Abstract
SCD (stearoyl-CoA desaturase) catalyses the conversion of saturated fatty acids into mono-unsaturated fatty acids, a critical step involved in lipid metabolism and various other biological functions. In the present study, we report the identification and characterization of a human gene that encodes a novel SCD enzyme (hSCD2). The hSCD2 gene codes for a 37.5-kDa protein that shares 61% and 57% sequence identity with the human SCD1 and mouse SCD2 enzymes respectively. The recombinant hSCD2 enzyme expressed in mammalian and Sf9 insect cells efficiently catalysed desaturation of both stearoyl- and palmitoyl-CoAs to the corresponding mono-unsaturated fatty acids. In comparison with the hSCD1 gene that is predominantly expressed in liver, hSCD2 is most abundantly expressed in pancreas and brain. Additionally, hSCD2 transcripts from adult and foetal tissues exhibit different sizes because of alternative splicing in the non-coding region, suggesting that hSCD2 expression is developmentally regulated. The recombinant human SCD2 and SCD1 transiently expressed in COS-7 cells exhibited as oligomeric proteins that consist of homodimers and oligomers when resolved by SDS/PAGE. The complex formation was independent of SCD protein expression levels, as supported by a relatively constant ratio of the level of dimers and oligomers to that of the monomers from COS-7 cells transiently transfected with different amounts of SCD expression vectors. Furthermore, treatment of intact COS-7 cells with a cross-linking reagent resulted in dose-dependent increases in the levels of SCD protein and activity, suggesting that oligomerization may play an important role in regulating the stability of SCD enzymes.
Keywords: cross-linking, desaturation, homodimer, palmitoyl-CoA, stearoyl-CoA desaturase
Abbreviations: DSP, dithiobis(succinimidylpropionate); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SCD, stearoyl-CoA desaturase
INTRODUCTION
SCD (stearoyl-CoA desaturase) is a microsomal fatty acid mono-desaturase, also commonly known as Δ9-desaturase, which catalyses the committed step in the biosynthesis of mono-unsaturated fatty acids from saturated fatty acids [1,2]. This oxidative reaction introduces a cis-double bond to a spectrum of methylene-interrupted fatty acyl-CoAs [2], catalysed by a set of microsomal electron-transport proteins composed sequentially of NADH-cytochrome b5 reductase, cytochrome b5 and the terminal SCD [1]. Mono-unsaturated fatty acids play diverse and crucial roles in living organisms as the major components of membrane phospholipids, triacylglycerols and cholesterol esters. Apart from being components of lipids, mono-unsaturated fatty acids have been implicated as mediators in signal transduction, apoptosis, and differentiation of neurons and other cell types [3,4]. In addition, oleate is the principal form of mono-unsaturated fatty acid in human adipose tissue, the major depot of surplus energy in mammals. Mono-unsaturated fatty acids are also known to regulate food intake in the brain [5] and islet β-cell function [6,7].
A number of SCD genes have been cloned and studied in various species, including yeast [8], Caenorhabditis elegans [9], sheep [10], hamster [11], rat [12], mice [13–16] and humans [17,18]. Four SCD genes (SCD1, SCD2, SCD3 and SCD4) have been identified and characterized in mice [13–16]. The expression of the mouse isoforms varies among tissues. SCD1 and SCD2 are the main isoforms expressed in mouse liver and brain respectively [13,14]. SCD3 is expressed exclusively in skin, whereas SCD4 is expressed predominantly in the heart [15]. In some tissues of mouse, such as adipose and eyelids, multiple SCD isoforms are expressed, and all four of the isoforms are expressed in skin [18,19]. The physiological relevance for having multiple SCD isoform expression in the same tissue remains elusive.
SCD expression and activity are regulated by a complex network of hormonal and physiological events, including dietary lipids, steroids and development [20,21]. SCD1 is a major peripheral target of the signalling events mediated by leptin, a key adipokine that regulates energy homoeostasis by sensing the body's fat reserve in adipose tissues and signalling to the satiety centre in the brain [22]. Dysregulation of SCD has been implicated in non-alcoholic fatty liver disease, hyperlipidaemia and metabolic diseases [23–26]. The involvement of SCD in regulating lipid metabolism has recently been corroborated in mice with targeted deletion of SCD1 gene and in asebia mice that carry natural mutations in the SCD1 gene. Mice deficient in the SCD1 gene exhibit reduced body adiposity, increased insulin and leptin sensitivity, and are resistant to diet-induced obesity, as a result of increased lipid oxidation [22,27,28]. Perhaps one of the most intriguing pathophysiological events associated with SCD1 deficiency is the development of skin abnormality as a result of reduced level of oleate [29], which is not rescued by dietary supplementation of mono-unsaturated fatty acids [29], or by compensation by other SCD isoforms, even though all of the four isoforms are expressed in skin. Because of its involvement in regulating energy metabolism, SCD1 has been suggested to be a potential drug target for human obesity [30].
Despite their involvement in regulating lipid metabolism and diseases, hSCD1 has been the only human SCD gene identified and characterized to date [13,14,17], and this has hindered efforts in investigation of the tissue distribution and physiological functions of different human SCD isoforms. The existence of additional human SCD genes has been suggested, but never confirmed. In the present study, we identified and characterized a new member of the human SCD gene family, which is designated as hSCD2, based on its tissue distribution and sequence homology with the mouse SCD2 gene. We also demonstrated that both human SCD2 and SCD1 were oligomeric proteins in intact cells. Furthermore, the level of protein and activity of SCD enzymes were significantly enhanced by oligomerization, suggesting a novel regulatory step for SCD activity.
MATERIALS AND METHODS
Cloning of the full-length hSCD1 and hSCD2 cDNAs
The human SCD2 gene was identified in GenBank® (accession number AF389338) based on its sequence identity with the human SCD1 and multiple isoforms of mouse SCD genes. A full-length cDNA sequence was cloned by PCR amplification using primer pairs designed from the 5′- and 3′-flanking sequences of the predicted open reading frame of the hSCD2 cDNA (forward, 5′-CAAGATCTGCATCCCTAGG-3′; reverse, 5′-AACGGCAGACATGTGGGATGGCTGT-3′) and Marathon-Ready cDNAs prepared from human foetal brain (BD Biosciences Clontech, Palo Alto, CA, U.S.A.). Amplification was performed by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA, U.S.A.) and a programme of 35 cycles of 94 °C for 30 s, 62 °C for 30 s and 72 °C for 2 min. The same PCR amplification condition was also used to amplify the human SCD1 cDNA using Marathon-Ready cDNA from human liver and primer pairs designed from the 5′- and 3′-untranslated sequences of the human SCD1 gene (forward, 5′-GTTGCCAGCTCTAGCCTTTA-3′; reverse, 5′-CAGCTCAAGGAAAGAGAGTT-3′). The PCR products of hSCD1 and hSCD2, 1.6- and 1.2-kb respectively, were each subcloned into the pPCR-script Amp SK(+) vector (Stratagene) and sequenced.
