Abstract
Pi class GSTs (glutathione S-transferases) are a member of the vertebrate GST family of proteins that catalyse the conjugation of GSH to electrophilic compounds. The expression of Pi class GST genes can be induced by exposure to electrophiles. We demonstrated previously that the transcription factor Nrf 2 (NF-E2 p45-related factor 2) mediates this induction, not only in mammals, but also in fish. In the present study, we have isolated the genomic region of zebrafish containing the genes gstp1 and gstp2. The regulatory regions of zebrafish gstp1 and gstp2 have been examined by GFP (green fluorescent protein)-reporter gene analyses using microinjection into zebrafish embryos. Deletion and point-mutation analyses of the gstp1 promoter showed that an ARE (antioxidant-responsive element)-like sequence is located 50 bp upstream of the transcription initiation site which is essential for Nrf 2 transactivation. Using EMSA (electrophoretic mobility-shift assay) analysis we showed that zebrafish Nrf 2–MafK heterodimer specifically bound to this sequence. All the vertebrate Pi class GST genes harbour a similar ARE-like sequence in their promoter regions. We propose that this sequence is a conserved target site for Nrf 2 in the Pi class GST genes.
Keywords: antioxidant-responsive element (ARE), gene transcription, glutathione S-transferase (GST), green fluorescent protein (GFP), Phase 2 detoxification enzyme, Pi class glutathione S-transferase
Abbreviations: ARE, antioxidant-responsive element; DEM, diethylmaleate; ef1α, elongation factor 1α; EMSA, electrophoretic mobility-shift assay; EST, expressed sequence tag; GPEI, Pi class glutathione S-transferase enhancer I; GST, glutathione S-transferase; MAPEG, membrane-associated proteins in eicosanoid and glutathione; Neh, Nrf 2-ECH homology; Nrf 2, NF-E2 p45-related factor 2; 5′-RACE, 5′-rapid amplification of cDNA ends; RT-PCR, reverse transcriptase-PCR; TRE, PMA (‘TPA’)-responsive element
INTRODUCTION
The transcription factor Nrf 2 (NF-E2 p45-related factor 2) induces the expression of Phase 2 detoxification and antioxidant genes in response to electrophilic insults [1,2]. The importance of Nrf 2 in this induction has been demonstrated by a number of experiments using Nrf 2-deficient mice [3]. The electrophile response is regulated through a cis-acting element called the ARE (antioxidant-responsive element) which is found within the regulatory region of each gene [4]. Nrf 2 binds to the ARE sequence as a heterodimeric complex with small Maf proteins using a basic region leucine-zipper domain [5,6]. Under non-induced conditions, Nrf 2 is retained in the cytoplasm by interaction with the actin-binding protein Keap1 [7,8]. The Nrf 2–Keap1 interaction facilitates the degradation of Nrf 2 by a proteasome-mediated mechanism [9,10]. Electrophiles cause Nrf 2 to escape from Keap1, allowing Nrf 2 to accumulate in the nucleus and thereby enhance the expression of target genes [7].
Recently, we demonstrated that the Nrf 2–Keap1 system is conserved in zebrafish and that the Nrf 2-dependent induction of Phase 2 detoxification and antioxidant genes is conserved among vertebrates [11,12]. Zebrafish Nrf 2 protein shares six highly conserved domains, Neh (Nrf 2-ECH homology) domains, with mammalian Nrf 2 proteins, which are considered to play critical functions in Nrf 2 regulation [5]. High sequence identities in each Neh domain between zebrafish and mouse Nrf 2 imply a common regulatory mechanism among vertebrate Nrf 2s [11]. This finding provided us with a powerful tool to analyse the molecular basis of the Nrf 2–Keap1 system in zebrafish. In particular, the use of random mutagenesis and mutant-screening techniques enables us to perform a genetic search of molecules that may regulate the Nrf 2–Keap1 system. An additional advantage of the zebrafish system is the ease of detecting transactivation of endogenous genes by forced expression of Nrf 2 that is difficult in cultured cells. The analysis of endogenous genes will be significant for the study gene-specific elements important for Nrf 2 transactivation.
Among the known endogenous target genes of zebrafish Nrf 2, the Pi class GSTs (glutathione S-transferases) showed relatively strong induction in both electrophile-treated larvae and Nrf 2-overexpressing embryos [11]. GSTs are a multigene family of detoxification enzymes that catalyse the conjugation of GSH with a variety of endogenous and exogenous electrophilic compounds [13]. GSTs are widely distributed in nature, being found in all eukaryotes and in many prokaryotes. In mammals, GSTs can be divided into distinct classes: Alpha, Mu, Pi, Theta, Sigma, Zeta, Omega, Kappa and four subgroups of MAPEG (membrane-associated proteins in eicosanoid and glutathione) enzymes, according to their homologies and properties [14,15]. The sequence identity of GSTs in the same class is greater than 60%, whereas between classes the identity is less than 30%. Members of each class exhibit overlapping yet distinct substrate specificities.
Pi class GSTs are the most abundant GST family member found in many normal and malignant tissues [13]. There has been considerable interest in the properties of Pi class GSTs, particularly in relationship to their role in carcinogenesis and human cancer. Pi class GSTs are highly efficient in the glutathione-conjugation of carcinogenic benzo[a]pyrene derivatives [16] and is effective in the detoxification of electrophilic α,β-unsaturated carbonyl compounds [17]. In addition, studies using rats harbouring a GSTP1 transgene have shown that over-expression of Pi class GSTs inhibits the early phase of chemical carcinogenesis in rat liver [18]. Likewise, ablation of GSTP1 and GSTP2 in mice resulted in increased susceptibility to skin tumorigenesis induced by chemical carcinogens [19]. In humans, genetic polymorphisms within the GSTP1 locus have been identified that are associated with susceptibility to bladder, testicular and prostate cancer [20]. Therefore, the expression level of Pi class GST is regarded as one of an important determinant for the protection against various chemical insults.
