of public database
The raw data of RNA-seq of pan-tumor and their adjacent normal tissues were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) database. The sequencing data were extracted as Transcripts Per Million (TPM) format, and transformed with “log2(value + 1)” for further analyses. Comparison between cancer tissues and adjacent normal tissues were made via Mann-Whitney U test. The groups with insufficient samples (<3) were excluded for comparison. Furthermore, the data of multiple lymphoma cell lines were accessed from Human Protein Atlas (https://www.proteinatlas.org) database. Finally, the survival data of lymphoma and renal cancer patients and their association with CD70 transcriptional level were accessed and analyzed via GEPIA database (http://gepia.cancer-pku.cn/).40 The cutoff point was set as the median of CD70 expression within the respective patients’ cohort.
Transcriptome sequencing was accessed from three GEO datasets, including GSE182861 (cutaneous T cell lymphoma),41 GSE122703 (cutaneous T cell lymphoma),42 GSE222703 (clear cell renal cancer).43 The sequencing libraries were analyzed using the Illumina HiSeq 2000 platform. The matrix files of each dataset were retrieved from GEO and imported via the Seurat package. Cells that expressed fewer than 200 genes were filtered in each sample, as well as genes expressed in fewer than three cells. The Louvain algorithm was employed to cluster the neighborhood graph of the cells in Seurat. The common gene markers (CD3D, CD4, CD8A, FOXP3, LYZ) were utilized for manual annotation. To identify the tumor cell in T cells, two datasets (GSE122703 and GSE222703) were obtained from download files, while GSE182861 was identified by Copykat package. The clusters in GSE182861 were considered as the tumor cell if they meet two conditions. One condition is that more than 50% aneuploid cell identified by Copykat and the other is that more than 50% cells derived from one patient.
Gene editing on cell lines
Tumor cell lines were engineered to stably express both luciferase and green fluorescent protein (GFP). CD70-KO Raji cells were generated via use of the guide RNA targeting CD70 gene (‘5-GCCCGCAGGACGCACCCATA-3’), and the CD70-negative population was sorted to high purity by flow cytometry for functional assays. K562 cell line was transduced with plasmid containing HLA-E construct by piggyBac transposon system, and the HLA-E positive population of K562 cell was sorted and expanded for functional assays.
Multi-gene editing on human iPSCs
The plasmid contained the sequences of IL15RF, hnCD16 and anti-CD70 CAR (Figure 2A). Specifically, the CAR molecule was consisted of a CD8 leader, followed by CD70-targeting single-chain variable region, the CD8α hinge, CD8 transmembrane domain, 4-1BB costimulatory domain and CD3zeta chain. IL15RF and hnCD16 were constructed as previously described.44,45 To generate mutiplex-gene-engineered iPSCs, Cas9 protein (Invitrogen) and CD70 targeted gRNA (‘5-GCCCGCAGGACGCACCCATA-3’) were mixed and incubated for 15 min to form ribonucleoprotein, followed by an addition of the plasmid, piggyBac plasmid and iPSCs. Transfection was performed using a Lonza electroporator. After transfection, PCR gel electrophoresis and fragment analyses were performed to assess the efficiency of CD70 deletion on iPSCs. Then, iPSCs were stained with fluorescently conjugated antibodies against IL-15 (Thermo Fisher), CD16 (BD Biosciences), and FITC-Labeled Human CD70 Protein (Acrobiosystems). iPSC individual clones was isolated using a SONY SH800S cell sorter. Thereafter, Sanger sequencing as well as fragment analyses were utilized to validate the frameshift mutations in both alleles of CD70 gene, and flow cytometry analyses were used to confirm the stemness and stability of transgene expression.
NK cell derivation and expansion from human iPSCs
Adopting previously used protocols, we established an NK cell production platform derived from hiPSCs which incorporates procedures of embryoid body (EB) formation, NK cell differentiation and NK cell expansion (Figure 1A).46 Human iPSCs were cultured in StemFit Basic03 medium (AJINOMOTO, Basic03) supplemented with human bFGF, and surface markers including TRA-1-81 (BD Biosciences) and SSEA-4 (BD Biosciences) were stained to determine their pluripotency. The derivation of NK cells from hiPSCs has been previously described.47,48 Differentiated iNK cells were expanded by co-culturing with artificial antigen-presenting cells (aAPCs). The aAPCs were made by γ-irradiated K562 cells expressing membrane-bound IL-21 and CD137 ligand. Expanded iNK cells were harvested for experiments. Mock iNK cells were referred to as iPSC-derived NK cells without further genetic modification, and backbone iNK cells were referred to as iPSC-derived NK cells with IL15RF and hnCD16 expression and CD70-KO.
