Abstract
In iron-starved cells, IRP1 (iron regulatory protein 1) binds to mRNA iron-responsive elements and controls their translation or stability. In response to increased iron levels, RNA-binding is inhibited on assembly of a cubane [4Fe-4S] cluster, which renders IRP1 to a cytosolic aconitase. Phosphorylation at conserved serine residues may also regulate the activities of IRP1. We demonstrate that Ser-711 is a phosphorylation site in HEK-293 cells (human embryonic kidney 293 cells) treated with PMA, and we study the effects of the S711E (Ser-711→Glu) mutation on IRP1 functions. A highly purified preparation of recombinant IRP1S711E displays negligible IRE-binding and aconitase activities. It appears that the first step in the aconitase reaction (conversion of citrate into the intermediate cis-aconitate) is more severely affected, as recombinant IRP1S711E retains approx. 45% of its capacity to catalyse the conversion of cis-aconitate into the end-product isocitrate. When expressed in mammalian cells, IRP1S711E completely fails to bind to RNA and to generate isocitrate from citrate. We demonstrate that the apparent inactivation of IRP1S711E is not related to mutation-associated protein misfolding or to alterations in its stability. Sequence analysis of IRP1 from all species currently deposited in protein databases shows that Ser-711 and flanking sequences are highly conserved in the evolutionary scale. Our results suggest that Ser-711 is a critical residue for the control of IRP1 activities.
Keywords: aconitase, ferritin, iron metabolism, iron regulatory protein, iron-responsive element, transferrin receptor
Abbreviations: DFO, desferrioxamine; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; HEK-293 cells, human embryonic kidney 293 cells; IRE, iron-responsive element; IRP, iron regulatory protein; 2-ME, 2-mercaptoethanol; Ni-NTA, Ni2+-nitrilotriacetic acid; PKC, protein kinase C
INTRODUCTION
Iron is essential for a number of cellular functions, but when present in excess, it catalyses the generation of reactive oxygen species, and leads to tissue damage [1]. Consequently, iron metabolism has to be tightly controlled. Cells of higher eukaryotes adapt to alterations in iron levels by the IRE (iron-responsive element)/IRP (iron regulatory protein) regulatory system (reviewed in [2,3]). The expression of proteins of iron uptake, transport, storage and utilization is post-transcriptionally regulated by binding of iron regulatory proteins, IRP1 and IRP2, to mRNA IREs. The IRE–IRP interactions co-ordinately control the translation or stability of IRE-containing mRNAs of crucial homoeostatic proteins, such as the transferrin receptor 1 and ferritin, which mediate cellular iron uptake and storage respectively [4].
Both IRP1 and IRP2 belong to the family of iron-sulphur cluster isomerases, which includes the mitochondrial aconitase, but they sense intracellular iron levels by distinct mechanisms [2,3]. In iron-rich cells, IRP1 assembles a cubane [4Fe-4S] cluster that converts the RNA-binding protein into a cytosolic aconitase, whereas IRP2 undergoes proteasomal degradation. The [4Fe-4S] cluster is disassembled in response to iron deficiency and apoIRP1 acquires RNA binding activity. Under these conditions, IRP2 expression is stimulated by stabilization and de novo synthesis. The [4Fe-4S] cluster of IRP1 is subjected to regulation by additional iron-independent signals, such as nitric oxide and oxidative stress; in addition, IRP2 stability is regulated by nitric oxide and hypoxia.
Earlier experiments with PMA-treated HL-60 cells have suggested that IRP1 and IRP2 are targets of phosphorylation, most probably by PKC (protein kinase C) [5]. In vitro phosphorylation assays of synthetic oligopeptides with PKC provided indirect evidence that the conserved Ser-138 and Ser-711 of IRP1 are, probably, physiologically relevant phosphorylation sites. The interest in a potential regulatory role for Ser-138 phosphorylation has been spurred by results obtained with phosphomimetic IRP1 mutants expressed in an aconitase-deficient yeast strain (aco1). Wild-type IRP1 complements aerobic growth of aco1 cells; however, S138D (Ser-711→Asp) or S138E substitutions fail to do so, due to defects in the [4Fe-4S] cluster assembly pathway [6,7]. We have demonstrated that IRP1S138E expressed in mammalian cells not only fails to assemble the [4Fe-4S] cluster but, furthermore, undergoes iron-dependent degradation [8] in a fashion akin to the regulation of IRP2.
The potential role of Ser-711 phosphorylation in IRP1 regulation has not received much attention so far, despite the fact that the oligopeptides containing Ser-711 appeared to be better substrates for PKC (Km=0.6 mM) compared with those containing Ser-138 (Km=1.2–1.8 mM) [5]. To address this issue, we utilize epitope-tagged wild-type IRP1, or modified versions bearing phosphorylation-deficient S138A and/or S711A mutations, and demonstrate that Ser-711 is the actual phosphorylation site in transfected HEK-293 (human embryonic kidney 293) cells treated with PMA. Furthermore, we analyse the function of a phosphomimetic IRP1S711E and a control IRP1S711A mutant in vitro and in transfected cells. The phosphomimetic S711E substitution is associated with a severe impairment of both aconitase and IRE-binding activities of IRP1, thus suggesting that Ser-711 is a critical site for regulation.
MATERIALS AND METHODS
Materials
Haemin, PMA, bovine liver rhodanese, citrate, DL-isocitrate, cis-aconitate and antibodies against FLAG (M2-FLAG) and β-actin were purchased from Sigma (St. Louis, MO, U.S.A.). DFO (desferrioxamine) was from Novartis (Dorval, Canada).
