Abstract
The stoned locus of Drosophila produces a dicistronic transcript and encodes two proteins, stoned-A (STNA) and stoned-B (STNB). Both proteins are located at synaptic terminals. The STNB protein contains a domain that has homology with the μ-subunit of the AP (adaptor protein) complex, as well as a number of NPF (Asp-Pro-Phe) motifs known to bind EH (Eps15 homology) domains. Mutations at the stoned locus interact synergistically with mutations at the shibire (dynamin) locus and alter synaptic vesicle endocytosis. The STNB protein has also been shown to interact with synaptic vesicles via synaptogamin-I. We initiated an investigation of the possible interaction of DAP-160 (dynamin-associated protein of 160 kDa), a Drosophila member of the intersectin family, with the STNB protein. We show here that both of the viable stoned alleles interacted with a genetic construct that reduces DAP-160 levels to 25% of normal. One of these stoned alleles contains a substitution resulting in a stop codon in the open reading frame encoding STNB. This allele also shows markedly reduced levels of both DAP-160 and dynamin. As anticipated, the NPF motifs in STNB are found to be high-affinity binding motifs for the EH domains of DAP-160. One of the SH3 (Src homology 3) domains of DAP-160 also interacts with STNB. Finally, we show that immunoprecipitation of STNB from fly head extracts co-precipitates with DAP-160, and we conclude that the interaction of the STNB protein with both synaptotagmin I and DAP-160 may regulate synaptic vesicle recycling by recruiting dynamin to a pre-fission complex.
Keywords: Drosophila, dynamin-associated protein of 160 kDa (DAP-160), Eps15 homology domain (EH domain), intersectin, stoned-B protein, Src homology 3 domain (SH3 domain)
Abbreviations: AP, adaptor protein; Cy, Curly; DAP-160, dynamin-associated protein of 160 kDa; EH domain, Eps15 homology domain; GST, glutathione S-transferase; MBP, maltose-binding protein; NMJ, neuromuscular junction; NPF motif, Asp-Pro-Phe motif; ORF, open reading frame; SH domain, Src homology domain; STNA, stoned-A; STNB, stoned-B; SYT-I, synaptotagmin-I; TBS, Tris-buffered saline; μHD, μ-homology domain
INTRODUCTION
The general model for synaptic vesicle recycling involves the binding of a heterotetrameric adaptor protein complex (AP2) to the nascent recycling vesicle, and the recruitment of clathrin to the membrane patch to be endocytosed. The clathrin-coated bud requires other auxiliary proteins, including Eps15, synaptojanin, endophilin and the GTPase dynamin, to complete the fission reaction and release the vesicle into the cytoplasm. Other mechanisms for synaptic vesicle recycling have been suggested. Ceccarelli and Hurlbut [1] proposed a rapid clathrin-independent endocytic mechanism, known as ‘kiss and run’ (for review, see [2]). Recent studies indicate that rapid endocytic mechanisms may be more the rule than the exception in certain mammalian CNS (central nervous system) neurons [3,4]. At the Drosophila NMJ (neuromuscular junction) there are at least two distinct pools of synaptic vesicles [5], and two recycling pathways have been identified [6]. Recently, a putative ‘kiss-and-run’ type of vesicle recycling has been uncovered in endophilin null mutants of Drosophila [7]. Whereas the molecular mechanism and the various proteins involved in clathrin-mediated synaptic vesicle recycling have been intensely studied, there have been few clues as to the molecular mechanisms underlying alternative modes of vesicle recycling. One possible component of a specific synaptic vesicle recycling pathway is the Drosophila stoned-B (STNB) protein. STNB is derived from the second ORF (open reading frame) of a dicistronic transcript expressed only in the nervous system [8,9]. The C-terminal region of STNB contains a region of homology to the μ-subunits [μHD (μ-homology domain)] of the AP complexes known to be involved in endocytic processes [10]. Mutations at the stoned locus affect synaptic transmission [8,11,12], interact with mutations at the shibire (dynamin) locus in vivo [8], and alter the rates of synaptic vesicle recycling [13,14]. The STNB protein is found to be constitutively associated with a population of synaptic vesicles, and this association has been shown to be due to a direct interaction between the μHD of STNB and the C2B domain of SYT-I (synaptotagmin-I) [15]. Within the STNB N-terminal region there are seven NPF (Asp-Pro-Phe) motifs that have been reported to be the sequence recognized by EH (Eps-15 homology) domains [16], as well as a putative binding target for SH3 (Src homology 3) domains. The product encoded by the first stoned ORF (STNA) also contains a proline-rich domain, and a series of DPF (Asp-Pro-Phe) motifs, binding sites for the ear domain of α-adaptin [17] and clathrin [18].