Gene editing on iPSC derived NK cells
To preliminarily explore the function of CD70 on iNK cells, CD70 was directly knocked out on iNK cells via using the guide RNA targeting CD70 gene (“GGTACACATCCAGGTGACGCTGG”) (Figure S3A). NKG2A-KO was performed on iNK cells and CBNK cells in the same methods.
iNK cell fratricide assays
Mock iNK cells, backbone iNK cells, and CD70-/- iNK cells were stained with CellTrace CFSE (Invitrogen, C34554). And then unstained 70CAR-iNK cells were added as effectors to the co-culture system at an E:T ratio of 1:1 or 5:1. After 4-h co-culture, cells were stained with propidium iodide (Thermo Fisher) to differentiate fratricide-induced dead cells.
Mixed lymphocyte reaction
To generate primed alloT cells, peripheral blood mononuclear cells (PBMCs, Sailybio, Shanghai, China and Oricellbio Biotechnology, Jiangsu, China) from recipients were mixed with iNK cells at 1:1 ratio in KBM581 complete medium supplemented with 200 IU/mL human recombinant IL-2 (Beijing Four Rings Biopharmaceutical Co., Ltd., Beijing, China). The medium was changed every 2–3 days. Around 2 weeks later, T cells robustly expanded and were harvested for further analysis. The harvested cells were stained with CD3 (BD Biosciences), CD8 (BD Biosciences), CD56 (BD Biosciences) and CD70 (BD Biosciences) antibodies to validate the purity of T cells. The purity of CD3+ cells routinely exceeded 95% on day 14. Also, alloT cells were stained with CD70, CD25 (Biolegend), CD107a (BD Biosciences), CD69 (BD Biosciences), CD137 (Biolegend), and CD40 ligand (Thermo Fisher) antibodies to reflect the activation status. Furthermore, CFSE-labeled backbone iNK cells were co-cultured with alloT cells, and propidium iodide were added to mixtures after 6 or 12 h. Specific cytotoxicity was calculated as (% specific death - % spontaneous death)/(1- % spontaneous death) ×100%.
Tumor cell line and iNK cell phenotyping
To detect the phenotype of tumor cell lines, we used the fluorescently conjugated antibodies. including CD70, MICA/B (Thermo Fisher), ULBP1 (R&D Systems), ULBP2/5/6 (R&D Systems), ULBP3 (R&D Systems), B7H6 (Thermo Fisher), NKp44L (USBiological). Meanwhile, fluorescently conjugated antibodies, including CD56, CD16 (BD Biosciences), CD57 (BD Biosciences), NKG2D (BD Biosciences), NKp30 (Biolegend), NKp44 (BD Biosciences), NKp46 (BD Biosciences), DNAM1 (R&D Systems), FasL (BD Biosciences), NKG2C (R&D Systems), NKG2A (Biolegend), CD158a (Beckman Coulter), CD158b (Beckman Coulter), CD158e (Beckman Coulter), PD1 (Biolegend), TIM3 (Biolegend), LAG3 (Biolegend), TIGIT (Biolegend), CD94 (Thermo Fisher), pSTAT3 (BD Biosciences), and pSTAT5 (BD Biosciences), were to analyze the phenotype of iNK cells.
To detect surface markers, cells were stained with antibodies at room temperature. After 20 min, stained cells were washed and re-suspended in buffer (1% BSA in PBS). To detect intracellular phosphorylated proteins, cells were fixed using BD Phosflow Fix Buffer I (BD Biosciences) and then incubated with BD Phosflow Perm Buffer III (BD Biosciences). Thereafter, cells were washed and stained with Anti-Stat3 (pY705) or Anti-Stat5 (pY694) antibody. Flow cytometry was performed on CytoFLEX S flow cytometer (BD Biosciences).