Plasmid construction
The S711A and S711E point mutations were introduced by site-directed mutagenesis with the ExSite™ PCR-based method (Stratagene, La Jolla, CA, U.S.A.), according to the manufacturer's recommendations. The plasmid pSG5-hIRP1 [9] was utilized as template with the following oligonucleotides: (a) for S711A, 5′-GAATTCAACTCCTATGGCGCCCGCCGAGGTAATGACGCC-3′ and the reverse complement 5′-GGCGTCATTACCTCGGCGGGCGCCATAGGAGTTGAATTC-3′; and (b) for S711E, 5′-GAATTCAACTCCTATGGCGAACGCCGAGGTAATGACGCC-3′ and the reverse complement 5′-GGCGTCATTACCTCGGCGTTCGCCATAGGAGTTGAATTC-3′. All mutations were confirmed by sequencing. A 0.6 kb EcoRV–ApaI fragment from pSG5-hIRP1S711A was subcloned into the respective site of pSG5-hIRP1S138A [8], to yield pSG5-hIRP1S138A/S711A. For the generation of recombinant protein in bacteria, a 0.6 kb EcoRV–ApaI fragment from pSG5-hIRP1S711A or pSG5-hIRP1S711E was subcloned into the respective sites of pT7-hIRP1 [10], to yield pT7-hIRP1S711A and pT7-hIRP1S711E respectively. For expression of His-tagged proteins in eukaryotic cells, the 2.9 kb IRP1 cassette was excised from pT7-hIRP1, pT7-hIRP1S711A and pT7-hIRP1S711E with XbaI–HindIII and transferred into the BamH1 site of pUHD10-3 [9], after blunt-end formation with the Klenow fragment, to yield 10-3-His-hIRP1, 10-3-His-hIRP1S711A and 10-3-His-hIRP1S711E respectively. The constructs encoding IRP1S138A and IRP1S138E utilized in the present study were described in [8].
Expression and purification of recombinant IRP1
pT7-hIRP1, pT7-hIRP1S711A and pT7-hIRP1S711E were expressed in Escherichia coli BL21 (DE3) and recombinant human IRP1 was purified by affinity chromatography on Ni-NTA (Ni2+-nitrilotriacetic acid) beads, followed by a second purification step with anion-exchange chromatography on a Resource Q FPLC column (Pharmacia, Baie d'Urfé, Quebec, Canada) [8].
Iron-sulphur cluster reconstitution
The iron-sulphur cluster of human recombinant IRP1 (wild-type and mutants) was reconstituted by three different approaches: 5 μg of purified protein was incubated for 25 min at 25 °C either with 10 mM cysteine/HCl (pH 7.4) and 100 μM ferrous sulphate [10], or with 100 mM DTT (dithiothreitol), 1 mM thiosulphate, 400 μM ferric citrate and 10 μM bovine liver rhodanese (enzyme catalysing the transfer of sulphur between thiosulphate and a thiophilic anion) [11], or with 70 mM DTT, 800 μM sodium sulphide and 350 μM ferric citrate [12].
Aconitase assay
Aconitase activity was determined by the reduction of NADP+ at 340 nm in a coupled reaction with isocitrate dehydrogenase [10], with 200 μM citrate or cis-aconitate as substrate. The reverse aconitase activity was measured in a direct assay with 20 mM isocitrate as substrate. The formation of cis-aconitate was monitored spectrophotometrically at 240 nm, using a molar absorption coefficient ε of 3600 M−1·cm−1 [13]. One unit of aconitase is defined as the amount that converts 1 μmol substrate/min at pH 7.4 at 25 °C.
Cell culture and transfections
Murine B6 fibroblasts and HEK-293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin and 0.1 ng/ml streptomycin. The stable B6 and HEK-293 clones, expressing N-terminally His-tagged wild-type IRP1 or C-terminally FLAG-tagged IRP1S138E respectively were described earlier [8]. Similarly, stable HEK-293 clones expressing pSG5-hIRP1S711E, and B6 clones expressing either 10-3-His-hIRP1S711A or 10-3-His-hIRP1S711E, were obtained as described in [8] for the respective Ser-138 mutants. Transient transfections were performed with the Lipofectamine Plus™ reagent (Gibco BRL, Burlington, Ontario, Canada).
Metabolic labelling with [32P]orthophosphate and analysis of phosphorylated IRP1
Parent and transfected HEK-293 cells were washed with phosphate-free media and metabolically labelled with 32PO43− (1 mCi/107 cells). Cell lysates containing phosphatase inhibitors (50 mM NaF and 50 mM β-glycerophosphate) were subjected to immunoprecipitation with the M2-FLAG antibody (Sigma), as described in [9]. Immunoprecipitated material was analysed by SDS/PAGE (8% polyacrylamide) and visualized by autoradiography.
Western blotting and EMSA (electrophoretic mobility-shift assay)
Analysis of IRP1 and control β-actin expression by Western blotting, and assessment of IRE-binding activity by EMSA were performed as in [8].
Generation of 711[pS] phospho-specific antibodies and analysis of phosphorylated IRP1
Polyclonal 711[pS] phospho-specific antibodies were obtained by immunizing a rabbit against NSYG[pS]RRGND, corresponding to IRP1707–716 (ResGen, Invitrogen). Specificity was assessed by dot-blot analysis against the non-phosphorylated control peptide NSYGSRRGND and the phosphorylated NSYG[pS]RRGND antigen.
CD spectroscopy
CD spectra of purified recombinant IRP1 (wild-type and mutants) were recorded over the 180–320 nm range with a Jasco 810 spectropolarimeter (Jasco, Hachioji City, Tokyo, Japan) at a scanning speed of 5 nm/min, with a bandwidth of 4 nm and a time response of 2 s [14]. Multiple spectra (between 3 and 10) were summed to improve the signal-to-noise ratio. Protein solutions were prepared in 10 mM Tris/HCl buffer with 50 mM KCl at pH 8.0. They were kept in argon-filled quartz cuvettes of 0.1 cm optical path in a nitrogen-flushed sample holder.