Human homologues of STNB, hSTNB and stonins 1 and 2, have been isolated [19,20]. However, these mammalian STNB-like proteins are expressed in both neuronal and non-neuronal tissues [19,20], whereas there is no evidence for stoned expression outside of the nervous system in Drosophila [8–10]. The mammalian stonin-2 protein also binds SYT-I, as well as EH-domain-containing proteins, such as Eps15 and intersectins [19–21]. These interactions have been shown to be dependent on the two NPF motifs present in stonin-2 [19]. In Drosophila a novel intersectin, DAP-160 (dynamin-associated protein of 160 kDa), was isolated due to its ability to bind to the proline-rich region of dynamin [22]. DAP-160 contains both SH3 and EH domains, is found, similarly to STNB, at synaptic terminals [22], and may therefore link the STNB protein to dynamin and explain the stoned–shibire genetic interaction [8]. In the present study, we show that a viable stoned mutant interacts in vivo with a Dap-160 hypomorph, that this mutation dramatically reduces the levels of the STNB protein without affecting the levels of STNA, and that it also reduces the in vivo levels of both DAP-160 and dynamin. We further show that the STNB protein can bind DAP-160 in vitro via both the DAP-160 EH domains, and one of the SH3 domains, and that DAP-160 can be co-immunoprecipitated with STNB from a synaptic vesicle-containing fraction.
EXPERIMENTAL
Drosophila strains and crosses
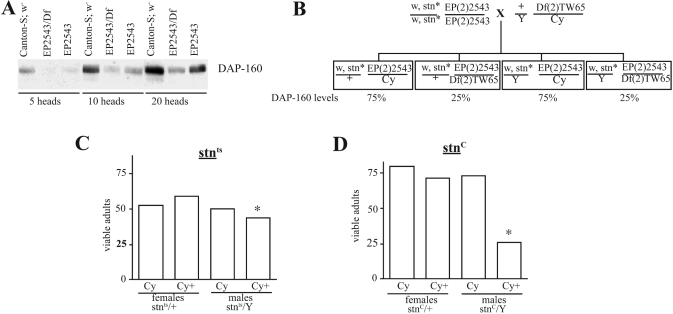
All Drosophila stocks were kept at 20 °C in a 12-h light–dark cycle. The stoned-temperature-sensitive (stnts) and stoned-C (stnC) mutant alleles have been described previously [8,15,23]. Both stoned strains carried the white-eyed (w) mutation, and the w1118 strain was used as controls in all but the EP strain experiments where a white-eyed strain in the Canton-S genetic background was used. The EP strain, EP(2)2543, inserts the EP transposon in the 5′-untranslated region encoding the second exon of the Dap-160 transcript [24]. Df(2L)TW65 extends across the cytological region 38A1–39F1 and hence deletes the Dap-160 locus at 39A4. Both of these strains were obtained from Sean Sweeney (Department of Biology, University of York, York, U.K.). To generate the stnts and stnC flies in a EP(2)2543/Df(2L)TW65 background, w, stnts and w, stnC flies were first crossed to EP(2)2543 individuals and homozygous w stnts; EP(2)2543 and w stnC; E (2)2543 strains generated. These were then crossed to males carrying the Df(2L)TW65 balanced over a chromosome carrying the dominant marker Curly (Cy). The progeny resulting from this cross are shown in Figure 1, along with the anticipated DAP-160 levels in each class of progeny.
Figure 1. Generation of mutant stoned flies with a reduction in the levels of DAP-160.