In vitro cytotoxicity assays
For flow cytometry-based cytotoxicity assay, live tumor cells were determined and calculated as propidium iodide−, CD56−. For luciferase-based cytotoxicity assay, tumor cell lines were engineered to express luciferase and co-cultured with iNK cells at multiple E:T ratios, and residual tumor cells were detected using luciferase assay. In brief, luciferin (Promega) was mixed with the co-culture system, and protected from light for 10 min. The absorbance was detected using a Synergy LX (Biotek). Imaging-based cytotoxicity assays were performed using GFP-expressing tumor cells and the Incucyte S3 system. All green fluorescence data were normalized to the initial scan data. As for serial killing, an equivalent count of target cells were added to the mixtures at an interval of 24 h. Readouts of cell index were measured every 3–4 h, and the plotted readouts of cell index were normalized to time point zero.
Activation status of NK cells during co-culture was detected by surface CD107a expression and cytokine release. After co-incubation, mixture was stained with CD107a (BD Biosciences) antibody for around 4 h, followed by staining with CD56 and live/dead staining. Finally, the percentage of CD107a positive NK cells was measured by flow cytometry. Besides, supernant of mixture was collected for the analysis of cytokines via ELISA, including IL-2, IFN-γ, TNF-α, and granzyme B.
Animal experiments
A mouse xenograft model of lymphoma was established by intravenous injection of 2×105 Raji cells or 1×106 MT-4 cells labeled by luciferase through the tail vein. Renal cancer model was built by intraperitoneally injection of 1×106 Caki-1 cells. Mice were randomly administered with mock iNK, backbone iNK, 70CAR-iNK cells or left untreated. After tumor inoculation, mice were left untreated or injected with three doses of 5×106 70CAR-iNK cells in 200 μL 2% human serum albumin at day 0, day 3, and day 7. Tumor burden was weekly monitored by bioluminescent imaging (BLI), and luminescent images were acquired and analyzed using the living imaging software (PerkinElmer) or photon imager optima (Biospace Lab). Moreover, blood was weekly collected from mice tail vein, and peripheral NK cell counts was determined by flow cytometry using fluorescently labeled antibodies against CD45 and CD56.
To test the in vivo persistence of 70CAR-iNK in the presence of allogeneic T cells, we intravenously inoculated NOG murine with 1 × 106 T cells pre-activated with CD3/CD28 dynabeads (Thermo Fisher).19 On the next day, mice were left untreated or injected with three doses of 5×106 backbone iNK or 70CAR-iNK cells at day 0, day 3, and day 7. Blood was weekly collected from mice tail vein, and peripheral NK cell count was determined by flow cytometry using fluorescently labeled antibodies against CD45 and CD56.
Quantification and statistical analysis
Statistical analyses were performed via Prism 9 (GraphPad software) or R software. Continuous variables were described as means ± standard error of mean (SEM), whereas categorical variables were described as percentages. Mann-Whitney U test or t test (two-tailed, unpaired or paired as indicated) was used to compare continuous variables between two groups, while one-way ANOVA analysis was used for the comparison among multiple groups. Data grouped by two independent categorical variables were analyzed using a two-way ANOVA. Survival was graphed as Kaplan-Meier curves, and the log rank test was used to determine statistical significance. And p-values <0.05 were considered significant for all analyses. Specific methods of statistical analyses can be found in corresponding figure legends.
Published: January 9, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101889.
Contributor Information
Dongrui Wang, Email: dongrui-wang@zju.edu.cn.
Yongxian Hu, Email: 1313016@zju.edu.cn.
Luhan Yang, Email: luhan.yang@qihanbio.com.
He Huang, Email: huanghe@zju.edu.cn.