Northern blot analysis
To analyse the expression level and the tissue distribution pattern of the human SCD1 and SCD2 mRNAs, multiple tissue polyadenylated RNA Northern blots (BD BioSciences Clontech) were hybridized with [α-32P]dCTP (3000 Ci/mmol; ICN Radiochemicals, Irvine, CA, U.S.A.) labelled probes prepared from the full-length cDNA of hSCD1 and hSCD2 respectively, using a random primer DNA-labelling system (Invitrogen, Carlsbad, CA, U.S.A.). Hybridization was carried out in ULTRAhyb (Ambion, Austin, TX, U.S.A.) at 50 °C overnight, followed by one wash at 50 °C in 0.1×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) and 0.1% (w/v) SDS, and two washes in 2×SSC buffer containing 0.1% (w/v) SDS and 1 mM EDTA. Blots were stripped with boiling 1% (w/v) SDS to remove radiolabelled probes, and re-hybridized with radiolabelled β-actin probe as an internal control. The blots were exposed to Bio-Max MR film or a PhosphorImager screen to visualize the signals, and quantified using ImageQuant (Amersham Biosciences). The results were presented as relative activity after normalization using the expression level of β-actin as the internal control of mRNA loading.
Expression of hSCD2 and hSCD1 in mammalian cells
Mammalian expression vectors for hSCD1 and hSCD2 cDNAs were engineered by unidirectional subcloning from the pPCR-script Amp SK(+) vector into pcDNA3.1(+)/Hygro mammalian expression vector (Invitrogen) at the EcoRV and NotI sites for hSCD2 cDNA, or KpnI and NotI sites for hSCD1 cDNA. For transient expression in mammalian cells, COS-7 cells were maintained under the conditions recommended by the American Tissue Culture Collection (A.T.C.C., Manassas, VA, U.S.A.). At 1 day before transfection, 2×106 cells were subcultured on to a 100 mm×20 mm plate, resulting in approx. 70% confluence. Cells were transfected with 10 μg of DNA, premixed with FuGENE 6 (Roche Molecular Biochemicals, Indianapolis, IN, U.S.A.) according to the manufacturer's instructions. At 48 h after the transfection, cells were harvested in ice-cold PBS, pelleted by centrifugation at 2000 g for 10 min, lysed and assayed immediately, or frozen in liquid nitrogen for later use.
Expression of hSCD2 and hSCD1 in insect cells
Expression of the human SCD enzymes in Sf9 insect cells was performed by using a Bac-to-Bac Baculovirus Expression System (Invitrogen). The full-length cDNA of hSCD2 and hSCD1 were each subcloned from the pPCR-script Amp SK(+) vector described above into the SalI and NotI sites of the pFastBac vector (Invitrogen), which was subsequently transformed into DH10Bac™ Escherichia coli cells to generate a recombinant bacmid containing each of the target genes. High-titre recombinant baculoviruses were generated by transfecting the bacmid DNA into Sf9 insect cells with several rounds of amplification. After 65 h of infection with the recombinant baculoviruses, Sf9 insect cells were harvested in ice-cold PBS, pelleted by centrifugation at 2000 g for 10 min, homogenized and assayed immediately for enzyme activity, or frozen in liquid nitrogen for later use. The protein concentration in homogenates was determined using a BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, U.S.A.), according to the manufacturers' instructions.
In vitro assays for desaturase activity
Desaturase activities were determined by conversion of radiolabelled saturated fatty acyl-CoAs into the corresponding mono-unsaturated fatty acids, and resolved by TLC. The fatty acyl-CoAs substrates used in the enzyme assays included palmitoyl-CoA (C16:0) and stearoyl-CoA (C18:0). Cell pellets were homogenized in 100 mM potassium phosphate buffer (pH 7.2) supplemented with 1×Complete™ protease inhibitors (Roche Molecular Biochemicals) for 15 s using a Brinkman polytron homogenizer. Protein homogenates from Sf9 cells infected with recombinant baculoviruses expressing the SCD cDNAs, or from COS-7 cells transfected with the SCD expression vectors, were used as the sources of SCD enzymes. Desaturase activities were determined at room temperature (21 °C) for 5 min in a final volume of 200 μl of reaction mixture that contained 100 μg of cell homogenate, 2 μM NADH and 6 μM of 14C-labelled fatty acyl-CoA (50 mCi/mmol). The reaction was terminated by addition of 200 μl of 2.5 M KOH in 75% ethanol, and incubated at 85 °C for 1 h to saponify the sample. After being cooled down to room temperature and acidified with 280 μl of methanoic (formic) acid, the lipids were extracted with 700 μl of hexane and separated on a 10% AgNO3-impregnated TLC plate with chloroform/methanol/ethanoic (acetic) acid/water (90:8:1:0.8). The TLC plate was exposed to an X-ray film or a PhosphorImager screen to visualize and quantify the incorporation of 14C into the enzymatic products.
In vivo cross-linking analysis of human SCD1 and SCD2
To facilitate detection of the recombinant hSCD1 and hSCD2 enzymes by Western blot analysis, FLAG-tagged versions of hSCD1 and hSCD2 were each engineered by PCR amplification using the full-length cDNAs as templates and anchored primers (forward, 5′-TGCAAAGCTTGCCACCATGGATTACAAGGATGACGACGATAAGATCCCGGCCCACTTGCTGCAGGACGATATC-3′ and reverse, 5′-TCGAGCGGCCGCTCAGCCACTCTTGTAGTTTC-3′ for hSCD1; and forward, 5′-GCTTGGCGCGCCGGCCACCATGGATTACAAGGATGACGACGATAAGATCCCAGGCCCGGCCACCGACGCG-3′ and reverse, 5′-TCGAGCGGCCGCTCAAGCACTGCTGTCTCCAGTCCT-3′ for hSCD2) to attach the FLAG sequence in-frame to the 5′-coding region of each SCD cDNA. Amplified cDNA fragments were sub-cloned into the HindIII and NotI sites or AscI and NotI sites of mammalian expression vector XenoFLIS for FLAG–hSCD1 and FLAG–hSCD2 respectively. COS-7 cells were transiently transfected with the expression vectors to express the recombinant FLAG-tagged hSCD1 and hSCD2 proteins. At 40 h post-transfection, cells were cultured with fresh medium for 1 h, and treated with 0, 0.5 and 1.0 mM respectively DSP [dithiobis(succinimidylpropionate)], dissolved in DMSO (Pierce), for 30 min at 37 °C. The treated cells were quenched with 1 mM Tris/HCl, pH 7.5, for 30 min at room temperature, washed with PBS and pelleted by centrifugation at 2000 g for 10 min. The cell pellets were immediately processed for Western blot analysis or frozen at −80 °C for later use.