(A) Quantitative estimates of DAP-160 levels were determined for the various strains. 5, 10 and 20 heads from a white-eyed Canton-S ‘wild-type’ strain, homozygous EP(2)2543 flies and EP(2)2543/Df(2)TW65 heterozygotes were homogenized directly into SDS/PAGE sample buffer, centrifuged to remove particulates, and loaded on to SDS/PAGE (7% gel). The resulting Western blot was then probed with anti-DAP-160 antibody showing that EP(2)2543 reduced DAP-160 levels by 50% and EP(2)2543/Df(2)TW65 to 25% of the wild-type levels. (B) A diagram of the crosses performed to produce flies that carry a stoned mutation and have different levels of DAP-160. Only the males exhibit the stoned phenotypes and one group of these males will also have 25% of the normal DAP-160 levels. (C) The relative viability of the progeny from the cross outlined in (B) using female parents that are w stnts; EP(2)2543 homozygotes were determined. The asterisked group of flies are the w stnts/Y; EP(2)2543/Df(2)TW65 males. (D) The relative viability of the progeny from the cross outlined in (B) using female parents that are w stnC; EP(2)2543 homozygotes. The asterisked group of flies are the w stnC/Y; EP(2)2543/Df(2)TW65 males.
The comparison of the phenotype of the males that do not exhibit the Cy phenotype (Cy+), as compared with those that carry the Cy chromosome, is indicative of any interaction that might be occurring between the stoned allele and the reduced level of DAP-160.
Preparation of head extracts, Western blotting and overlays
Heads were collected from flies frozen in liquid N2 and stored at −80 °C until required.
The P1 (1000 g pellet), P2 (25000 g pellet), P3 (125000 g pellet) and S3 (final supernatant) fractions were prepared from Drosophila head homogenates as described previously [15] using Hepes buffer (10 mM Hepes, pH 7.4, 1 mM EGTA, 0.1 mM MgCl2 and 1 mM PMSF). The buffer-to-tissue ratio was 10:1(v/w). The P1 and P2 pellets were resuspended in half of the original homogenization volume and the P3 pellet in one quarter of the original volume. The resulting fractions all had approximately equal protein contents. Samples from different strains were matched and if the protein levels for any particular fraction differed by more than ±5% they were discarded. Fractions were loaded on to SDS/PAGE in the proportion 2:1:4 (P1 and P2/P3/S3). The total amount of any protein identified on Western blotting therefore could be represented as the simple sum of that protein in each of the fractions. The levels of protein identified by Western blotting were quantified by scanning the film and using MacBAS program (version 2.1, Fuji Photo Film Co.). Glycerol gradients were also prepared and run as described previously [15] using an S1 (1000 g supernatant) fraction loaded on to the gradient. Where individual fly heads were required, the flies were briefly anaesthetized using carbon dioxide. The heads were removed individually with a scalpel and collected in a 1.5-ml Eppendorf tube on ice. They were immediately homogenized in SDS/PAGE loading buffer in the same tube, using a plastic pestle, placed in a boiling water bath for 5 min and then centrifuged at 13000 g for 5 min. The resulting supernatant was loaded on to SDS/PAGE (7% gels). Western blots were generated by electroblotting on to nitrocellulose. The nitrocellulose was blocked in 5% skimmed milk in TBS (Tris-buffered saline) for 1 h followed by addition of the primary antibodies and incubation at 4 °C overnight. The blots were washed in TBS/0.5% Nonidet P40 as described previously [15], and secondary horseradish-peroxidase-linked anti-rabbit antibodies (Promega) were added for 1 h at room temperature. The blots were developed using the ECL® method (Pharmacia Biotech) according to the manufacturer's instructions.
The overlay method was as described previously [22]. The GST (glutathione S-transferase)–DAP-160(EH2) or GST–DAP-160(SH3C) fusion proteins (5 μg/ml) were added to nitrocellulose blots that had been blocked with 5% skimmed milk in TBS and incubated at 4 °C overnight. The blot was washed and then probed first with rabbit anti-GST antibodies (Chemicon) at room temperature for 1 h, and finally horseradish-peroxidase-linked anti-rabbit antibodies. The blots were developed as described above.