RESULTS
Treatment of HEK-293 cells with PMA leads to phosphorylation of IRP1 at Ser-711
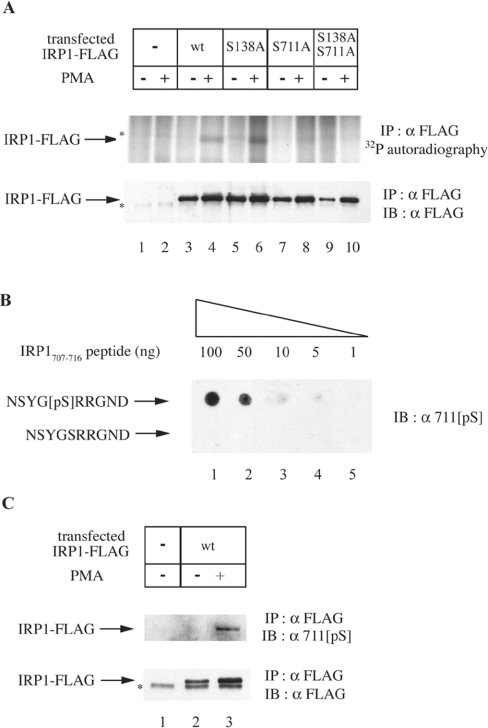
IRP1 is subjected to phosphorylation in HL-60 cells treated with PMA, and in vitro experiments with oligopeptides suggested that Ser-138 and Ser-711 are potential phosphorylation sites [5]. To map the phosphorylation site(s) of IRP1 in vivo, we utilized cDNA constructs encoding C-terminally FLAG epitope-tagged wild-type or mutant IRP1, bearing phosphorylation-deficient single S138A, S711A, or double S138A/S711A substitutions. The constructs were transiently transfected into HEK-293 cells and analysed for phosphorylation after metabolic labelling with [32P]orthophosphate and treatment with or without PMA. Immunoprecipitation with the FLAG antibody (Figure 1A) reveals that transfected wild-type IRP1 and IRP1S138A undergo phosphorylation in response to PMA treatment (upper panel, lanes 3–6). In contrast, no IRP1-specific signal is evident in FLAG immunoprecipitates from parent cells (lanes 1–2) and from cells transfected with IRP1S711A or with the double mutant IRP1S138A/S711A (lanes 7–10). Analysis of the immunoprecipitated material by Western blotting shows the recovery of transfected IRP1 (lower panel). The utilization of 711[pS], a Ser-711 phospho-specific antibody (Figure 1B), confirms that Ser-711 is a site of in vivo phosphorylation (Figure 1C). Taken together, these results suggest that PMA triggers IRP1 phosphorylation at Ser-711.
Figure 1. Ser-711 is the site of IRP1 phosphorylation in cells treated with PMA.
(A) HEK-293 cells were transiently transfected with constructs encoding FLAG-tagged wild-type (wt) or mutant (S138A, S711A or S138A/S711A) IRP1. Parent and transfected cells were metabolically labelled with [32P]orthophosphate. After 30 min, 0.2 μM PMA (dissolved in DMSO) or solvent alone were added in the radioactive medium, and the treatment was continued for 90 min. To assess the phosphorylation status of transfected wild-type IRP1, IRP1S138A, IRP1S711A and IRP1S138A/S711A, cytoplasmic extracts (1000 μg) were subjected to quantitative immunoprecipitation (IP) with a FLAG antibody (8.8 μg). The immunoprecipitated material was analysed by SDS/PAGE (8% polyacrylamide). Phosphorylated proteins were visualized by autoradiography (upper panel) and the recovery of transfected IRP1 was analysed by immunoblotting (IB) with 1:1000 diluted FLAG antibody (lower panel). (B) The indicated amounts of NSYG[pS]RRGND and NSYGSRRGND peptides were spotted on a nitrocellulose filter and analysed by IB with 1:1000 diluted 711[pS], a phospho-specific antibody raised against NSYG[pS]RRGND. (C) HEK-293 cells were transiently transfected with a construct encoding FLAG-tagged wild-type IRP1 and treated with 0.2 μM PMA or DMSO alone for 90 min. To assess phosphorylation of IRP1 at Ser-711, cytoplasmic extracts (1000 μg) were subjected to quantitative IP with the FLAG antibody (8.8 μg) and subsequently analysed by IB with 1:1000 diluted 711[pS] antibody (upper panel). The recovery of transfected IRP1 was analysed by immunoblotting with the FLAG antibody (lower panel). The asterisks denote non-specific bands.
Purified recombinant IRP1S711E displays minimal aconitase and IRE-binding activities
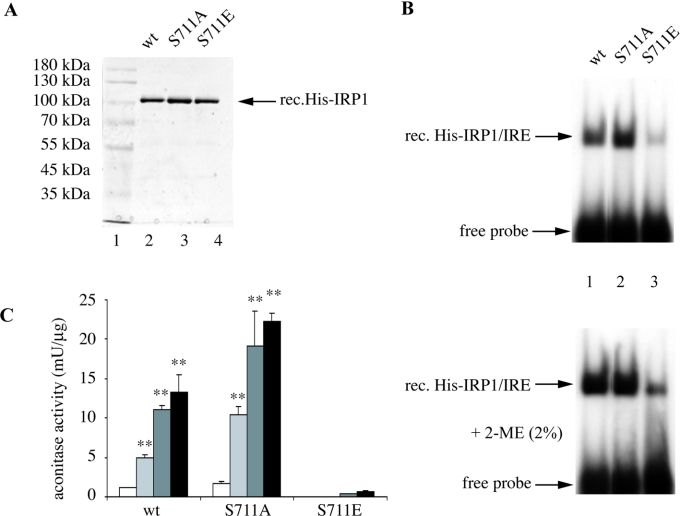
Having established that Ser-711 is a biologically relevant phosphorylation site, we utilized recombinant wild-type and mutated versions of human IRP1, including the phosphorylation-deficient IRP1S711A and the phosphomimetic IRP1S711E mutants, to investigate further the role of this residue in IRP1 function. The proteins were tagged with an N-terminal His6 epitope, expressed in E. coli and purified by affinity and ion exchange chromatography. The purity of the preparations was very high, as evaluated by SDS/PAGE and staining with Coomassie Brilliant Blue (Figure 2A). Aliquots of the purified material (80 ng) were analysed for IRE-binding activity by EMSA (Figure 2B). As expected, wild-type IRP1 and IRP1S711A were active in IRE-binding (lanes 1 and 2). Treatment with 2-ME (2-mercaptoethanol), known to induce latent [4Fe-4S]-IRP1 for IRE-binding [15], only slightly stimulated the activity of wild-type IRP1 and IRP1S711A (lower panel), suggesting that the analysed proteins were almost fully active to bind to RNA after the purification. Interestingly, the IRE-binding activity of IRP1S711E was minimal (∼10% of wild-type, lane 3), even after 2-ME treatment (lower panel), indicating that this response is not due to the presence of a [4Fe-4S] cluster. Similar results were obtained with different quantities of recombinant proteins (results not shown).