Western blot analysis
The cell pellets were thawed on ice and resuspended in 50 μl of Bug Buster HT buffer (Novagen, Madison, WI, U.S.A.), supplemented with 1×Complete™ protease inhibitors. The cell suspension was incubated at room temperature for 10 min, followed by addition of 5 μl of 10% (w/v) SDS and incubation at room temperature for 10 min. The cell lysate was centrifuged at 14000 g for 6 min at 4 °C to remove nucleus and cell debris. The supernatant was used to detect the expression of SCD enzymes by Western blot analyses as previously described [31], using anti-FLAG or anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibodies.
RESULTS
Identification and cloning of hSCD2 cDNA
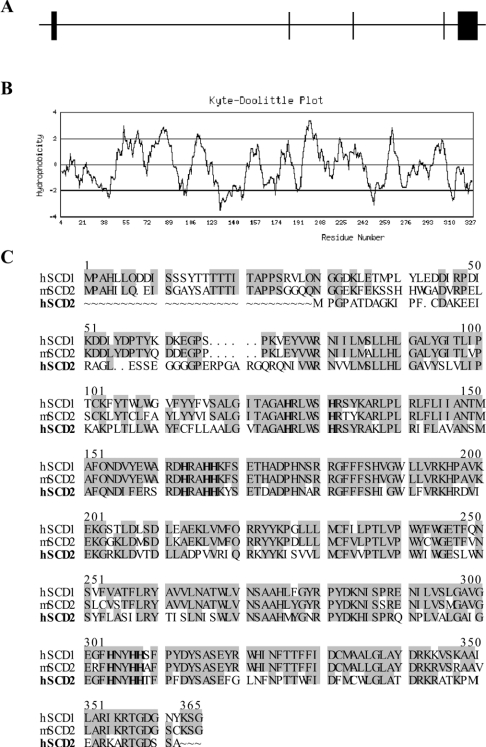
A human cDNA clone (GenBank® accession number AF389338) was identified the GenBank® databases based on its sequence homology with the human and mouse SCD genes, and was designated hSCD2. A 1.2-kb cDNA fragment encoding a putative hSCD2 enzyme was cloned by PCR amplification using a cDNA library from human foetal brain and primers designed from the 5′- and 3′-untranslated regions of the predicted human hSCD2 open reading frame. Sequence analysis of the 1.2-kb cDNA fragment suggested a full-length coding region for the enzyme as evidenced by an in-frame stop codon located 171 bp upstream of the predicted AUG start codon. The full-length hSCD2 encodes a 37.7 kDa protein that shares 61% and 57% sequence identity with the human SCD1 and the mouse SCD2 respectively (Figure 1C). The human SCD2 also shares close similarity in hydrophobicity profile (Figure 1B) with the mouse SCD2 enzyme, and possesses all the three histidine rich motifs, HRLWSH, HRAHH and HNYHH, highly conserved among SCD proteins (Figure 1C) [15,16]. The human SCD2 gene is organized in five exons (Figure 1A) that span a 15-kb region localized on chromosome 4 at 4q21.3 adjacent to SEC31L1 that encodes an endoplasmic-reticulum-associated protein required for vesicle budding from endoplasmic reticulum [32].
Figure 1. Genomic structure and predicted peptide sequences of the hSCD2 gene.
(A) The genomic structure of the hSCD2 gene. (B) The predicted hydrophobicity profile of the hSCD2 protein. (C) Homology between hSCD2 with the hSCD1 and the mouse SCD2 (mSCD2) protein sequences. The shaded residues represent identical sequences. The conserved histidine residues essential for SCD activity are highlighted in bold.
Distinct tissue distribution of hSCD2 mRNAs from hSCD1
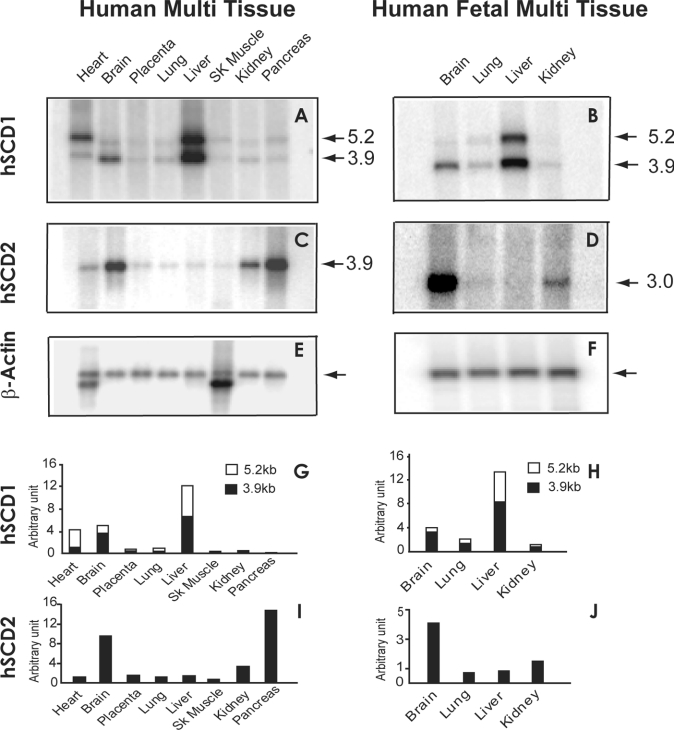
The hSCD2 expression and tissue distribution were analysed by Northern blotting using a radiolabelled cDNA probe, and compared with those of hSCD1. We also compared hSCD2 with hSCD1 expression in human foetal tissues, since oleate is believed to play a role in foetal development. As a result, a major transcript of 3.9 and 3.0 kb of the hSCD2 gene was detected in most adult and foetal human tissues respectively (Figures 2C and 2D). The differences in transcript size were caused by alternative splicing of the non-coding region during development, which was confirmed by PCR amplification and sequence comparison of the coding region of hSCD2 cDNAs from foetal and adult brains. In contrast with the hSCD1 gene that is predominantly expressed in human livers (Figures 2A and 2B, quantified in Figures 2G and 2H), hSCD2 is most abundantly expressed in pancreas and brain, as well as in human foetal brain (Figures 2C and 2D, quantified in Figures 2I and 2J), when normalized to the level of β-actin (Figures 2E and 2F), an internal control for mRNA loading.