In mice, the administration of electrophilic agents induced expression of Pi class GSTs. This induction was shown to be drastically impaired when the Nrf 2 gene is disrupted [3,21,22]. The electrophile-induction of GSTP1 expression was also observed in the rat liver epithelial RL34 cells [23,24]. Studies in these cells identified an element known as GPEI (Pi class glutathione S-transferase enhancer I), located in a 2.5-kbp region upstream of the transcriptional initiation site, that is responsible for this regulation and shown to be a strong enhancer element for hepatocarcinogenesis [23]. GPEI contains an ARE-like sequence capable of binding Nrf 2 protein [24]. The results suggest that the GPEI is a target for the Nrf 2 transcription factor and that is important for the induced expression of GSTP1 by electrophilic compounds. However, a similar GPEI has not been found in the promoter of Pi class GST genes from other species. Ikeda et al. [25] have demonstrated that Nrf 2 binds to an ARE-like sequence located on the promoter region of the mouse GSTP1 gene and that this region is important for transactivation by Nrf 2. This ARE-like sequence is conserved in the promoter region of human, rat and mouse Pi class GST genes, and seems to be common target site for Nrf 2 among vertebrates, although it was shown to be inactive in the rat cells [26].
As described above, Pi class GST genes are also strongly induced in zebrafish. It is of interest to identify the specific target sites for Nrf 2 in the zebrafish genome and to compare the regulatory mechanism between fish and mammalian genes. In the present study, we have isolated whole genomic regions of the zebrafish Pi class GST genes and identified their transcriptional regulatory regions. The results indicate that an ARE-like sequence is also conserved in the promoter region of fish Pi class GST genes and that it is a functional element for Nrf 2-dependent transactivation.
EXPERIMENTAL
Isolation of genomic DNA and full-length cDNA of zebrafish Pi class GST genes
A zebrafish EMBL3 SP6/T7 genomic library (Clontech) was screened with a probe containing a partial cDNA of zebrafish gstp1 [11]. Probes were labelled using the AlkPhos Direct DNA labelling kit, and the positive plaques on the membrane filters were detected with CDP-Star as substrate, according to the manufacturer's instruction (Amersham Pharmacia Biotech). The DNA inserts of the positive clones were subcloned into pBluescript II SK. A 0.61-kbp fragment of genomic DNA including the gstp2 promoter was prepared by PCR using zebrafish genomic DNA and specific primers (5′-CCGTCGACACAGCAAGAAGGTCACTGG-3′ and 5′-GGGGATCCTCTGTGAAGTTGCTGCTCCTGAAATGTGTAG-3′). The full-length cDNAs were isolated by RT (reverse transcriptase)-PCR of total RNA from 4-day-old embryos treated with 100 μM DEM (diethylmaleate) using specific primers, and subcloned into pGEM-T Easy (Promega). Total zebrafish RNA was prepared using RNAzol B (TEL-TEST). The primers have the following sequences: 5′-CTAGGAGCAGCTTTGAAACGCAC-3′ and 5′-CGTTGTTGGAGAATGTTGTACCGACG-3′ (gstp1); 5′-CACATTTCAGGAGCAGCAACTTCACAGAC-3′ and 5′-CATTTGAGAACGTTGTATCAACG-3′ (gstp2).
Inducer treatment
Zebrafish embryos and larvae were obtained by natural mating. For the induction studies, larval and adult fish were placed in culture dishes or plastic chambers respectively, and treated with DEM.
Expression analyses by RT-PCR
Expression of Pi class GST genes was analysed by RT-PCR as described previously [11] using following primers: gstp1, 5′-CTAGGAGCAGCTTTGAAACGCAC-3′ and 5′-CGTTGTTGGAGAATGTTGTACCGACG-3′; gstp2, 5′-CACTCTCACATACTTCGCTATC-3′ and 5′-AATATTTTCAAATGGTTTGAACTC-3′; ef1α (elongation factor 1α), 5′-GCCCCTGCCAATGTA-3′ and 5′-GGGCTTGCCAGGGAC-3′.
Radiation hybrid mapping
Radiation hybrid mapping using panel LN54 was performed as described in Hukriede et al. [27] using two sets of primers specific to each Pi class GST gene. Sequences of each primer are following: 5′-TTGACCAAGGAAGACGTGG-3′ and 5′-CTGTGATTGGCAGACTTGAC-3′ (gstp1); 5′-CAACCACCTCAAATGTTTGAAAATG-3′ and 5′-CGTTGTTGGAGAAGTTGTACCGACG-3′ (gstp1); 5′-GAGAGCTGAATCAGCACTTG-3′ and 5′-CAGCAACTTTGCCTGGTATG-3′ (gstp2); 5′-CATTTGAGAACGTTGTATCAACG-3′ and 5′-GTCGTTCTTTCCATACGCAC-3′ (gstp2).
5′-RACE (5′-rapid amplification of cDNA ends) assays
5′-RACE assays were carried out using the 5′-RACE System (Gibco BRL) as described previously [28]. Briefly, 4 μg each of total RNA from 4-day-old embryos treated with 100 μM DEM was reverse transcribed using the antisense primer 5′-GCAGATCTTTGATGAACGC-3′, which corresponds to the sequences in the sixth exon of gstp1. The product was further amplified using the 5′-RACE abridged anchor primer and a gstp1 fifth exon-specific antisense primer 5′-CAGCTTTATGTACTTCAGGCG. DNA fragments of gstp2 were also amplified using these primers. 5′-RACE products were subcloned into pBluescript II SK and their sequences were determined.
Plasmid construction
DNA fragments corresponding to the 3.5- or 0.61-kbp upstream region of the transcriptional initiation site of zebrafish gstp1 or gstp2 and the EGFP (enhanced green fluorescent protein) fragment of pCS2-eGFP [28] were ligated together into pBluescript IISK, and the resulting plasmids were named p3.5gstp1GFP and p0.61gstp2GFP respectively. For constructing 5′-deleted mutants, p3.5gstp1GFP was linearized with ApaI and SalI, and incubated with ExoIII, followed by blunting with mung bean nuclease and self-ligation. Selected constructs were sequenced and named p1.3gstp1GFP and p0.12gstp1GFP, according to the positions of their 5′ ends from the transcriptional initiation site. For construction of p3.5gstp1m1GFP, mutations in the proximal ARE-like sequence were introduced into p3.5gstp1GFP by PCR. Other plasmids used in this study were as described previously [11,12].