Figure 2. Recombinant IRP1S711E possesses minimal aconitase and IRE-binding activities.
(A) SDS/PAGE of purified human recombinant His-tagged wild-type IRP1, IRP1S711A and IRP1S711E. The proteins were visualized by staining with Coomassie Brilliant Blue. (B) Purified recombinant proteins (80 ng) were analysed by EMSA with a 32P-labelled IRE probe in the absence (upper panel) or presence (lower panel) of 2% (w/v) 2-ME. The positions of specific His–IRP1–IRE complexes and excess free probe are indicated by arrows. (C) Aconitase assay before (white bars) and after iron-sulphur cluster reconstitution with one among ferrous sulphate/cysteine (light grey bars), ferric citrate/rhodanese/thiosulphate (dark grey bars) or ferric citrate/sulphide (black bars). Values correspond to means of triplicate samples; the aconitase activity is expressed in terms of m-units/μg of recombinant protein. **P<0.01 versus untreated control (Student's t test).
The purified proteins were further analysed for aconitase activity. Consistent with the view that our preparations of wild-type IRP1 and IRP1S711A are mostly in the apo-form, these proteins exhibited relatively low levels of aconitase activity (Figure 2C, white bars), which depends on the presence of the [4Fe-4S] cluster [16]. Three different approaches were employed to reconstitute the [4Fe-4S] cluster of the purified proteins in vitro, involving a treatment with one among ferrous sulphate/cysteine [10], ferric citrate/rhodanese/thiosulphate [11] or ferric citrate/sulphide [12]. The aconitase activities of wild-type IRP1 and IRP1S711A were significantly (P<0.01) stimulated by all cluster reconstitution treatments. The proteins treated with ferric citrate/sulphide displayed the highest activity, which was induced approx. 13-fold more than that of untreated controls. Thus it appears that, under the above experimental conditions, ferric citrate/sulphide promotes the most efficient [4Fe-4S] cluster reconstitution of recombinant IRP1. Notably, untreated IRP1S711E displayed no aconitase activity at all. Moreover, all cluster reconstitution treatments merely resulted in a minimal aconitase activity, which at best reached approx. 5% of that of wild-type IRP1. These results suggest that IRP1S711E may be defective in the assembly or the maintenance of its [4Fe-4S] cluster.
This apparent inactivation of IRP1S711E as an IRE-binding protein and as an aconitase could be a result of distortions in the protein structure, due to the introduction of a negative charge by the S711E mutation. To address this possibility, the purified proteins were analysed by CD spectroscopy. However, all wild-type IRP1, IRP1S711A and IRP1S711E, but also the IRP1S138A and IRP1S138E mutants [8], yielded similar spectra (results not shown), indicating that the phenotype of recombinant IRP1S711E depicted in Figures 2(B) and 2(C) is not due to improper folding.
The phosphomimetic S711E substitution primarily affects the first step of the IRP1 aconitase reaction
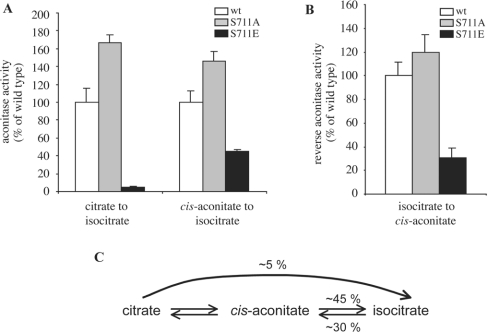
Aconitases catalyse the isomerization of citrate to isocitrate through the intermediate cis-aconitate, in a two-step reaction [16]. To gain mechanistic insights on the aconitase inactivation by the phosphomimetic S711E mutation, purified recombinant preparations of wild-type IRP1, IRP1S711A and IRP1S711E were subjected to [4Fe-4S] cluster reconstitution with ferric citrate/sulphide and analysed for enzymatic activity with different substrates. Although IRP1S711E displayed minimal (∼5% of wild-type) capacity to catalyse the conversion of citrate into isocitrate, it retained sizeable enzymatic activity (∼45% of wild-type) when the intermediate cis-aconitate was used as substrate (Figure 3A). Accordingly, IRP1S711E also displayed partial (∼30% of wild-type) enzymatic activity in the reverse reaction, the conversion of isocitrate into cis-aconitate (Figure 3B). Taken together, these results (summarized in Figure 3C) suggest that the phosphomimetic S711E substitution strongly impairs the first step of the aconitase reaction, and modestly inhibits the second step.
Figure 3. IRP1S711E displays a profound defect in the first step of the aconitase catalytic reaction.
Purified recombinant wild-type IRP1 (white bars), IRP1S711A (grey bars) or IRP1S711E (black bars) (5 μg each) were analysed for aconitase activity after treatment with ferric citrate/sulphide to reconstitute the [4Fe-4S] cluster. (A) The conversion of citrate or cis-aconitate into isocitrate was measured by the coupled isocitrate dehydrogenase reaction [10]. (B) The formation of cis-aconitate from isocitrate was monitored spectrophotometrically at 240 nm [13]. (C) Schematic representation of the aconitase reaction, with the relative efficiency of IRP1S711E to catalyse each step, compared with wild-type IRP1.