Figure 2. Comparison of tissue distribution profiles between human SCD2 and SCD1 in adult as well as foetal tissues by Northern blot analyses.
Human adult and foetal multiple tissue blots containing 2 μg of polyadenylated RNAs were hybridized with radiolabelled probes prepared from cDNAs of the human SCD1 (A and B) and SCD2 genes (C and D) respectively. Sizes (in kb) are indicated by arrows. The hybridized blots were stripped of residual radioactivity, and re-probed with a radiolabelled β-actin cDNA probe as an internal control (E and F). The arrows indicate the β-actin mRNA band. The expression level of each gene was quantified by normalization with that of β-actin, and is expressed as arbitrary unit (G–J). (G) and (H) represent quantitative data from blots (A) and (B), whereas (I) and (J) represent quantitative data from blots (C) and (D) respectively. SK Muscle, skeletal muscle.
Analysis of substrate specificity of hSCD2 expressed in mammalian and Sf9 insect cells
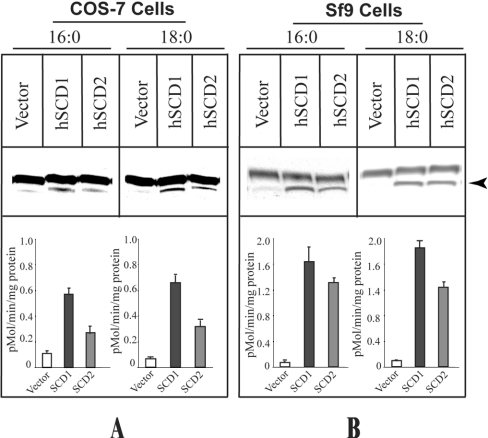
To determine whether the protein encoded by hSCD2 cDNA possesses SCD activities, hSCD2 was subcloned into the mammalian expression vector pcDNA3.1(+)/Hygro and was transiently expressed in COS-7 cells for measurement of Δ9-desaturase activity using the human SCD1 transiently expressed in COS-7 cells as a positive control for the enzyme assay. The experiments were conducted under conditions where 50 μM of each of the 14C-labelled fatty acyl-CoAs was incubated with 2 μM NADH for 5 min at room temperature in the presence of 100 μg of protein lysate as a source of the recombinant hSCD enzymes. The recombinant hSCD2 expressed in COS-7 cells catalysed efficiently the conversion of both palmitoyl-CoA and stearoyl-CoA into the corresponding mono-unsaturated fatty acids (Figure 3, indicated by an arrowhead), as evidenced by a significant increase in SCD activity from COS-7 cells transfected with either the SCD1 or SCD2 expression vector relative to the negative control (COS-7 cells transfected with empty vector). To provide additional information that hSCD2 encodes a Δ9-desaturase, we also engineered a baculovirus expression system to achieve higher expression of hSCD2 enzymes for the assay. As shown in Figure 3(B), the recombinant hSCD2 and hSCD1, expressed in Sf9 cells, exhi-bited strong SCD activities with both palmitoyl- and stearoyl-CoAs relative to that observed in Sf9 cells infected with the wild-type baculovirus, thus confirming further that the hSCD2 gene encodes an SCD enzyme.
Figure 3. Enzymatic analyses of the recombinant human SCD2 expressed in COS-7 and Sf9 cells.
(A) Analysis of SCD activity from the recombinant human SCD1 and SCD2 enzymes transiently expressed in COS-7 cells. COS-7 cells transiently transfected with either the hSCD1 expression vector, the hSCD2 expression vector, or empty vector were lysed with cell-harvesting buffer containing multiple protease inhibitors (1×Complete™) after 48 h of transfection, and were processed for SCD enzyme assays. (B) Analysis of SCD activity from the recombinant human SCD1 and SCD2 enzymes expressed in Sf9 insect cells. Sf9 cells infected with the hSCD1 or the hSCD2 recombinant baculovirus, as well as wild-type baculovirus were harvested, after 72 h of infection, in the same harvesting buffer used for COS-7 cells, and processed for SCD activity assays. Desaturase activities were determined by conversion of radiolabelled palmitoyl-CoA (C16:0) and stearoyl-CoA (C18:0) into the corresponding mono-unsaturated fatty acids (indicated by an arrowhead). The SCD enzyme activities were quantified by PhosphorImaging from three independent experiments, and are expressed as pmol/min per mg of protein.
Analysis of subunit composition of SCD1 and SCD1 enzymes, and the effects of SCD expression levels on complex formation
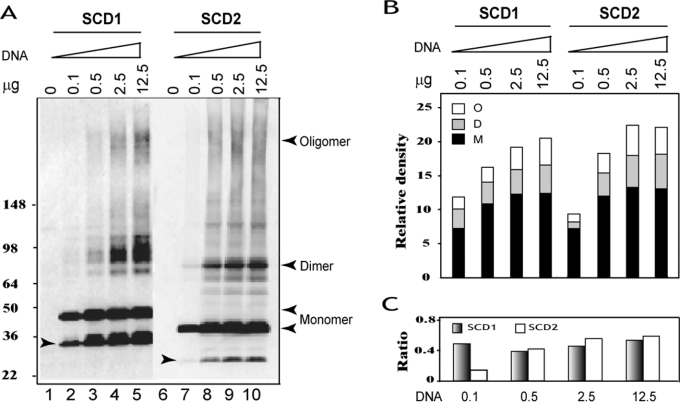
To facilitate detection of hSCD proteins expressed in mammalian cells by Western blot analysis, we engineered mammalian expression vectors by attaching a FLAG sequence in-frame to the 5′-coding region of each of the human SCD cDNAs. The attachment did not affect SCD enzyme activity, since both SCD enzymes catalysed efficiently the conversion of stearoyl-CoA into oleoyl-CoA (see below). Using the FLAG-tagged version of the human SCD1 and SCD2, we analysed the subunit composition of the SCD enzymes and the effect of SCD expression levels on SCD complex formation by transfecting COS-7 cells with different amounts of the expression vectors. As shown in Figure 4(A) (quantified in Figure 4B), the recombinant hSCD1 and hSCD2 enzymes expressed in COS-7 cells exhibited mainly as 42- and 38-kDa proteins respectively, when resolved by SDS/PAGE (Figure 4A, lanes 2 and 7, indicated by arrowheads), which is consistent with the predicted molecular mass of each enzymes. The recombinant SCD proteins expressed in COS-7 cells were not stable, even in the presence of multiple protease inhibitors, as evidenced by the presence of a major 32- and 26-kDa protein product (Figure 4A, indicated by left-hand arrowheads inside the Figure) resulting from protein degradation of the recombinant SCD1 and SCD2 proteins respectively.