Microinjection of zebrafish embryos
Reporter DNAs were linearized by digesting the vector with SacI (gstp1) or SalI (gstp2). Digested DNA was resuspended in water and injected into the blastomere of early one-cell stage embryos (see Figure 5A). Synthetic capped RNA was made with the SP6 mMESSAGE mMACHINE™ in vitro transcription kit (Ambion) using linearized DNA of the pCS2 derivatives described above. GFP expression was examined under a GFP Plus (480 nm excitation, 505 nm emission) filter on a MZFLIII microscope (Leica) equipped with a 600CL-CU digital camera (Pixera).
Figure 5. Induction of the GFP-reporter genes by Nrf 2 overexpression.
(A) Scheme showing the analysing method. (B) GFP expression of the 3.5gstp1GFP and 0.61gstp2GFP constructs. 3.5gstp1GFP (20 pg) or 0.61gstp2GFP (100 pg) was injected with or without 40 pg of Nrf 2 mRNA into blastomere of one-cell stage embryos. GFP expression was analysed in 8-h embryos. Values indicate the percentages of embryos that showed more than ten GFP-positive cells. The number of embryos observed for each construct are indicated in parentheses.
EMSAs (electrophoretic mobility-shift assays)
EMSAs were carried out as described previously [12]. Zebrafish Nrf 2 and MafK proteins were prepared by transcription and translation reactions in vitro using TNT® wheat germ extract (Promega). An oligonucleotide containing the proximal ARE-like sequence (5′-GGGGCGCGTGCATGACTCATCAAAAACGCTGAG-3′) was prepared by annealing synthetic oligonucleotides and labelled with using 32P Rediprime II DNA Labelling System (Amersham Biosciences). A 40- or 100-fold molar excess of unlabelled oligonucleotide was added to the reactions for specific competitors.
Antibodies
Anti-Nrf 2 antibody was produced by immunizing rabbits with N-terminal residues of mouse Nrf 2 (Neh2 domain, amino acids 1–X98) as an antigen (Sawady Technology, Tokyo, Japan). The Neh2 protein was purified from recombinant Escherichia coli cells as described previously [29]. Anti-MafK antibody has been described previously [30].
RESULTS
Identification of two Pi class GST genes in zebrafish
We previously isolated a partial cDNA for the zebrafish Pi class GST and showed that its expression is induced in larvae by electrophilic compounds and that the induction is dependent on the Nrf 2–Keap1 system [11]. To exclude the possibility that the isolated clone is a pseudogene, we tried to obtain a full-length cDNA clone of the Pi class GST gene. In the process, we found a second related Pi class GST gene in zebrafish. After carrying out RT-PCR on total RNA from DEM-treated 4-day-old larvae using primers specific to the two Pi class GST genes, we isolated full-length cDNAs for both the gstp1 and gstp2 genes. Primers for each gene were designed on the basis of a combination of information from the EST (expressed sequence tag) (http://www.ncbi.nlm.nih.gov/dbEST/index.html) and the zebrafish genomic DNA (http://www.ensembl.org/Danio_rerio/) databases and our own preliminary data on a partial clone of gstp1, the Pi class GST cDNA described in our previous paper [11].
The percentage identity of the coding region sequence between gstp1 and gstp2 is 90.4%, and that of the encoding amino acid sequence is 87.0%. The amino acid identities between rat GSTP1 and zebrafish GSTP1 or GSTP2 proteins are 59.0% and 57.1% respectively. The identity relative to other classes of rat GST proteins is less than 30%, indicating that the isolated clones are zebrafish homologues of Pi class GST. Figure 1(A) shows a phylogenetic tree of the vertebrate Pi class GST proteins. To date, three full-length sequences of Pi class GST genes have been identified in non-mammalian animals, Xenopus laevis [31], Oncorhynchus nerka (sockeye salmon) [32] and Anguilla anguilla (European eel) (DDBJ, EMBL and GenBank® accession number AY530199). As expected, zebrafish GSTP1 and GSTP2 proteins share a higher similarity with these non-mammalian proteins compared with the mammalian Pi class GST proteins. Interestingly, the similarity between zebrafish GSTP1 and GSTP2 is higher than that other fish Pi class GSTs.
Figure 1. Comparison of the vertebrate Pi class GST proteins.
(A) Phylogenetic tree of Pi class GST proteins. The tree was constructed using the Clustal W program in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/search/clustalw-j.html). Scale bar, genetic distance. (B) Sequence alignment of various Pi class GST proteins. Conserved amino acids among Pi class GST proteins are highlighted in grey. Closed and open circles denote G- and H-sites respectively. Abbreviations: z, zebrafish; s, salmon; e, eel; x, Xenopus; b, bovine; h, human; r, rat; m, mouse.
Figure 1(B) shows the multiple alignment of Pi class GST proteins. The Pi class GST protein contains two ligand-binding sites: the glutathione-binding site (G-site) and electrophilic substrate-binding site (H-site) [33–35]. The residues in the G-site (Tyr8, Arg14, Trp39, Lys45, Gln52, Leu53, Gln65 and Ser66) are conserved in both GSTP1 and GSTP2, with the exception of Arg45 in GSTP2 (Figure 1B, closed circles). The residues in the H-site (Tyr8, Phe9, Val11, Arg14, Ile105, Tyr109, Asp203 and Gly204) are also conserved, apart from Ile11 in GSTP2 (Figure 1B, open circles). It is interesting to find that residues in position 11 also vary between the mouse GSTP1 and GSTP2 proteins. In humans, variant forms of GSTP1, Ile105 and Val105, exist, and they have been shown to have a different substrate preference [36,37]. Isoleucine is found at position 105 in both zebrafish GSTP1 and GSTP2 proteins, suggesting that their substrate specificies are related to the Ile105-type human GSTP1.