IRP1S711E expressed in mammalian cells exhibits properties of a null mutant
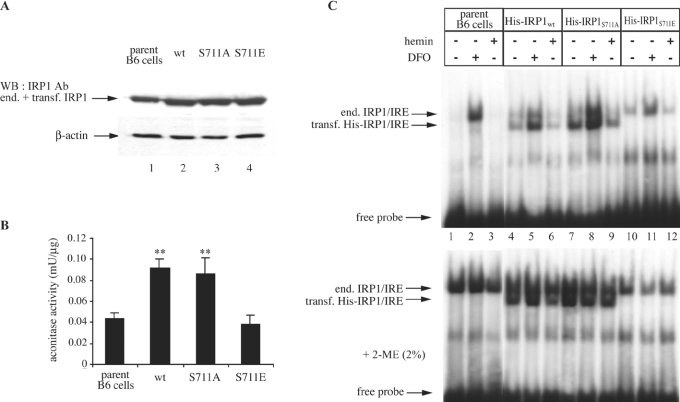
To evaluate further the role of Ser-711 on IRP1 function, B6 murine fibroblasts were transfected with constructs encoding His-tagged wild-type IRP1, IRP1S711A or IRP1S711E. Stable transfectants were selected and representative clones were analysed for expression of transfected IRP1 by Western blotting (Figure 4A, upper panel). Commercial His antibodies did not yield reliable signals and thus, a polyclonal IRP1 antibody, which does not distinguish between endogenous and transfected IRP1, was utilized for detection. Under these conditions, the IRP1 signal intensity was increased approx. 2-fold in all transfectants (lanes 2–4) compared with parent cells (lane 1), suggesting an approx. 1:1 ratio between endogenous and transfected IRP1. The immunoblot was also probed with an antibody against β-actin (lower panel) to control equal loading. The transfected cells were further analysed for cytosolic aconitase activity (Figure 4B). As shown previously [8], this activity increased approx. 2-fold (P<0.01) in cells expressing wild-type IRP1 compared with parent cells, and a similar effect was observed for IRP1S711A. In contrast, the expression of IRP1S711E failed to increase total cytosolic aconitase activity (with citrate as substrate). Thus, in agreement with the results obtained with purified recombinant IRP1S711E (Figures 2C and 3A), it appears that IRP1S711E expressed in B6 cells is not capable of catalysing the conversion of citrate into isocitrate.
Figure 4. IRP1S711E expressed in B6 cells exhibits a phenotype of a null mutant.
Wild-type IRP1, IRP1S711A or IRP1S711E, tagged with a His6 epitope at their N-termini, were stably transfected into B6 cells. (A, B) Cytosolic extracts, devoid of mitochondrial aconitase [20], of non-transfected parent cells and of clones expressing wild-type IRP1, IRP1S711A or IRP1S711E were analysed by Western blotting (A) with antibodies against IRP1 (upper panel) and β-actin (lower panel) and for aconitase activity by the coupled isocitrate dehydrogenase reaction with citrate as substrate (B). Values in (B) correspond to triplicate samples; the aconitase enzymatic activity is expressed in terms of m-units/μg of total proteins in the cytosolic lysate. (C) Cells were treated overnight with 100 μM DFO or haemin. Cytoplasmic extracts of non-transfected parent cells (lanes 1–3) and of clones expressing wild-type IRP1 (lanes 4–6), IRP1S711A (lanes 7–9) or IRP1S711E (lanes 10–12) were analysed by EMSA with a 32P-labelled IRE probe in the absence (upper panel) or presence (lower panel) of 2% 2-ME. The positions of excess free probe and of specific IRP1–IRE complexes, corresponding to endogenous murine IRP1 and transfected human His–IRP1, are indicated by arrows. **P<0.01 versus control (Student's t test).
The transfected proteins were further assessed for their IRE binding properties. Parent cells and transfectants were subjected to iron manipulations and cytoplasmic extracts were analysed by EMSA (Figure 4C, upper panel). In this experimental setting, IRE complexes with transfected human IRP1 display a different electrophoretic mobility from those with endogenous mouse IRP1 [8]; thus, the IRE-binding activities of transfected and endogenous proteins can be readily separated (IRP2–IRE complexes are not visible in the absence of DTT in the buffer). The extracts from parent cells gave rise to a single band (lanes 1–3), corresponding to IRE complexes with endogenous IRP1, the intensity of which was strongly induced after an overnight treatment with the iron chelator DFO (lane 2). The extracts of wild-type IRP1 and IRP1S711A transfectants yielded an additional, faster-migrating band (lanes 4–9), corresponding to IRE complexes with transfected IRP1. As with endogenous IRP1, the IRE-binding activities of transfected IRP1 and IRP1S711A were induced by iron chelation (lanes 5 and 8) and suppressed after a treatment with haemin (lanes 6 and 9). Notably, the extracts from IRP1S711E transfectants did not give rise to a fast-migrating band, suggesting that IRP1S711E is not active in IRE-binding. Mainly, no detectable exogenous IRE-binding activity could be restored in extracts of IRP1S711E-expressing cells treated with 2-ME, although, as expected, this treatment was effective for wild-type IRP1 and IRP1S711A (lower panel). Thus IRP1S711A behaves essentially as wild-type IRP1; however, IRP1S711E lacks any apparent biochemical activity.