Figure 4. Analysis of subunit composition of SCD enzymes and the effects of expression level on SCD complex formation.
COS-7 cells were transiently transfected with increasing amounts of each FLAG-tagged SCD expression vector (0, 0.1, 0.5, 2.5 or 12.5 μg) or empty vector, and were analysed by Western blot using anti-FLAG antibodies. (A) Detection of different SCD moieties by Western blot analyses. The position of different moieties of each enzyme is indicated by left-hand arrowheads, and degraded products by right-hand arrowheads. (B) Quantification of different moieties of hSCD1 and hSCD2, including oligomers (O), dimers (D) and monomers (M) from (A). (C) The ratio of SCD homodimers and oligomers to monomers of each SCD enzyme from data shown in (B).
In addition to the full-length SCD molecules and the degradation products, both SCD1 and SCD2 proteins were also found as homodimers and oligomers when resolved under denaturing conditions, as shown by the presence of protein bands that coincide with the sizes of homodimers and oligomers of each SCD enzyme (Figure 4A, indicated with right-hand arrowheads). In comparison with SCD homodimers, the SCD oligomers were less stable, as evidenced by poor resolution of the high-molecular-mass SCD complexes under denaturing conditions. The formation of homodimers and oligomers was independent of SCD protein expression levels, since the complexes were also detected in COS-7 cells transfected with a very low level of vector DNA (0.1 μg) (Figure 4B), with which the presence of homodimers and oligomers were below detection limit by Western blot analysis. The relative abundance of all SCD moieties, including degradation products, increased proportionally with SCD expression levels. When normalized with the level of monomers, the ratio of homodimers and oligomers to monomers of SCD1 protein remained constant with increasing levels of SCD1 protein expression (Figure 4C). In contrast, the relative abundance of SCD2 complexes increased proportionally with SCD2 expression levels (Figure 4C), possibly reflecting the fact that SCD2 proteins were more stable, as shown by the low abundance of the 26-kDa degradation product. The natural SCD complexes were not totally disrupted when resolved by denaturing SDS/PAGE under which non-specific protein–protein interactions would have been disrupted, suggesting that the formation of homodimers and oligomers is an intrinsic property of SCD proteins.
Increased protein stability and enzyme activity of human SCD1 and SCD2 by oligomerization in intact COS-7 cells
To investigate SCD subunit composition in intact cells, we next performed cross-linking analysis of the SCD enzymes transiently expressed in COS-7 cells. COS-7 cells transiently transfected with each of the SCD expression vectors or empty vector were treated with increasing doses of DSP, a cross-linking agent with high selectivity and membrane permeability, followed by Western blot analyses. As demonstrated in Figure 5, treatment of COS-7 cells with DSP resulted in dose-dependent increases in the level of homodimers and oligomers relative to the monomers when quantified by PhosphorImaging (Figure 5B). The SCD oligomers showed good resolution as a result of cross-linking which stabilized SCD complexes, in contrast with poor resolution of the natural SCD oligomers shown in Figure 4(A). However, the cross-linking treatment also resulted in detection of heterogeneous SCD1 dimers, as shown by the presence of multiple SCD1 dimer bands. This is unlikely to be caused by overloading of protein samples, as judged by the resolution of GAPDH protein that was used as an internal control to show equal loading of the protein samples. Since SCD1 proteins are unstable and are subject to rapid degradation, the multiple SCD1 dimer bands probably reflect dimerization of the degraded hSCD1 products with the full-length SCD1 molecules or with each other. This is supported by the observation that the SCD2 homodimers are more homogeneous in size, which is consistent with lower levels of 26-kDa degradation products (Figure 5A, indicated by an arrowhead).
Figure 5. Cross-linking analyses of human SCD1 and SCD2 expressed in COS-7 cells.
COS-7 cells were transiently transfected with each of the FLAG-tagged version of hSCD expression vector or empty vector, and were treated with 0, 0.5 and 1.0 mM of DSP respectively for 30 min at 37 °C, followed by Western blot analyses using anti-FLAG antibodies, or enzyme assay using [14C]stearoyl-CoA as a substrate. (A) Detection of different SCD moieties by Western blot analyses. The same blot was stripped and re-probed with anti-GAPDH antibodies to provide an internal control for protein loading. The position of different moieties of each enzyme is indicated by arrows, and degraded product is shown by arrowheads. A sample from mock-transfected COS-7 cells was used as a negative control (C). (B) Quantification of different moieties of hSCD1 and hSCD2, including oligomers (O), dimers (D) and monomers (M) from (A). (C) Analysis of SCD activity of the recombinant SCD enzymes used for Western blot analysis shown in (A). Data were collected from at least three independent experiments, and were expressed as means±S.E.M. Student's t test was performed to analyse differences in enzymatic activity between the DSP-treated and the non-cross-linked samples of each SCD enzymes, and significant differences (P<0.05) are indicated with an asterisk (*).
Unexpectedly, the level of SCD monomers was not significantly reduced as a result of cross-linking, even at high doses of DSP, in contrast with what is often observed from cross-linking experiments [33,34]. Instead, treatment of COS-7 cells with DSP dose-dependently increased the level of all the SCD moieties, including the degradation products (Figure 5A compare lane 2 with lanes 3 and 4, and lane 5 with lanes 6 and 7). This is further confirmed by quantitative analysis of each moieties as shown in Figure 5(B). Consistent with the increased levels of the total SCD proteins, treatment with DSP significantly (P<0.05) increased the SCD enzyme activities when tested with [14C]stearoyl-CoA as a substrate (Figure 5C, compare lane 2 with lanes 3 and 4, and lane 5 with lanes 6 and 7), which also demonstrated dose-dependency with the level of DSP used in the cross-linking experiments. The increases in SCD expression and enzyme activity were not caused by changes in global protein expression in COS-7 cells as a result of DSP treatment, since the treatment did not alter the level of GAPDH expression (Figure 5A, lower panel), suggesting that dimerization and oligomerization may play an important role in maintaining the stability of the SCD enzymes.