Nrf 2 can induce the expression of both gstp1 and gstp2
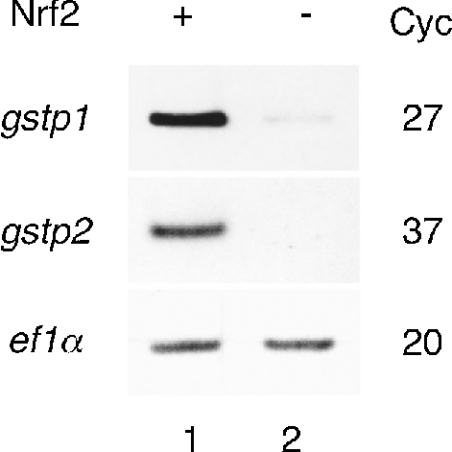
The high level of similarity in nucleotide sequences between gstp1 and gstp2 suggests that the electrophile induction of zebrafish Pi class GST expression that we observed previously, by whole mount in situ hybridization and Northern blot analysis [11], may be the combined contribution of the two related genes. Therefore, to investigate the inducibility of the genes independently we carried out RT-PCR analysis using sets of primers specific to each gene. The specificity of each primer set was demonstrated by PCR using cloned cDNA of gstp1 and gstp2 as templates (Figure 2A). Total RNA from whole body of 5-day-old larvae treated with or without 100 μM DEM for 6 h was analysed. Both gstp1 and gstp2 were strongly induced by DEM treatment (Figure 2B). These inductions were greatly reduced when we knocked-down the expression of Nrf 2 by injecting antisense morpholino oligonucleotide (nrf2-MO). These results indicated that electrophiles induce both gstp1 and gstp2 genes and that Nrf 2 mediates this induction. We next examined the induction of Pi class GST expression in adult gills. Electrophile-induced expression was observed for both gstp1 and gstp2 (Figure 2C). Next, we analysed the expression in male and female fish, since male-specific expression of GSTP1 has been shown in mice [21,38]. As shown in Figure 2(C), we could not observe any sex-specific differences in the expression of either gstp1 or gstp2. All these results were confirmed by RT-PCR using different sets of primers specific for gstp1 and gstp2 (results not shown). Our findings indicated that electrophiles induce gstp1 and gstp2 in both larvae and adults and that these two genes may be regulated by a similar molecular mechanism.
Figure 2. Nrf 2-dependent induction of gstp1 and gstp2 in zebrafish larvae.
(A) Confirmation of the primer specificity for RT-PCR. PCR was carried out using plasmid DNAs containing gstp1 (gstp1) or gstp2 (gstp2) cDNA as templates. Numbers indicate reaction cycles (Cyc). (B) Effect of nrf2-MO on the DEM-induced expression of gstp1 and gstp2. RT-PCR analysis of nrf2-MO (9 ng)- or mock-injected larvae, treated (+) or not treated (−) with 100 μM DEM for 6 h at day 5. The amounts of cDNA were standardized by the expression of ef1α. Each experiment was carried out at least three times and resulted in similar expression patterns. (C) Expression of the Pi class GST genes in the adult gills. RT-PCR analysis of the gills of adult male or female fish, treated (+) or not treated (−) with 100 μM DEM for 6 h.
We next examined the effect of exogenous Nrf 2 on the expression of gstp1 and gstp2. RT-PCR was carried out using 8-h-old embryos, injected either with or without zebrafish Nrf 2 mRNA at the one-cell stage. Remarkable induction of gstp1 and gstp2 expression was observed in the Nrf 2-overexpressing embryos, but not in the control embryos (Figure 3). The induction was confirmed by RT-PCR using different sets of primers specific for gstp1 and gstp2 (results not shown). These results indicate that both gstp1 and gstp2 are endogenous target genes for Nrf 2.
Figure 3. Expression of gstp1 and gstp2 in Nrf 2-overexpressing embryos.
RT-PCR analysis using total RNA isolated from embryos 8-h post-injection with or without 40 pg of Nrf 2 mRNA. Amounts of cDNA were standardized by the expression of ef1α.
Identification of the genomic locus containing gstp1 and gstp2
To analyse the regulatory mechanism of electrophile-induction of zebrafish gstp1 and gstp2 expression, genomic DNA fragments containing these genes were cloned and characterized. For gstp1, a λ-phage genomic library (2×106) was screened using a partial cDNA of zebrafish gstp1 [11], and two positive clones were isolated (Figure 4, #2 and #6). After characterizing these clones by restriction site mapping, a DNA fragment of the most 5′-extended clone (Figure 4, #6), containing about 11-kbp of the gstp1 locus, was subcloned into a cloning vector for further analysis. In the case of gstp2, we carried out PCR on genomic DNA using a primer set whose design was based on the information from the zebrafish genomic DNA database (Figure 4, contigs NA9028 and NA575). A 0.7-kbp DNA fragment that included a 0.61-kbp region upstream of the transcriptional initiation site was isolated.
Figure 4. Structure of the zebrafish gstp1 and gstp2 genes.
Maps of contigurated sequences and λ phage clones containing gstp1 or gstp2 are displayed. E, X and S indicate enzyme sites for EcoRI, XhoI and SacI respectively. Black and white boxes denote the coding and non-coding regions of the Pi class GST cDNAs respectively. Nucleotide sequence data of 3.5-kbp regions of gstp1 and 0.61-kbp regions of gstp2 used for the GFP-reporter constructs have been deposited in the DDBJ, EMBL and GenBank® databases with accession numbers AB194129 and AB194130 respectively.
As shown in Figure 4, the information of the zebrafish genomic DNA database suggests that gstp1 and gstp2 are arranged in tandem on the same chromosome. We carried out the genomic PCR around the junction site of gstp1 and gstp2 and confirmed this tandem arrangement (results not shown). Furthermore, we mapped gstp1 and gstp2 on the linkage map by using a LN54 hybrid panel [27], and demonstrated that both genes mapped to a linkage group 14, 124.37 cR (centiray) from the most terminal marker. The approximate sizes of the introns for both genes were determined by PCR, and it was shown that gstp1 and gstp2 span about 6.8- and 7.5-kbp respectively (Figure 4). These gene lengths are quite large in comparison with the mammalian Pi class GST genes such as mouse GSTP1 and GSTP2 [39,40]. To investigate the transcription initiation sites, a 5′-RACE assay was performed. The results indicated that the transcriptional initiation site of both gstp1 and gstp2 are about 60 bp upstream of translation initiation sites. These positions are quite similar to the mammalian Pi class GST genes [40–42].