The expression of IRP1S711E is not affected by iron
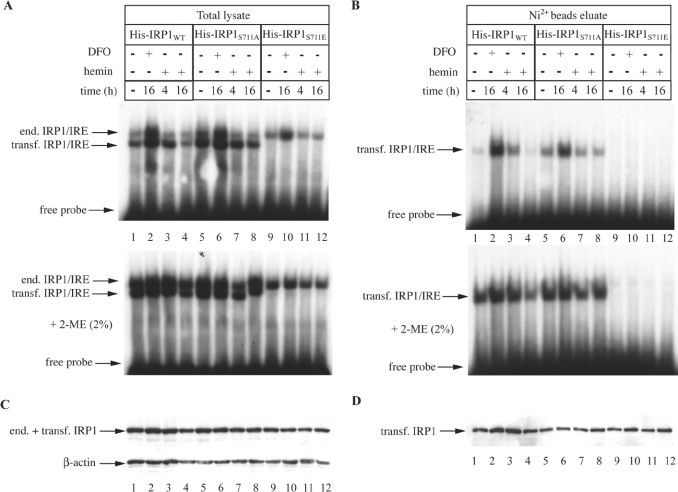
To characterize better this unusual phenotype, all His-tagged proteins from transfected cells were purified from the crude cytoplasmic extracts by affinity chromatography on Ni-NTA beads (Figure 5). Analysis of the total lysate (Figure 5A) and of the purified His-tagged proteins (Figure 5B) by EMSA confirmed that IRP1S711E lacks any apparent IRE-binding activity, independently of the iron status of the cells, and even after 2-ME treatments. The aconitase activity of transfected His-tagged IRP1 appears to be unstable to dilution after affinity purification [8], and thus, an aconitase assay with the preparations of all affinity-purified proteins yielded negative results. The crude lysate and the purified material were subsequently analysed by Western blotting with the IRP1 antibody. In agreement with the results shown in Figure 4(A), the analysis of the crude lysate (Figure 5C, upper panel) confirms that the expression levels of all His-tagged IRP1 constructs were in the same range. Moreover, this assay shows that the expression of wild-type IRP1, IRP1S711A and IRP1S711E are largely unresponsive to iron manipulations. The immunoblot with β-actin (lower panel) serves as a loading control. Similar results were obtained with the affinity-purified material (Figure 5D).
Figure 5. The expression of N-terminally His-tagged IRP1S711E in B6 cells is not affected by iron.
Cells expressing human wild-type IRP1, IRP1S711A or IRP1S711E were treated with 100 μM DFO or haemin for the indicated time intervals. His-tagged IRP1 was purified from cytoplasmic extracts by affinity chromatography with Ni-NTA beads. (A, B) Total lysates (A) or affinity-purified transfected IRP1 (B) were analysed by EMSA with a 32P-labelled IRE probe in the absence (upper) or presence (lower panel) of 2% 2-ME. The positions of excess free probe and of specific IRP1–IRE complexes, corresponding to endogenous murine IRP1 and transfected human His–IRP1, are indicated by arrows. (C, D) Western blotting of total lysates (C) or affinity-purified transfected IRP1 (D) with antibodies against IRP1 and β-actin (lower panel in C).
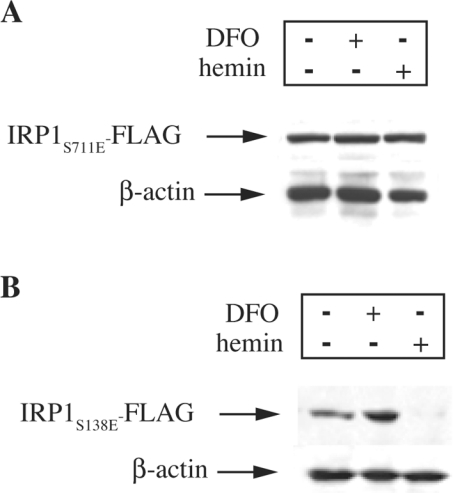
We have previously established that IRP1 bearing a phosphomimetic S138E substitution apparently fails to assemble a [4Fe-4S] cluster and undergoes proteasomal degradation in iron-replete cells [8]. To characterize better the response of IRP1S711E to iron and to directly compare it with that of IRP1S138E, constructs encoding the respective mutants, tagged with a FLAG epitope at their C-termini, were stably transfected into HEK-293 cells. The expression levels of FLAG-tagged proteins were analysed by Western blotting after iron perturbations of the transfected cells by an overnight treatment with haemin or DFO (Figure 6). Iron manipulations had no effect on the expression of IRP1S711E (Figure 6A, upper panel) or on control β-actin (lower panel). However, under the same experimental conditions, the treatment with haemin diminished the expression of IRP1S138E (Figure 6B, upper panel), as expected on the basis of our previous observations [8]. We conclude that the inactivation of IRP1S711E is not due to iron-dependent alterations in protein expression.
Figure 6. The expression of C-terminally FLAG-tagged IRP1S711E in HEK-293 cells is not affected by iron.
The cells were stably transfected with constructs encoding C-terminal FLAG epitope-tagged human IRP1S711E (A) or IRP1S138E (B). After an overnight treatment with 100 μM DFO or haemin, cell lysates were analysed by Western blotting with antibodies against FLAG (upper panel) or β-actin (lower panel).
Ser-711 is highly conserved in IRP1 molecules
Ser-711 and Ser-138 were earlier identified as conserved PKC phosphorylation sites within IRP1 [5]. By analysing a set of IRP1 homologues we showed that Ser-138 appears to be conserved only in IRP1 molecules of vertebrates, but not of plants or lower organisms [8]. A multiple sequence alignment among all IRP1 molecules currently deposited in the SwissProt database was performed to evaluate the conservation of Ser-711 (Figure 7, upper panel). This analysis reveals that, with the exception of the IRP1 homologue of the malaria parasite (Plasmodium falciparum), Ser-711 and flanking sequences are highly conserved in all other IRP1 or IRP1-like molecules from mammals (Homo sapiens, Oryctolagus cuniculus, Rattus norvegicus and Mus musculus), chicken (Gallus gallus), crayfish (Pacifastacus leniusculus), insects (Drosophila melanogaster, Anopheles gambiae and Manduca sexta) and plants (Arabidopsis thaliana, Citrus limon and Curcubita maxima). Interestingly, despite the significant conservation of flanking sequences, Ser-711 appears to be replaced with an alanine residue in IRP2 (Figure 7, lower panel), but a larger number of samples would be required to draw a firm conclusion. Taken together, these results suggest that Ser-711 and the domain surrounding it are highly conserved in IRP1 molecules from a broad spectrum of species.
Figure 7. Multiple sequence alignment of amino acids 702–721 in IRP1 (upper panel) and the respective amino acids in IRP2 (lower panel) molecules from all species currently deposited in the SwissProt database.