DISCUSSION
In comparison with four SCD genes identified in mice [13–16], only one SCD gene has been identified and characterized in humans thus far, and the existence of additional human SCD genes has not been confirmed. In the present study, we identified a novel human SCD gene, designated hSCD2. The hSCD2 cDNA encodes a 37.5-kDa protein that shares 61% and 57% sequence identity with the human SCD1 and murine SCD2 enzymes respectively. The human SCD2 contains all three of the histidine-rich motifs which are highly conserved among all the mammalian SCD enzymes [13,14,16,18], and which are believed to be essential for Δ9-desaturase activity [35]. Consistent with the conserved features and sequence homology with other SCD isoforms, the recombinant hSCD2 enzyme expressed both in mammalian and Sf9 cells efficiently catalysed the desaturation of C18:0 and C16:0 fatty acyl-CoAs to the corresponding mono-unsaturated fatty acids.
In contrast with the hSCD1 gene that is predominantly expressed in liver, hSCD2 is most abundantly expressed in pancreas. The hSCD2 gene shares a close similarity in tissue distribution profile to that of the murine SCD2 among all the four murine SCD genes. The mouse SCD2 gene was initially identified from 3T3-L1 adipocytes, and its mRNA level was induced more than 10-fold during 3T3-L1 pre-adipocyte differentiation [13]. Like SCD1, murine SCD2 expression was induced significantly by a fat-free diet, and suppressed by dietary polyunsaturated fatty acids [13,36]. Although the physiological significance of hSCD2 expression in pancreas remains to be elucidated, lipid composition is known to affect insulin secretion both in vivo and in vitro in isolated islets and in cultured pancreatic β-cell lines [37–39]. Thus saturated fatty acids (palmitate and stearate) were shown to be more effective than unsaturated (palmitoleate and linoleate) in stimulating insulin secretion from isolated rat islets [38]. Additionally, saturated fatty acids were reported to exacerbate postprandial insulinaemia in obese patients with non-insulin-dependent (Type II) diabetes mellitus [39]. Moreover, lipid composition was also shown to affect pancreatic β-cell proliferation and apoptosis [6,7]. Exposure of islets to saturated fatty acids resulted in a significant increase in β-cell DNA fragmentation, which could be circumvented by the addition of mono-unsaturated palmitoleic acid. In support of a role for SCD2 in regulating islet β-cell function, leptin was shown to suppress SCD2 expression in mice [16] and insulin secretion from the INS-1 β-cell line [40], and to prevent β-cell apoptosis in cultured rat islets [7].
Human SCD2 is also abundantly expressed in adult and foetal brains, and its expression appears to be developmentally regulated, as shown by the different size of transcripts between adult and foetal human tissues. However, such a difference does not involve the coding region of the gene, as confirmed by PCR cloning and sequence analysis of hSCD2 cDNA clones from foetal and adult human brains. Consistent with a role for SCD enzymes in regulating brain function, murine SCD2 gene expression is induced during the neonatal myelinating period [4]. Moreover, oleic acid is a major component of sciatic nerves, and is believed to be required for the normal postnatal development of the tissue. A decreased level of SCD2 mRNA and activity during development was associated with the onset of trembler mouse phenotype [4], a mouse model of dysmyelination.
Like SCD1, the recombinant SCD2 expressed in COS-7 cells was unstable and subject to rapid degradation, even in the presence of multiple protease inhibitors, as shown by the presence of a 26-kDa moiety resulting from a major degradation site at the C-terminus. SCD degradation is believed to be a highly conserved process that was also observed in yeast [41], and has been subject to intensive investigation [20,42–46]. In contrast with a major degradation site at the N-terminus, which has been well characterized [20,42–46], a 32-kDa hSCD1 degradation product identified in the present study was caused by a major cleavage site at the C-terminus, thus representing a novel degradation product. It is possible that the two SCD degradation products originated from a common degradation site at the C-terminal of the two enzymes, based on the cleavage profile by a plasminogen-like SCD degradation protein [46]. Accordingly, a predicted cleavage site (KNISPRENI) at position 283 of SCD1, which is highly conserved among SCD enzymes, would generate a 32-kDa and a 26-kDa polypeptide from hSCD1 and hSCD2 respectively. Although the physiological significance of rapid degradation of SCD enzymes remains to be investigated, the short half-life of SCD enzymes may prevent the detrimental effect of excessive accumulation of mono-unsaturated fatty acids on membrane structures [47,48].
The recombinant SCD2 expressed in COS-7 cells displayed a propensity for complex formation, as shown by the presence of natural homodimers and oligomers when resolved by denaturing SDS/PAGE. These complexes are unlikely to be caused by non-specific association of overexpressed SCD proteins, since non-specific protein–protein interactions are normally disrupted when resolved by denaturing SDS/PAGE. Furthermore, the SCD proteins also formed dimers and oligomers in COS-7 cells expressing very low levels of SCD proteins, and the SCD1 complexes remained at a constant ratio to monomers in COS-7 cells transiently transfected with different amounts of SCD1 expression vectors. Using cross-linking analysis of FLAG-tagged SCD enzymes transiently expressed in COS-7 cells, we demonstrated that both hSCD2 and hSCD1 were oligomeric proteins in intact cells. Treatment with DSP resulted in significant enhancement of dimeric and oligomeric complexes of each hSCD enzymes. The cross-linking also stabilized the SCD oligomers, as shown by good resolution of the high-molecular-mass SCD complexes by SDS/PAGE under the denaturing conditions. Furthermore, the subunit composition of the hSCD1 dimers was also affected by protein degradation that resulted in heterodimerization of truncated SCD polypeptides with each other or with the full-length SCD enzymes, as shown by the presence of multiple hSCD1 dimeric moieties on SDS/PAGE. The presence of multiple SCD1 dimeric moieties was unlikely to be caused by protein overloading, as judged by well-resolved GAPDH protein that was used as an internal control for protein loading. The results suggest that hSCD1 dimerization may not require the integrity of the C-terminus of the SCD1 enzyme, which remains to be confirmed by deletion analyses.