Isolation of gene regulatory regions of gstp1 and gstp2
To identify the gene regulatory regions, we prepared GFP reporter constructs that are fused to DNA fragments containing the 3.5- and 0.61-kbp regions of the transcriptional initiation sites of gstp1 and gstp2 respectively (Figure 4, 3.5gstp1GFP and 0.61gstp2GST). An examination of promoter activity was made using zebrafish embryos. These constructs were microinjected into one-cell-stage embryos and expression of the GFP reporter genes monitored using a fluorescence microscope (Figure 5). Only low GFP expression was observed during development in the case of both genes, suggesting that basal promoter activities are quite weak (Figure 5B, −Nrf 2). We next tested whether Nrf 2 can transactivate the activity of these reporter genes. GFP-reporter DNAs with synthetic capped RNA of zebrafish Nrf 2 were co-injected into zebrafish embryos at the one-cell stage (Figure 5A) and the GFP activity was examined at the 8-h stage. GFP expression was dramatically enhanced in embryos over-expressing Nrf 2 compared with embryos injected with the reporter gene alone (Figure 5B, +Nrf 2). These results suggest that the Nrf 2-responsive element is present in both the 3.5- and 0.61-kbp regions of gstp1 and gstp2 respectively.
A proximal ARE-like sequence is essential for Nrf 2 transactivation
To determine the exact target sites for Nrf 2 in the 3.5-kbp region of gstp1, a series of deletion constructs of 5′-flanking region fused to the GFP gene were prepared and injected into one-cell embryos along with Nrf 2 mRNA. The induction of GFP expression by Nrf 2 was maintained despite deleting the promoter to a region only −122 bp upstream of the transcription initiation site (Figure 6A, 0.12 k). This result indicated that the target site for Nrf 2 is located within a 122-bp region. Examination of this region identified an ARE-like sequence (TGACTCATC), located between basepairs −53 and −45, which contains a single mismatch from the core ARE (TGAC/GnnnGC) [43]. To determine whether this ARE-like sequence is essential for the transactivation, we introduced point mutations into the ARE-like sequence of the 0.12gstp1GFP construct (Figure 6B). The mutation eliminated GFP induction activity (0.12gstp1m1), suggesting that the ARE-like sequence is essential for induction by Nrf 2 (Figure 6A).
Figure 6. Difference in GFP induction by Nrf 2 among various gstp1 constructs.
(A) Transcriptional activity of deletion and mutation constructs. GFP expression was analysed in 8-h embryos that were injected with 40 pg of each construct and 40 pg of Nrf 2 mRNA. More than 30 embryos were examined for each construct. (B) Structure of the proximal ARE-like sequence in the gstp1 promoter and its mutation construct. Numbers indicate nucleotides from the transcriptional initiation site.
Other than the ARE-like sequence in the promoter region, five sequences containing the core ARE can be identified in the remaining 3.5-kbp region as shown in Figure 6(A). It is possible that some of these core ARE regions can substitute for the activity of the proximal ARE-like sequence. To characterize the promoter activity of these additional ARE motifs, we generated the construct 3.5gstp1GFPm1, a derivative of 3.5gstp1GFP with point mutations in the proximal ARE-like sequence. The mutation completely eliminated the GFP induction activity of Nrf 2, confirming that the proximal ARE-like sequence, but not other ARE motifs, in the 3.5-kbp regions is required for Nrf 2-dependent activation (Figure 6A).
Nrf 2 can bind the proximal ARE-like sequence
To investigate whether Nrf 2 has the capability of binding the proximal ARE-like sequence, we carried out EMSA experiments using an oligonucleotide containing the region −68 to −40 of gstp1 gene as a probe (Figures 6 and 7). To bind DNA, it is necessary for Nrf 2 to form a heterodimeric complex with the small Maf family proteins [5]. Therefore, we prepared zebrafish Nrf 2 and MafK proteins using the wheat germ in vitro translation system. MafK is one of four zebrafish small Maf proteins that are known to heterodimerize with Nrf 2, and thereby activate its DNA binding and transactivation activity [12]. As shown in Figure 7, a binding complex was observed when we added both Nrf 2 and MafK to the reaction mixture (lane 4, arrowhead), but not when the proteins were supplied individually (lanes 2 and 3). Since addition of anti-Nrf 2 or anti-MafK antibodies resulted in a gel super-shift or reduction of the binding complex (Figure 7, lanes 5 and 6), we concluded that the complex is an Nrf 2–MafK heterodimer.
Figure 7. Binding of Nrf 2–MafK heterodimers to the ARE-like sequence.
In vitro-translated Nrf 2 and MafK proteins were incubated with the 32P-labelled oligonucleotide probe containing the ARE-like sequence in gstp1. Arrowheads indicate the positions of shifted bands containing the Nrf 2–MafK heterodimer, which were super-shifted (arrow) or reduced with the addition of anti-Nrf 2 (lane 5) or anti-MafK (lane 6) antibodies (Ab). Shifted complexes, including Nrf 2–MafK heterodimers, were specifically competed by the addition of a 25- or 100-fold molar excess of unlabelled oligonucleotide (lanes 8 and 9). Mutations (Mut) in the ARE-like sequence, as shown in Figure 6(B), eliminated the competition (lanes 10 and 11).
To verify the importance of the ARE-like sequence for the binding of the Nrf 2–MafK complex to the DNA probe, competition assays were performed. Unlabelled oligonucleotides containing the wild-type ARE-like sequence or its mutation were added to the reaction mixture. As shown in Figure 7 (lanes 7–11) wild-type, but not mutant, competitor reduced the formation of the shifted complex by the Nrf 2–MafK heterodimer. These results indicated that Nrf 2 specifically binds to the proximal ARE-like sequence.