Supplemental information
References
- 1.Baker D.J., Arany Z., Baur J.A., Epstein J.A., June C.H. CAR T therapy beyond cancer: the evolution of a living drug. Nature. 2023;619:707–715. doi: 10.1038/s41586-023-06243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June C.H., Sadelain M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June C.H., O'Connor R.S., Kawalekar O.U., Ghassemi S., Milone M.C. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 4.Dimitri A., Herbst F., Fraietta J.A. Engineering the next-generation of CAR T-cells with CRISPR-Cas9 gene editing. Mol. Cancer. 2022;21:78. doi: 10.1186/s12943-022-01559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias J., Yu J., Varshney M., Inzunza J., Nalvarte I. Hematopoietic stem cell- and induced pluripotent stem cell-derived CAR-NK cells as reliable cell-based therapy solutions. Stem Cells Transl. Med. 2021;10:987–995. doi: 10.1002/sctm.20-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Depil S., Duchateau P., Grupp S.A., Mufti G., Poirot L. 'Off-the-shelf' allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020;19:185–199. doi: 10.1038/s41573-019-0051-2. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs J., Deschoolmeester V., Zwaenepoel K., Rolfo C., Silence K., Rottey S., Lardon F., Smits E., Pauwels P. CD70: An emerging target in cancer immunotherapy. Pharmacol. Ther. 2015;155:1–10. doi: 10.1016/j.pharmthera.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y., Chen J., Feng M., Wang Y., Liao W., Wu Q., Wen F., Li Q. The correlation of CD70 in immune characteristics and drug therapy of pan-cancer. Hum. Cell. 2023;36:476–479. doi: 10.1007/s13577-022-00786-2. [DOI] [PubMed] [Google Scholar]
- 9.Wajant H. Therapeutic targeting of CD70 and CD27. Expert Opin. Ther. Targets. 2016;20:959–973. doi: 10.1517/14728222.2016.1158812. [DOI] [PubMed] [Google Scholar]
- 10.Flieswasser T., Van den Eynde A., Van Audenaerde J., De Waele J., Lardon F., Riether C., de Haard H., Smits E., Pauwels P., Jacobs J. The CD70-CD27 axis in oncology: the new kids on the block. J. Exp. Clin. Cancer Res. 2022;41:12. doi: 10.1186/s13046-021-02215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zain D.R., Iyer S.P., Sica R.A., Ho P.J., Hu B., Prica A., Weng W.K., Kim Y.H., Khodadoust M.S., Palomba M.L., et al. The COBALT-LYM study of CTX130: a phase 1 dose escalation study of CD70-targeted allogeneic CRISPR-Cas9-engineered CAR T cells in patients with relapsed/refractory (R/R) T-cell malignancies. Europ. J. Cancer. 2022;173:S21. doi: 10.1016/S1470-2045(24)00508-4. [DOI] [PubMed] [Google Scholar]
- 12.Srour S., Kotecha R., Curti B., Chahoud J., Drakaki A., Tang L., Goyal L., Prashad S., Szenes V., Norwood K., Pal S. A phase 1 multicenter study (TRAVERSE) evaluating the safety and efficacy of ALLO-316 following conditioning regimen in pts with advanced or metastatic clear cell renal cell carcinoma (ccRCC) Cancer Res. 2023;83:CT011. doi: 10.1158/1538-7445.Am2023-Ct011. [DOI] [Google Scholar]
- 13.Cichocki F., Bjordahl R., Gaidarova S., Mahmood S., Abujarour R., Wang H., Tuininga K., Felices M., Davis Z.B., Bendzick L., et al. iPSC-derived NK cells maintain high cytotoxicity and enhance in vivo tumor control in concert with T cells and anti-PD-1 therapy. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aaz5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimasaki N., Jain A., Campana D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020;19:200–218. doi: 10.1038/s41573-019-0052-1. [DOI] [PubMed] [Google Scholar]
- 15.Lamers-Kok N., Panella D., Georgoudaki A.M., Liu H., Özkazanc D., Kučerová L., Duru A.D., Spanholtz J., Raimo M. Natural killer cells in clinical development as non-engineered, engineered, and combination therapies. J. Hematol. Oncol. 2022;15:164. doi: 10.1186/s13045-022-01382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamin R., Graham C., Yallop D., Jozwik A., Mirci-Danicar O.C., Lucchini G., Pinner D., Jain N., Kantarjian H., Boissel N., et al. Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: results of two phase 1 studies. Lancet. 2020;396:1885–1894. doi: 10.1016/S0140-6736(20)32334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Zhou Y., Zhang M., Ge W., Li Y., Yang L., Wei G., Han L., Wang H., Yu S., et al. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2021;27:2764–2772. doi: 10.1158/1078-0432.CCR-20-3863. [DOI] [PubMed] [Google Scholar]