One surprising observation from the cross-linking experiments is the effect of the cross-linking reagent on the level of SCD protein and enzyme activity. Treatment of COS-7 cells with DSP not only increased the detection of dimers and oligomers in a dose-dependent manner, but also increased the total level of SCD proteins, including the monomers and the degradation products, as detected by Western blot analysis. Consistent with the increased level of SCD proteins, the treatment also dose-dependently increased SCD enzyme activity. Such increases were unlikely to have resulted from changes in global protein expression in COS-7 cells from cross-linking reactions, as evidenced by a lack of any significant changes in GAPDH expression before and after the DSP treatment. Additionally, as a homo-bifunctional and amine-reactive cross-linking reagent commonly used to investigate protein–protein interactions, DSP has not been reported to affect the transcription or protein expression of any specific genes. Thus the effect of cross-linking on SCD expression and activity is likely to be due to an improved stability of SCD homodimers and oligomers by DSP. Therefore our current data suggest that dimerization and oligomerization of SCD proteins may play an important role in regulating the half-life of the SCD enzymes, thus representing a novel regulatory mechanism for SCD enzymes, in addition to the transcriptional and post-translational regulations previously reported [20,24].
Acknowledgments
We thank Dr Jian Wang for technical assistance and Dr Eric Su for help with bioinformatics.
References
- 1.Ntambi J. M. The regulation of stearoyl-CoA desaturase (SCD) Prog. Lipid Res. 1995;34:139–150. doi: 10.1016/0163-7827(94)00010-j. [DOI] [PubMed] [Google Scholar]
- 2.Enoch H. G., Catala A., Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase: studies of the substrate specificity, enzyme–substrate interactions, and the function of lipid. J. Biol. Chem. 1976;251:5095–5103. [PubMed] [Google Scholar]
- 3.Velasco A., Tabernero A., Medina J. M. Role of oleic acid as a neurotrophic factor is supported in vivo by the expression of GAP-43 subsequent to the activation of SREBP-1 and the up-regulation of stearoyl-CoA desaturase during postnatal development of the brain. Brain Res. 2003;977:103–111. doi: 10.1016/s0006-8993(03)02772-0. [DOI] [PubMed] [Google Scholar]
- 4.Garbay B., Boiron-Sargueil F., Shy M., Chbihi T., Jiang H., Kamholz J., Cassagne C. Regulation of oleoyl-CoA synthesis in the peripheral nervous system: demonstration of a link with myelin synthesis. J. Neurochem. 1998;71:1719–1726. doi: 10.1046/j.1471-4159.1998.71041719.x. [DOI] [PubMed] [Google Scholar]
- 5.Obici S., Feng Z., Morgan K., Stein D., Karkanias G., Rossetti L. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271–275. doi: 10.2337/diabetes.51.2.271. [DOI] [PubMed] [Google Scholar]
- 6.Maedler K., Spinas G. A., Dyntar D., Moritz W., Kaiser N., Donath M. Y. Distinct effects of saturated and monounsaturated fatty acids on β-cell turnover and function. Diabetes. 2001;50:69–76. doi: 10.2337/diabetes.50.1.69. [DOI] [PubMed] [Google Scholar]
- 7.Shimabukuro M., Wang M. Y., Zhou Y. T., Newgard C. B., Unger R. H. Protection against lipoapoptosis of β cells through leptin-dependent maintenance of Bcl-2 expression. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9558–9561. doi: 10.1073/pnas.95.16.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stukey J. E., McDonough V. M., Martin C. E. The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 9.Watts J. L., Browse J. A palmitoyl-CoA-specific Δ9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 10.Ward R. J., Travers M. T., Vernon R. G., Salter A. M., Buttery P. J., Barber M. C. The ovine stearyl-CoA desaturase gene: cloning and determination of gene number within the ovine genome. Biochem. Soc. Trans. 1997;25:S673. doi: 10.1042/bst025s673. [DOI] [PubMed] [Google Scholar]
- 11.Ideta R., Seki T., Adachi K., Nakayama Y. The isolation and characterization of androgen-dependent genes in the flank organs of golden Syrian hamsters. Dermatology. 1998;196:47–50. doi: 10.1159/000017865. [DOI] [PubMed] [Google Scholar]
- 12.Thiede M. A., Ozols J., Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J. Biol. Chem. 1986;261:13230–13235. [PubMed] [Google Scholar]
- 13.Kaestner K. H., Ntambi J. M., Kelly T. J., Jr, Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: a second differentially expressed gene encoding stearoyl-CoA desaturase. J. Biol. Chem. 1989;264:14755–14761. [PubMed] [Google Scholar]
- 14.Ntambi J. M., Buhrow S. A., Kaestner K. H., Christy R. J., Sibley E., Kelly T. J., Jr, Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: characterization of a differentially expressed gene encoding stearoyl-CoA desaturase. J. Biol. Chem. 1988;263:17291–17300. [PubMed] [Google Scholar]
- 15.Zheng Y., Prouty S. M., Harmon A., Sundberg J. P., Stenn K. S., Parimoo S. Scd3 – a novel gene of the stearoyl-CoA desaturase family with restricted expression in skin. Genomics. 2001;71:182–191. doi: 10.1006/geno.2000.6429. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki M., Jacobson M. J., Man W. C., Cohen P., Asilmaz E., Friedman J. M., Ntambi J. M. Identification and characterization of murine SCD4, a novel heart-specific stearoyl-CoA desaturase isoform regulated by leptin and dietary factors. J. Biol. Chem. 2003;278:33904–33911. doi: 10.1074/jbc.M304724200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Ge L., Parimoo S., Stenn K., Prouty S. M. Human stearoyl-CoA desaturase: alternative transcripts generated from a single gene by usage of tandem polyadenylation sites. Biochem. J. 1999;340:255–264. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Ge L., Tran T., Stenn K., Prouty S. M. Isolation and characterization of the human stearoyl-CoA desaturase gene promoter: requirement of a conserved CCAAT cis-element. Biochem. J. 2001;357:183–193. doi: 10.1042/0264-6021:3570183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki M., Kim H. J., Man W. C., Ntambi J. M. Oleoyl-CoA is the major de novo product of stearoyl-CoA desaturase 1 gene isoform and substrate for the biosynthesis of the Harderian gland 1-alkyl-2,3-diacylglycerol. J. Biol. Chem. 2001;276:39455–39461. doi: 10.1074/jbc.M106442200. [DOI] [PubMed] [Google Scholar]