DISCUSSION
Nrf 2 targets the proximal ARE-like sequence
In this study, we have demonstrated that Nrf 2 binds directly to a proximal ARE-like sequence located −50 bp upstream of the transcription initiation site of the zebrafish gstp1 gene, and that Nrf 2 activates gene expression through this element in zebrafish embryos. The ARE-like sequence harbours a ‘TC’ in place of a ‘GC’ motif previously reported to form the core ARE sequence (TGAC/GnnnGC). Though the GC dinucleotide is an important motif in the ARE for the interaction to the extended homology region of small Maf proteins [44], we speculate that a TC sequence is functionally equivalent to GC for the following reasons: (i) Nrf 2–MafK heterodimer can specifically bind the proximal ARE-like sequence (Figure 7); (ii) Kataoka et al. [45] have shown that chicken Nrf 2 has the ability to bind the GATGACTCATC which has the sequence TC instead of GC; and (3) Nioi et al. [46] mutated every nucleotide across the ARE of the mouse NQO1 [NAD(P)H:quinone oxidoreductase] gene and produced clear evidence that mutation of 5′-TGAGTCGGC-3′ to 5′-TGAGTCGTC-3′ did not result in loss of ARE-driven induction by sulphoraphane or transactivation by mouse Nrf 2.
In the upstream region of gstp1, in addition to the proximal ARE-like sequence, five distinct core ARE sequence can be identified. Reporter analysis showed that these distal AREs are dispensable for the transactivation, even though they all comprise GC motifs. This raises the question of what differences exist between the proximal ARE-like sequence and the distal AREs. The Nrf 2–MafK heterodimer may prefer to bind the proximal ARE-like sequence. It is important to remember that AREs are also the target sites for small Maf homodimers, which are known to negatively regulate transcription [47,48]. To distinguish heterodimer- and homodimer-favouring ARE sequences, we recently developed a new method to predict binding sequences for Nrf 2–small Maf heterodimer using surface plasmon resonance imaging technique (T. Yamamoto, H. Motohashi and M. Yamamoto, unpublished work, and [49]). According to this prediction, the proximal ARE-like sequence was classified into preferable ARE sequences for the Nrf 2–small Maf heterodimer. Indeed, it is of note that zebrafish MafK homodimers could not bind to a probe containing the proximal ARE-like sequence (Figure 7, lane 2). An alternative hypothesis is that an increase of distance between ARE sequences and the core promoter region can reduce the transactivation ability of Nrf 2. Numerous studies have mapped a functional ARE to within 700 bp of the transcriptional initiation site. An ARE located far from the core promoter may be inefficient, although some functional AREs have been found to exist more than 1 kbp upstream of the transcriptional initiation sites.
A proximal ARE-like sequence is conserved among vertebrates
The positions and sequence of the proximal ARE-like motif are conserved among vertebrate Pi class GST genes (Figure 8) [40,41,50], all being located immediately upstream of the TATA box. Since amino acid sequences of the basic DNA-binding domains (RDIRRRGKNKVAAQNCRKRK) [5] are completely identical between zebrafish Nrf 2 and its vertebrate orthologues, it is reasonable to assume all the vertebrate Nrf 2 proteins will bind the conserved ARE-like sequence. In mice, Nrf 2 was demonstrated to target this proximal ARE-like sequence and thereby activate gene expression [25]. Taken together, these previous reports and our present data suggest that the proximal ARE-like sequence is a conserved target site for Nrf 2 among vertebrates. It has been reported that in case of the rat GSTP1, the proximal ARE-like sequence does not function as an Nrf 2 target site [26]. We suppose that this discrepancy arises from the existence of a GPEI in the rat GSTP1, which may have a much higher affinity for Nrf 2 in comparison with the proximal ARE-like sequence. Indeed, Kawamoto et al. [24] have demonstrated that a response element to Phase 2 detoxification inducers is localized in a region between −140 and +59 bp of the rat GSTP1 which contains the proximal ARE-like sequence but not GPEI, suggesting that GPEI is not the only target site for Nrf 2 in the rat gene. We could not find GPEI-related regions in the zebrafish Pi class GST genes, and consider that the proximal ARE-like sequence is the major target sites for Nrf 2 in zebrafish.
Figure 8. Alignment of various Pi class GST promoters.
ARE-like sequences and the TATA box are highlighted in grey. Numbers indicate nucleotides from the transcriptional initiation site.
Interestingly, the TRE [PMA (‘TPA’)-responsive element] sequence (TGAG/CTCA) embedded in the proximal ARE-like sequence is completely conserved among vertebrate Pi class GST genes, implicating the Jun and Fos family of transcription factors in the universal regulation of Pi class GSTs. Indeed, members of the Jun and Fos family have been shown to bind this conserved TRE sequence [51,52]. In many human cell lines, however, the TRE sequence in human GSTP1 is unresponsive both to phorbol esters and to forced expression of Jun and Fos proteins [53,54]. This is a similar situation in zebrafish larvae in which PMA treatment did not induce expression of gstp1 or gstp2 (Y. Takagi, M. Kobayashi and M. Yamamoto, unpublished work). Of particular note, Borde-Chiché et al. [52] have demonstrated PMA-induced GSTP1 expression in human K562 leukaemia cells and importance of the conserved TRE sequence in this induction. Relationship between Jun/Fos proteins and Nrf 2 on Pi class GST regulation should be elucidated in future.
Identification of gstp1 and gstp2
We isolated two zebrafish Pi class GST genes, gstp1 and gstp2, and revealed that they are arranged in tandem in the chromosome. Their close proximity and high degree of similarity suggest that they arose as a consequence of a recent gene duplication event. Intriguingly, two mouse genes of Pi class GST also exist whose protein products differ by only 6 amino acids. However, no such duplication of Pi class GST genes has been observed in rat [41]. Similarly, identification of multiple forms of Pi class GST proteins in crab-eating macaque (Macaca fascicularis) [55] suggested that some monkeys harbour two Pi class GST genes, unlike humans which have a single gene [50]. Since all previously identified Pi class GST genes in fish seems to be single genes, we suggest that the duplications of Pi class GST genes of zebrafish, mouse and monkey occurred independently. Phylogenetic analysis of vertebrate Pi class GST proteins (Figure 1A) supports this idea. The homology between salmon Pi class GST [32] and zebrafish GSTP1 and GSTP2 proteins (68% and 62% respectively) is lower than that between the zebrafish GSTP1 and GSTP2 proteins (87%). The result indicates that gene amplification of zebrafish Pi class GST occurred after divergence of Euteleostei. Furthermore, we found a Pi class GST gene from catfish in the EST database (DDBJ, EMBL and GenBank® accession number CK405822) whose homology with zebrafish GSTP1 or GSTP2 was lower than 87% (71% and 65% respectively). Catfish belongs to Ostariophysi, the same superorder as zebrafish, suggesting that gene amplification of gstp1 and gstp2 took place after emergence of cypriniformes.