- 18.Myers J.A., Miller J.S. Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021;18:85–100. doi: 10.1038/s41571-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo F., Watanabe N., McKenna M.K., Hicks M.J., Srinivasan M., Gomes-Silva D., Atilla E., Smith T., Ataca Atilla P., Ma R., et al. Engineered off-the-shelf therapeutic T cells resist host immune rejection. Nat. Biotechnol. 2021;39:56–63. doi: 10.1038/s41587-020-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Wang L., Shao M., He X., Yue Y., Zhou Y., Yang L., Huang H., Hu Y. Off-the-Shelf, Multiplexed-Engineered iPSC-Derived CD33 CAR-NK Cells for Treatment of Acute Myeloid Leukemia. Blood. 2022;140:12685. doi: 10.1182/blood-2022-166420. [DOI] [Google Scholar]
- 21.Wang Y.Y., He X.J., Wang L.Q., Shao M., Yue Y.A., Gao Y.B., Church G., Huang H., Yang L.H. "Super NK cells" - natural killer cells derived from engineered hiPSC with enhanced NK receptor expression demonstrate better anti-tumor effects for solid tumors. Cancer Res. 2022;82:3155. [Google Scholar]
- 22.Ferrari G., Thrasher A.J., Aiuti A. Gene therapy using haematopoietic stem and progenitor cells. Nat. Rev. Genet. 2021;22:216–234. doi: 10.1038/s41576-020-00298-5. [DOI] [PubMed] [Google Scholar]
- 23.Nie M., Ren W., Ye X., Berglund M., Wang X., Fjordén K., Du L., Giannoula Y., Lei D., Su W., et al. The dual role of CD70 in B-cell lymphomagenesis. Clin. Transl. Med. 2022;12 doi: 10.1002/ctm2.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertrand P., Maingonnat C., Penther D., Guney S., Ruminy P., Picquenot J.M., Mareschal S., Alcantara M., Bouzelfen A., Dubois S., et al. The costimulatory molecule CD70 is regulated by distinct molecular mechanisms and is associated with overall survival in diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2013;52:764–774. doi: 10.1002/gcc.22072. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z.Z., Novak A.J., Ziesmer S.C., Witzig T.E., Ansell S.M. CD70+ non-Hodgkin lymphoma B cells induce Foxp3 expression and regulatory function in intratumoral CD4+CD25 T cells. Blood. 2007;110:2537–2544. doi: 10.1182/blood-2007-03-082578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adotevi O., Galaine J. Antitumor CAR T-cell Screening Platform: Many Are Called, but Few Are Chosen. Cancer Res. 2022;82:2517–2519. doi: 10.1158/0008-5472.CAN-22-1739. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J., Zhao Y., Hu H., Tang L., Zeng Y., Deng X., Ding S., Guo A.Y., Li Q., Zhu X. Revealing the impact of CD70 expression on the manufacture and functions of CAR-70 T-cells based on single-cell transcriptomics. Cancer Immunol. Immunother. 2023;72:3163–3174. doi: 10.1007/s00262-023-03475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivori S., Pende D., Quatrini L., Pietra G., Della Chiesa M., Vacca P., Tumino N., Moretta F., Mingari M.C., Locatelli F., Moretta L. NK cells and ILCs in tumor immunotherapy. Mol. Aspects Med. 2021;80 doi: 10.1016/j.mam.2020.100870. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y., van Hoef V., Zhang X., Wennerberg E., Lorent J., Witt K., Masvidal L., Liang S., Murray S., Larsson O., et al. IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells. Blood. 2016;128:1475–1489. doi: 10.1182/blood-2016-02-698027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L., Mohanty V., Dou J., Huang Y., Banerjee P.P., Miao Q., Lohr J.G., Vijaykumar T., Frede J., Knoechel B., et al. Loss of metabolic fitness drives tumor resistance after CAR-NK cell therapy and can be overcome by cytokine engineering. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma S., Caligiuri M.A., Yu J. Harnessing IL-15 signaling to potentiate NK cell-mediated cancer immunotherapy. Trends Immunol. 2022;43:833–847. doi: 10.1016/j.it.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu E., Marin D., Banerjee P., Macapinlac H.A., Thompson P., Basar R., Nassif Kerbauy L., Overman B., Thall P., Kaplan M., et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020;382:545–553. doi: 10.1056/NEJMoa1910607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borst J., Hendriks J., Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr. Opin. Immunol. 2005;17:275–281. doi: 10.1016/j.coi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Dequeant M.L., Sagert J., Kalaitzidis D., Yu H., Porras A., McEwan B., Padalia Z., Tetteh P., Nguyen T., Dunn A., et al. CD70 knockout: A novel approach to augment CAR-T cell function. Cancer Res. 2021;81:1537. [Google Scholar]