With the exception of Plasmodium falciparum, Ser-711 (*) appears to be highly conserved in IRP1 or IRP1-like molecules from vertebrates, invertebrates, insects and plants. Despite the homology in flanking sequences, the equivalent position in IRP2 appears to be replaced with an alanine residue. The genInfo identifier (gi) numbers of all molecules are indicated on the right. The alignment was made by the ClustalW algorithm (MacVector software, version 7.2).
DISCUSSION
The evidence that Ser-138 and Ser-711 of IRP1 are potential sites of phosphorylation by PKC has solely been based on in vitro experiments with oligopeptides [5]. Thus, in the present study, we sought to map the actual phosphorylation site(s) of IRP1 in vivo. To this end, constructs encoding wild-type or mutant versions of FLAG-tagged IRP1 were transiently transfected into HEK-293 cells, the cells were metabolically labelled with [32P]-orthophosphate in the presence or absence of PMA, and FLAG immunoprecipitates were analysed for the presence of 32P-labelled IRP1. The results presented in Figure 1 demonstrate that Ser-711 is the only residue of IRP1 that undergoes phosphorylation in response to PMA, at least in HEK-293 cells. Similar conclusions were also reached in a recent publication employing HEK-293 cells [17]. Interestingly, no spontaneous phosphorylation of endogenous or transfected IRP1 was evident in the absence of PMA. We speculate that Ser-138 may be subjected to phosphorylation in response to other signals. In addition, we cannot exclude the possibility that the pattern of IRP1 phosphorylation may also depend on tissue or cell-specific factors.
To understand better the functional implications associated with IRP1 phosphorylation at Ser-711, we undertook a mutagenesis approach involving the replacement of this residue with a ‘phosphomimetic’ glutamate. The effects of the S711E substitution on IRP1 activity were examined in vitro and in transfected cells. An analogous approach revealed that a single S138E substitution alters significantly the response of IRP1 to iron and renders the protein sensitive to iron-dependent degradation [8].
A first set of experiments with highly purified preparations of recombinant wild-type and mutant IRP1 showed that IRP1S711E is completely inactive as an aconitase (Figure 2C). Various procedures for in vitro reconstitution of the [4Fe-4S] cluster profoundly stimulated the aconitase activity of wild-type IRP1 and IRP1S711A, but only marginally recovered the catalytic activity of IRP1S711E. These results exclude the possibility that the failure of IRP1S711E to function as aconitase is merely due to predominance of apoprotein in the purified preparation. Moreover, at a first glance, they also hint that the S711E substitution may impair the capacity of IRP1 to assemble its [4Fe-4S] cluster.
Further studies revealed that iron-loaded IRP1S711E retains a considerable aconitase activity when the intermediate cis-aconitate is used as substrate (Figure 3). The same holds true for the reverse conversion of isocitrate into cis-aconitate, suggesting that the S711E mutation primarily affects the first step in the aconitase reaction. Considering that the [4Fe-4S] cluster is indispensable for catalysis [16], these results imply that IRP1S711E can assemble a [4Fe-4S] cluster, at least to some extent. Pitula et al. [17] recently reported a selective inhibition in the ability of IRP1 bearing phosphomimetic substitutions at Ser-711, to convert citrate into isocitrate, and proposed that this could be a result of impaired binding of cis-aconitate to the ‘citrate mode’ of aconitase. Earlier studies with mitochondrial aconitase showed that the intermediate cis-aconitate must ‘flip’ from a ‘citrate to an isocitrate mode’ of binding during catalysis [16]. Our conclusion that the S711E mutation primarily affects the first step in the aconitase reaction is consistent with this view. The results presented in Figure 3 show that the S711E mutation also affects the other half reaction, albeit to a smaller extent. This may be due to interference of the phosphomimetic residue with the assembly and maintenance of the [4Fe-4S] cluster. None of the above scenarios are mutually exclusive.
The purified recombinant IRP1S711E was also examined for its capacity as an RNA-binding protein. In contrast with wild-type IRP1 or IRP1S711A, IRP1S711E displayed a very limited IRE-binding activity, which only moderately increased on treatment with 2-ME (Figure 2B). Purified recombinant IRP1S138E was similarly found to have reduced IRE-binding activity compared with controls, but this was efficiently recovered by 2-ME treatment [8]. On the basis of the CD analysis, these defects are unlikely to be related to significant perturbations in the overall secondary structure of IRP1. To understand why the S711E mutation associates with loss of IRE-binding activity, one can hypothesize that Ser-711 may lie in close proximity to bound RNA [18], so that the introduction of a negative charge may disrupt the RNA–protein interaction. On the other hand, it should be noted that all RNA-binding assays were performed only with a ferritin IRE-probe. Thus our results do not exclude the possibility that IRP1S711E may bind more efficiently to alternative IRE structures [19].
We finally evaluated the properties of IRP1S711E when expressed in mammalian cells. In agreement with the results obtained with the purified recombinant material, the analysis of crude extracts of B6 transfectants (Figure 4B) confirmed that IRP1S711E fails to catalyse the conversion of citrate into isocitrate. Furthermore, transfected IRP1S711E completely lacked any apparent IRE-binding activity, regardless of the iron status of the cells. This result was corroborated in crude cell extracts (Figures 4C and 5A) and in an affinity-purified preparation of transfected IRP1S711E (Figure 5B), even in the presence of 2-ME. The apparent inactivation of IRP1S711E in transfected cells was not associated with alterations in its expression levels (Figures 5C and 5D). Thus, despite some similarities (e.g. the loss of aconitase activity), the phenotype of transfected IRP1S711E differs from that of IRP1S138E, which undergoes iron-dependent degradation ([8] and Figure 6). In contrast, IRP1S711E is stable in iron-loaded cells and thus it exhibits the hallmarks of a null mutant, even though it most probably retains some capacity to convert isocitrate back into cis-aconitate. It has been proposed that this reverse aconitase activity may, under physiological conditions, promote ‘enhanced accumulation of citrate and/or depletion of isocitrate and possibly NADPH’ from the cytosol [17]. However, it is evident from the results presented in Figure 3(A) that IRP1S711E can catalyse the conversion of cis-aconitate into isocitrate with an efficiency similar to that of the reverse reaction. We speculate that in vivo this may result in a vicious cycle. More significantly, the profound inhibition in the first step of the aconitase reaction (Figure 3A) could be sufficient to elicit accumulation of citrate and concomitant depletion of isocitrate from the cytosol, without the contribution of reverse aconitase activity.