Acknowledgments
We thank Y. Nakayama for technical assistance in radiation hybrid mapping, T. Arai, M. Doi, M. Eguchi, A. Hayashi, T. Kinoshita, M. Oikawa, Y. Terashita and Y. Wada for help in fish maintenance, and T. Yamamoto, H. Motohashi, F. Katsuoka, K. Itoh, K. Nishikawa for help and discussion. We also thank V. P. Kelly for critical reading of the manuscript. This work was supported by Grants-in-Aid from the Japan Science and Technology Corporation (ERATO), the Japan Society for Promotion of Sciences (JSPS-RFTF), and the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.Itoh K., Tong K. I., Yamamoto M. Molecular mechanism activating Nrf 2–Keap1 pathway in regulation of adaptive response to electrophiles. Free Radical Biol. Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi M., Yamamoto M. Molecular mechanisms activating the Nrf 2–Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 3.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf 2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 4.Wasserman W. W., Fahl W. E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh K., Igarashi K., Hayashi N., Nishizawa M., Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motohashi H., Katsuoka F., Engel J. D., Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1–Nrf 2 regulatory pathway. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf 2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D. R., et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf 2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 9.Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf 2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf 2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf 2–Keap1 system. Genes Cells. 2002;7:807–20. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 12.Takagi Y., Kobayashi M., Li L., Suzuki T., Nishikawa K., Yamamoto M. MafT, a new member of the small Maf protein family in zebrafish. Biochem. Biophys. Res. Commun. 2004;320:62–69. doi: 10.1016/j.bbrc.2004.05.131. [DOI] [PubMed] [Google Scholar]
- 13.Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 14.Board P. G., Coggan M., Chelvanayagam G., Easteal S., Jermiin L. S., Schulte G. K., Danley D. E., Hoth L. R., Griffor M. C., Kamath A. V., et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J. Biol. Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 15.Hayes J. D., Flanagan J. U., Jowsey I. R. Glutathione transferases. Annu. Rev. Phamacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 16.Robertson I. G. C., Guthenberg C., Mannervik B., Jernström B. Differences in stereoselectivity and catalytic efficiency of three human glutathione transferases in the conjugation of glutathione with 7β,8α-dihydroxy-9α,10α-oxy-7,8,9,10-tetrahydrobenzo(a)pyrene. Cancer Res. 1986;46:2220–2224. [PubMed] [Google Scholar]
- 17.Berhane K., Widersten M., Engstrom A., Kozarich J. W., Mannervik B. Detoxication of base propenals and other α,β-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1480–1484. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakae D., Denda A., Kobayashi Y., Akai H., Kishida H., Tsujiuchi T., Konishi Y., Suzuki T., Muramatsu M. Inhibition of early-phase exogenous and endogenous liver carcinogenesis in transgenic rats harboring a rat glutathione S-transferase placental form gene. Jpn. J. Cancer Res. 1998;89:1118–1125. doi: 10.1111/j.1349-7006.1998.tb00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson C. J., Smith A. G., Ure J., Brown K., Bacon E. J., Wolf C. R. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5275–5280. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harries L. W., Stubbins M. J., Forman D., Howard G. C., Wolf C. R. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis. 1997;18:641–644. doi: 10.1093/carcin/18.4.641. [DOI] [PubMed] [Google Scholar]
- 21.Chanas S. A., Jiang Q., McMahon M., McWalter G. K., McLellan L. I., Elcombe C. R., Henderson C. J., Wolf C. R., Moffat G. J., Itoh K., et al. Loss of the Nrf 2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimitsu Y., Nakagawa Y., Hayashi K., Fujii H., Kumagai T., Nakamura Y., Osawa T., Horio F., Itoh K., Iida K., et al. A sulforaphane analogue that potently activates the Nrf 2-dependent detoxification pathway. J. Biol. Chem. 2002;277:3456–3463. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y., Kumagai T., Yoshida C., Naito Y., Miyamoto M., Ohigashi H., Osawa T., Uchida K. Pivotal role of electrophilicity in glutathione S-transferase induction by tert-butylhydroquinone. Biochemistry. 2003;42:4300–4309. doi: 10.1021/bi0340090. [DOI] [PubMed] [Google Scholar]
- 24.Kawamoto Y., Nakamura Y., Naito Y., Torii Y., Kumagai T., Osawa T., Ohigashi H., Satoh K., Imagawa M., Uchida K. Cyclopentenone prostaglandins as potential inducers of phase II detoxification enzymes. 15-deoxy-Δ(12,14)-prostaglandin J2-induced expression of glutathione S-transferases. J. Biol. Chem. 2000;275:11291–11299. doi: 10.1074/jbc.275.15.11291. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda H., Serria M. S., Kakizaki I., Hatayama I., Satoh K., Tsuchida S., Muramatsu M., Nishi S., Sakai M. Activation of mouse Pi-class glutathione S-transferase gene by Nrf 2 (NF-E2-related factor 2) and androgen. Biochem. J. 2002;364:563–570. doi: 10.1042/BJ20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda H., Nishi S., Sakai M. Transcription factor Nrf 2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem. J. 2004;380:515–521. doi: 10.1042/BJ20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hukriede N. A., Joly L., Tsang M., Miles J., Tellis P., Epstein J. A., Barbazuk W. B., Li F. N., Paw B., Postlethwait J. H., et al. Radiation hybrid mapping of the zebrafish genome. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi M., Nishikawa K., Yamamoto M. Hematopoietic regulatory domain of gata1 gene is positively regulated by GATA1 protein in zebrafish embryos. Development. 2001;128:2341–2350. doi: 10.1242/dev.128.12.2341. [DOI] [PubMed] [Google Scholar]