- 35.Cheng J., Ge T., Zhu X., Wang J., Zeng Y., Mu W., Cai H., Dai Z., Jin J., Yang Y., et al. Preclinical development and evaluation of nanobody-based CD70-specific CAR T cells for the treatment of acute myeloid leukemia. Cancer Immunol. Immunother. 2023;72:2331–2346. doi: 10.1007/s00262-023-03422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehniger T.A. CD70 turns on NK cells to attack lymphoma. Blood. 2017;130:238–239. doi: 10.1182/blood-2017-06-786244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Sayed M.F., Ruckstuhl C.A., Hilmenyuk T., Claus C., Bourquin J.P., Bornhauser B.C., Radpour R., Riether C., Ochsenbein A.F. CD70 reverse signaling enhances NK cell function and immunosurveillance in CD27-expressing B-cell malignancies. Blood. 2017;130:297–309. doi: 10.1182/blood-2016-12-756585. [DOI] [PubMed] [Google Scholar]
- 38.Lanza R., Russell D.W., Nagy A. Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 2019;19:723–733. doi: 10.1038/s41577-019-0200-1. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J., Lin Q., Song Y., Liu D. Universal CARs, universal T cells, and universal CAR T cells. J. Hematol. Oncol. 2018;11:132. doi: 10.1186/s13045-018-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaydosik A.M., Stonesifer C.J., Khaleel A.E., Geskin L.J., Fuschiotti P. Single-Cell RNA Sequencing Unveils the Clonal and Transcriptional Landscape of Cutaneous T-Cell Lymphomas. Clin. Cancer Res. 2022;28:2610–2622. doi: 10.1158/1078-0432.CCR-21-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borcherding N., Voigt A.P., Liu V., Link B.K., Zhang W., Jabbari A. Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression. Clin. Cancer Res. 2019;25:2996–3005. doi: 10.1158/1078-0432.CCR-18-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massenet-Regad L., Poirot J., Jackson M., Hoffmann C., Amblard E., Onodi F., Bouhidel F., Djouadou M., Ouzaid I., Xylinas E., et al. Large-scale analysis of cell-cell communication reveals angiogenin-dependent tumor progression in clear cell renal cell carcinoma. iScience. 2023;26 doi: 10.1016/j.isci.2023.108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing Y., Ni Z., Wu J., Higgins L., Markowski T.W., Kaufman D.S., Walcheck B. Identification of an ADAM17 cleavage region in human CD16 (FcgammaRIII) and the engineering of a non-cleavable version of the receptor in NK cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woan K.V., Kim H., Bjordahl R., Davis Z.B., Gaidarova S., Goulding J., Hancock B., Mahmood S., Abujarour R., Wang H., et al. Harnessing features of adaptive NK cells to generate iPSC-derived NK cells for enhanced immunotherapy. Cell Stem Cell. 2021;28:2062–2075.e5. doi: 10.1016/j.stem.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H., Kaufman D.S. An Improved Method to Produce Clinical-Scale Natural Killer Cells from Human Pluripotent Stem Cells. Methods Mol. Biol. 2019;2048:107–119. doi: 10.1007/978-1-4939-9728-2_12. [DOI] [PubMed] [Google Scholar]
- 47.Knorr D.A., Ni Z., Hermanson D., Hexum M.K., Bendzick L., Cooper L.J.N., Lee D.A., Kaufman D.S. Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl. Med. 2013;2:274–283. doi: 10.5966/sctm.2012-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng E.S., Davis R., Stanley E.G., Elefanty A.G. A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nat. Protoc. 2008;3:768–776. doi: 10.1038/nprot.2008.42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This study analyzed publicly available datasets at Gene Expression Omnibus. GEO accession numbers are listed in the key resources table.
-
•
All the original codes have been deposited at Zenodo. Accession website is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.