In conclusion, we demonstrate here that Ser-711 is the site of IRP1 phosphorylation by a PMA-dependent pathway in vivo, and that a phosphomimetic S711E substitution functionally inactivates the protein. The high conservation of Ser-711 and flanking sequences in the evolutionary scale (Figure 7) further support the idea that Ser-711 is critical for IRP1 regulation and function. Considering that the conservation of Ser-138 appears to be restricted to IRP1 sequences from vertebrates [8], it is probable that Ser-711 evolved earlier as a regulatory site and was possibly lost in IRP1 homologues that do not assemble a [4Fe-4S] cluster, such as IRP2. The physiological implications of IRP1 phosphorylation at Ser-711 in the control of iron homoeostasis remain to be established.
Acknowledgments
We thank D. Chahine and P. Segal for experimental contributions in the early phase of this work. K.P. and J.M.M. acknowledge travel support from the Centre Jacques Cartier. C.F. is a recipient of a post-doctoral fellowship from the Fonds de la Recherche en Santé du Québec. K.P. is supported by the Canadian Institutes of Health Research and the Canada Foundation for Innovation. This work was supported by a grant from the Canadian Institutes of Health Research.
References
- 1.Halliwell B., Gutteridge J. M. C. The role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 2.Hentze M. W., Muckenthaler M. U., Andrews N. C. Balancing acts; molecular control of mammalian iron metabolism. Cell (Cambridge, Mass.) 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 3.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N.Y. Acad. Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 4.Ponka P., Beaumont C., Richardson D. R. Function and regulation of transferrin and ferritin. Semin. Hematol. 1998;35:35–54. [PubMed] [Google Scholar]
- 5.Eisenstein R. S., Tuazon P. T., Schalinske K. L., Anderson S. A., Traugh J. A. Iron-responsive element-binding protein. Phosphorylation by protein kinase C. J. Biol. Chem. 1993;268:27363–27370. [PubMed] [Google Scholar]
- 6.Brown N. M., Anderson S. A., Steffen D. W., Carpenter T. B., Kennedy M. C., Walden W. E., Eisenstein R. S. Novel role of phosphorylation in Fe-S cluster stability revealed by phosphomimetic mutations at Ser-138 of iron regulatory protein 1. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15235–15240. doi: 10.1073/pnas.95.26.15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown N. M., Kennedy M. C., Antholine W. E., Eisenstein R. S., Walden W. E. Detection of a [3Fe-4S] cluster intermediate of cytosolic aconitase in yeast expressing iron regulatory protein 1: insights into the mechanism of Fe-S cluster cycling. J. Biol. Chem. 2002;277:7246–7254. doi: 10.1074/jbc.M110282200. [DOI] [PubMed] [Google Scholar]
- 8.Fillebeen C., Chahine D., Caltagirone A., Segal P., Pantopoulos K. A phosphomimetic mutation at Ser-138 renders iron regulatory protein 1 sensitive to iron-dependent degradation. Mol. Cell. Biol. 2003;23:6973–6981. doi: 10.1128/MCB.23.19.6973-6981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Pantopoulos K. Conditional de-repression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol. Cell. Biol. 2002;22:4638–4651. doi: 10.1128/MCB.22.13.4638-4651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray N. K., Quick S., Goossen B., Constable A., Hirling H., Kühn L. C., Hentze M. W. Recombinant iron regulatory factor functions as an iron-responsive element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 11.Cassanelli S., Moulis J. Sulfide is an efficient iron releasing agent for mammalian ferritins. Biochim. Biophys. Acta. 2001;1547:174–182. doi: 10.1016/s0167-4838(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 12.Campanella A., Levi S., Cairo G., Biasiotto G., Arosio P. Blotting analysis of native IRP1: a novel approach to distinguish the different forms of IRP1 in cells and tissues. Biochemistry. 2004;43:195–204. doi: 10.1021/bi035386f. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy M. C., Emptage M. H., Dreyer J. L., Beinert H. The role of iron in the activation–inactivation of aconitase. J. Biol. Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 14.Brazzolotto X., Timmins P., Dupont Y., Moulis J. M. Structural changes associated with switching activities of the human iron regulatory protein 1. J. Biol. Chem. 2002;277:11995–12000. doi: 10.1074/jbc.M110938200. [DOI] [PubMed] [Google Scholar]
- 15.Hentze M. W., Rouault T. A., Harford J. B., Klausner R. D. Oxidation–reduction and the molecular mechanism of a regulated RNA–protein interaction. Science. 1989;244:357–359. doi: 10.1126/science.2711187. [DOI] [PubMed] [Google Scholar]
- 16.Beinert H., Kennedy M. C., Stout C. D. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 1996;96:2335–2374. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 17.Pitula J. S., Deck K. M., Clarke S. L., Anderson S. A., Vasanthakumar A., Eisenstein R. S. Selective inhibition of the citrate-to-isocitrate reaction of cytosolic aconitase by phosphomimetic mutation of serine-711. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10907–10912. doi: 10.1073/pnas.0404308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaldy P., Menotti E., Moret R., Kühn L. C. Identification of RNA-binding surfaces in iron regulatory protein-1. EMBO J. 1999;18:6073–6083. doi: 10.1093/emboj/18.21.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson H. E., Theil E. C. Iron-response element (IRE) structure and combinatorial RNA regulation. In: Templeton D. M., editor. Molecular and Cellular Iron Transport. New York: Marcel Dekker; 2002. pp. 237–253. [Google Scholar]
- 20.Drapier J. C., Hirling H., Wietzerbin J., Kaldy P., Kühn L. C. Biosynthesis of nitric oxide activates iron regulatory factor in macrophages. EMBO J. 1993;12:3643–3649. doi: 10.1002/j.1460-2075.1993.tb06038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]