- 29.Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igarashi K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. Conditional expression of the ubiquitous transcription factor MafK induces erythroleukemia cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7445–7449. doi: 10.1073/pnas.92.16.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Luca A., Favaloro B., Carletti E., Sacchetta P., Di Ilio C. A novel amphibian Pi-class glutathione transferase isoenzyme from Xenopus laevis: importance of phenylalanine 111 in the H-site. Biochem. J. 2003;373:539–545. doi: 10.1042/BJ20030261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudo H., Ueda H., Mochida K., Adachi S., Hara A., Nagasawa H., Doi Y., Fujimoto S., Yamauchi K. Salmonid olfactory system-specific protein (N24) exhibits glutathione S-transferase class pi-like structure. J. Neurochem. 1999;72:1344–1352. doi: 10.1046/j.1471-4159.1999.721344.x. [DOI] [PubMed] [Google Scholar]
- 33.Mannervik B. The isoenzymes of glutathione transferase. Adv. Enzymol. 1985;57:357–417. doi: 10.1002/9780470123034.ch5. [DOI] [PubMed] [Google Scholar]
- 34.Dirr H., Reinemer P., Huber R. Refined crystal structure of porcine class Pi glutathione S-transferase (pGST P1-1) at Å resolution. J. Mol. Biol. 1994;243:72–92. doi: 10.1006/jmbi.1994.1631. [DOI] [PubMed] [Google Scholar]
- 35.Ji X., Tordova M., O'Donnell R., Parsons J. F., Hayden J. B., Gilliland G. L., Zimniak P. Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class π glutathione S-transferase. Biochemistry. 1997;36:9690–9702. doi: 10.1021/bi970805s. [DOI] [PubMed] [Google Scholar]
- 36.Board P. G., Webb G. C., Coggan M. Isolation of a cDNA clone and localization of the human glutathione S-transferase 3 genes to chromosome bands 11q13 and 12q13-14. Ann. Hum. Genet. 1989;53:205–213. doi: 10.1111/j.1469-1809.1989.tb01786.x. [DOI] [PubMed] [Google Scholar]
- 37.Ali-Osman F., Akande O., Antoun G., Mao J. X., Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J. Biol. Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 38.Hatayama I., Satoh K., Sato K. Developmental and hormonal regulation of the major form of hepatic glutathione S-transferase in male mice. Biochem. Biophys. Res. Commun. 1986;140:581–588. doi: 10.1016/0006-291x(86)90771-0. [DOI] [PubMed] [Google Scholar]
- 39.Bammler T. K., Smith C. A., Wolf C. R. Isolation and characterization of two mouse Pi-class glutathione S-transferase genes. Biochem. J. 1994;298:385–390. doi: 10.1042/bj2980385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X., Stambrook P. J. Two murine GSTpi genes are arranged in tandem and are differentially expressed. J. Biol. Chem. 1994;269:30268–30273. [PubMed] [Google Scholar]
- 41.Okuda A., Sakai M., Muramatsu M. The structure of the rat glutathione S-transferase P gene and related pseudogenes. J. Biol. Chem. 1987;262:3858–3863. [PubMed] [Google Scholar]
- 42.Cowell I. G., Dixon K. H., Pemble S. E., Ketterer B., Taylor J. B. The structure of the human glutathione S-transferase π gene. Biochem. J. 1988;255:79–83. doi: 10.1042/bj2550079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rushmore T. H., Morton M. R., Pickett C. B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 44.Kusunoki H., Motohashi H., Katsuoka F., Morohashi A., Yamamoto M., Tanaka T. Solution structure of the DNA-binding domain of MafG. Nat. Struct. Biol. 2002;9:252–256. doi: 10.1038/nsb771. [DOI] [PubMed] [Google Scholar]
- 45.Kataoka K., Igarashi K., Itoh K., Fujiwara K. T., Noda M., Yamamoto M., Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. Identification of a novel Nrf 2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 48.Motohashi H., Katsuoka F., Shavit J. A., Engel J. D., Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 49.Kyo M., Yamamoto T., Motohashi H., Kamiya T., Kuroita T., Tanaka T., Engel J. D., Kawakami B., Yamamoto M. Evaluation of MafG interaction with Maf recognition element arrays by surface plasmon resonance imaging technique. Genes Cells. 2004;9:153–164. doi: 10.1111/j.1356-9597.2004.00711.x. [DOI] [PubMed] [Google Scholar]
- 50.Morrow C. S., Cowan K. H., Goldsmith M. E. Structure of the human genomic glutathione S-transferase-π gene. Gene. 1989;75:3–11. doi: 10.1016/0378-1119(89)90377-6. [DOI] [PubMed] [Google Scholar]
- 51.Moffat G. J., McLaren A. W., Wolf C. R. Involvement of Jun and Fos proteins in regulating transcriptional activation of the human Pi class glutathione S-transferase gene in multidrug-resistant MCF7 breast cancer cells. J. Biol. Chem. 1994;269:16397–16402. [PubMed] [Google Scholar]
- 52.Borde-Chiché P., Diederich M., Morceau F., Wellman M., Dicato M. Phorbol ester responsiveness of the glutathione S-transferase P1 gene promoter involves an inducible c-jun binding in human K562 leukemia cells. Leuk. Res. 2001;25:241–247. doi: 10.1016/s0145-2126(00)00118-1. [DOI] [PubMed] [Google Scholar]
- 53.Dixon K. H., Cowell I. G., Xia C. L., Pemble S. E., Ketterer B., Taylor J. B. Control of expression of the human glutathione S-transferase π gene differs from its rat orthologue. Biochem. Biophys. Res. Commun. 1989;163:815–822. doi: 10.1016/0006-291x(89)92295-x. [DOI] [PubMed] [Google Scholar]
- 54.Morrow C. S., Goldsmith M. E., Cowan K. H. Regulation of human glutathione S-transferase π gene transcription: influence of 5′-flanking sequences and trans-activating factors which recognize AP-1-binding sites. Gene. 1990;88:215–225. doi: 10.1016/0378-1119(90)90034-o. [DOI] [PubMed] [Google Scholar]
- 55.Di Ilio C., Angelucci S., Pennelli A., Bucciarelli T., Petruzzelli R., Tiboni G. M., Melino S., Sacchetta P. Purification and characterization of three Pi class glutathione transferase from monkey (Macaca fascicularis) placenta. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1996;114:377–382. doi: 10.1016/0305-0491(96)00067-3. [DOI] [PubMed] [Google Scholar]