- 20.Heinemann F. S., Ozols J. Stearoyl-CoA desaturase, a short-lived protein of endoplasmic reticulum with multiple control mechanisms. Prostaglandins Leukotrienes Essent. Fatty Acids. 2003;68:123–133. doi: 10.1016/s0952-3278(02)00262-4. [DOI] [PubMed] [Google Scholar]
- 21.Ntambi J. M., Miyazaki M. Recent insights into stearoyl-CoA desaturase-1. Curr. Opin. Lipidol. 2003;14:255–261. doi: 10.1097/00041433-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Ntambi J. M., Miyazaki M., Stoehr J. P., Lan H., Kendziorski C. M., Yandell B. S., Song Y., Cohen P., Friedman J. M., Attie A. D. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki M., Ntambi J. M. Role of stearoyl-coenzyme A desaturase in lipid metabolism. Prostaglandins Leukotrienes Essent. Fatty Acids. 2003;68:113–121. doi: 10.1016/s0952-3278(02)00261-2. [DOI] [PubMed] [Google Scholar]
- 24.Ntambi J. M. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- 25.Cohen P., Ntambi J. M., Friedman J. M. Stearoyl-CoA desaturase-1 and the metabolic syndrome. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2003;3:271–280. doi: 10.2174/1568008033340117. [DOI] [PubMed] [Google Scholar]
- 26.Rahman S. M., Dobrzyn A., Dobrzyn P., Lee S. H., Miyazaki M., Ntambi J. M. Stearoyl-CoA desaturase 1 deficiency elevates insulin-signaling components and down-regulates protein-tyrosine phosphatase 1B in muscle. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11110–11115. doi: 10.1073/pnas.1934571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen P., Miyazaki M., Socci N. D., Hagge-Greenberg A., Liedtke W., Soukas A. A., Sharma R., Hudgins L. C., Ntambi J. M., Friedman J. M. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science. 2002;297:240–243. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- 28.Dobrzyn P., Dobrzyn A., Miyazaki M., Cohen P., Asilmaz E., Hardie D. G., Friedman J. M., Ntambi J. M. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6409–6414. doi: 10.1073/pnas.0401627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki M., Man W. C., Ntambi J. M. Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J. Nutr. 2001;131:2260–2268. doi: 10.1093/jn/131.9.2260. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y., Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat. Rev. Drug Discov. 2004;3:695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- 31.Cao J., Liu Y., Lockwood J., Burn P., Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 32.Tang B. L., Zhang T., Low D. Y., Wong E. T., Horstmann H., Hong W. Mammalian homologues of yeast sec31p: an ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER–Golgi transport. J. Biol. Chem. 2000;275:13597–13604. doi: 10.1074/jbc.275.18.13597. [DOI] [PubMed] [Google Scholar]
- 33.Yu C., Chen J., Lin S., Liu J., Chang C. C., Chang T. Y. Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. J. Biol. Chem. 1999;274:36139–36145. doi: 10.1074/jbc.274.51.36139. [DOI] [PubMed] [Google Scholar]
- 34.Cheng D., Meegalla R. L., He B., Cromley D. A., Billheimer J. T., Young P. R. Human acyl-CoA:diacylglycerol acyltransferase is a tetrameric protein. Biochem. J. 2001;359:707–714. doi: 10.1042/0264-6021:3590707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanklin J., Whittle E., Fox B. G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki M., Dobrzyn A., Man W. C., Chu K., Sampath H., Kim H. J., Ntambi J. M. Stearoyl-CoA desaturase gene 1 expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element binding protein-1c-dependent and independent mechanisms. J. Biol. Chem. 2004;279:25164–25171. doi: 10.1074/jbc.M402781200. [DOI] [PubMed] [Google Scholar]
- 37.Regazzi R., Li G. D., Deshusses J., Wollheim C. B. Stimulus-response coupling in insulin-secreting HIT cells: effects of secretagogues on cytosolic Ca2+, diacylglycerol, and protein kinase C activity. J. Biol. Chem. 1990;265:15003–15009. [PubMed] [Google Scholar]
- 38.Gravena C., Mathias P. C., Ashcroft S. J. Acute effects of fatty acids on insulin secretion from rat and human islets of Langerhans. J. Endocrinol. 2002;173:73–80. doi: 10.1677/joe.0.1730073. [DOI] [PubMed] [Google Scholar]
- 39.Christiansen E., Schnider S., Palmvig B., Tauber-Lassen E., Pedersen O. Intake of a diet high in trans-monounsaturated fatty acids or saturated fatty acids: effects on postprandial insulinemia and glycemia in obese patients with NIDDM. Diabetes Care. 1997;20:881–887. doi: 10.2337/diacare.20.5.881. [DOI] [PubMed] [Google Scholar]
- 40.Ahren B., Havel P. J. Leptin inhibits insulin secretion induced by cellular cAMP in a pancreatic B cell line (INS-1 cells) Am. J. Physiol. 1999;277:R959–R966. doi: 10.1152/ajpregu.1999.277.4.R959. [DOI] [PubMed] [Google Scholar]
- 41.Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. Role of the ubiquitin-selective CDC48UFD1/NPL4 chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozols J. Degradation of hepatic stearyl CoA Δ9-desaturase. Mol. Biol. Cell. 1997;8:2281–2290. doi: 10.1091/mbc.8.11.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinemann F. S., Ozols J. Degradation of stearoyl-coenzyme A desaturase: endoproteolytic cleavage by an integral membrane protease. Mol. Biol. Cell. 1998;9:3445–3453. doi: 10.1091/mbc.9.12.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mziaut H., Korza G., Benraiss A., Ozols J. Selective mutagenesis of lysyl residues leads to a stable and active form of Δ9 stearoyl-CoA desaturase. Biochim. Biophys. Acta. 2002;1583:45–52. doi: 10.1016/s1388-1981(02)00159-2. [DOI] [PubMed] [Google Scholar]
- 45.Heinemann F. S., Mziaut H., Korza G., Ozols J. A microsomal endopeptidase from liver that preferentially degrades stearoyl-CoA desaturase. Biochemistry. 2003;42:6929–6937. doi: 10.1021/bi034071x. [DOI] [PubMed] [Google Scholar]
- 46.Heinemann F. S., Korza G., Ozols J. A plasminogen-like protein selectively degrades stearoyl-CoA desaturase in liver microsomes. J. Biol. Chem. 2003;278:42966–42975. doi: 10.1074/jbc.M306240200. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y., Hao M., Luo Y., Liang C. P., Silver D. L., Cheng C., Maxfield F. R., Tall A. R. Stearoyl-CoA desaturase inhibits ATP-binding cassette transporter A1-mediated cholesterol efflux and modulates membrane domain structure. J. Biol. Chem. 2003;278:5813–5820. doi: 10.1074/jbc.M208687200. [DOI] [PubMed] [Google Scholar]
- 48.Hao M., Mukherjee S., Sun Y., Maxfield F. R. Effects of cholesterol depletion and increased lipid unsaturation on the properties of endocytic membranes. J. Biol. Chem. 2004;279:14171–14178. doi: 10.1074/jbc.M309793200. [DOI] [PubMed] [Google Scholar]