Fusion constructs
The MBP (maltose-binding protein)–STNB(C-terminus) fusion construct was prepared as described previously [15], and comprised residues 642–1262 of the STNB protein fused to MBP. Construction of the truncated C-terminal STNB protein (clone lk232) involved the insertion of the XbaI–PstI fragment of p47Z7 [10] into the pMALC2 vector (New England Biolabs). As the XbaI site in p47Z7 is out of frame with the MBP, this produces a 50-kDa fusion protein, but with no STNB amino acid sequence. To bring this back into frame the pMAL construct was then digested with XbaI and SpeI to remove the intervening 480-bp fragment and give the correct STNB reading frame to produce an MBP fusion that includes amino acid residues 642–692 of STNB (clone lk233). The MBP–STNB(N-terminus) protein was produced by PCR using a primer that inserted an EcoRI site immediately 5′ to the start codon of the STNB ORF (5′-GAATTCGAAATGGCGAATCCC-3′) and a second primer that covered the first XbaI site in the STNB ORF ending at nucleotide 4047 (5′-GGAAAATCTAGACCGGTG-3′). The PCR product was cloned into pGEM-T (Promega) and then the EcoRI–XbaI fragment was subcloned into pMalC2 (New England Biolabs). The fusion protein thus produced comprised amino acid residues 1–480 of STNB fused to MBP. The GST–Dap160(EH1) and GST–Dap160(EH2) constructs and the various GST–SH3 constructs [22] were kindly provided by Dr Jack Roos (Neurogenetics, Inc., La Jolla, CA, U.S.A.). Purification of the recombinant GST-fusion protein was carried out as described by the manufacturer (Amersham Pharmacia) and as described previously [22]. The GST protein was obtained from the pGEX 4T-1 vector (Amersham Pharmacia).
Protein–protein interaction experiments
All GST-fusion proteins were expressed according to the manufacturer's instructions and bound to glutathione resin (Amersham Pharmacia). Limiting amount of resin was added to the lysates to ensure saturation of the resin with the GST-fusion proteins. When the GST-fusion proteins were first purified, the resin-bound material was eluted with 20 mM glutathione and dialysed against TBS. The N- and C-terminal MBP–STNB fusion proteins were either left bound to the amylose resin or purified by eluting with 10 mM maltose, as suggested by the manufacturers (New England Biolabs), and dialysed against TBS. The integrity of the fusion proteins bound to resin was determined by running a fraction of the resin-bound fusion proteins on SDS/PAGE (10% gel) and staining the gel with Coomassie Blue. The amount of each fusion protein bound to the resin was quantified using the Bio-Rad protein determination system. The amounts of resin-bound protein and purified proteins used in the binding assays were as indicated in the Figure legends.
In the experiments in which the MBP–STN fusion proteins were pulled down from Escherichia coli lysates, the harvested cells were resuspended in TBS at one-tenth of the original culture volume. The cells were then lysed by freeze–thawing, sonicated and clarified by centrifugation at 25000 g for 15 min. The lysates were subjected to SDS/PAGE, Western blotted and probed with anti-MBP antibodies. Lysates were then diluted to give approximately equal amounts of MBP-cross-reacting material and 100 μl of the lysates was then added to the binding assay.
All binding was carried out in a total volume of 500 μl of TBS and, where indicated, either 1 mM EGTA or 100 μM CaCl2 was included. The binding assays were performed at 4 °C overnight on a rotating wheel. The resin was then collected by centrifugation and washed five times with 1 ml of cold TBS (or TBS/EGTA; TBS/CaCl2). The GST-fusion proteins were eluted with 50 μl of 20 mM glutathione, and the MBP-fusion proteins with 50 μl of 10 mM maltose. Samples (10 μl) were prepared and subjected to SDS/PAGE (10% gel), Western blotted and probed with either anti-MBP/anti-STNB sera or anti-GST antibodies.
Antibodies
The rabbit antiserum used in these experiments was raised against an MBP–STNB fusion protein. This comprised an N-terminal fragment generated from the insertion of an XhoI–XhoI fragment from p95Z7 [10] into pMALc2, resulting in the fusion of residues 53–352 of STNB to MBP. The antiserum was found to contain antibodies specific to the N-terminal region of STNB using a GST-fusion protein constructed using the same XhoI–XhoI fragment in pGEX-4T1, as well as anti-MBP antibodies (results not shown). The IgG fractions from this antiserum, as well as a non-specific antiserum, were affinity-purified using Protein A–Sepharose (Amersham Pharmacia). This antiserum, or the purified IgG fraction generated from it, was used in all of the Western blot analyses where the STNB protein was identified, and in all of the binding assays. Two other antibodies raised against STNB, a C-terminal specific antibody [10] or an antibody raised against a mixture of an N-terminal and C-terminal peptide [15], were used for the specific identification of the truncated STNB present in the stnC mutant extracts. The affinity-purified anti-dynamin, anti-DAP-160 and anti-SYT-I antibodies were as described previously [15,22], and were supplied by Jack Roos (Neurogenetics, Inc., La Jolla, CA, U.S.A.) and Regis B. Kelly (Institute of Quantitative Biomedical Research, CA, U.S.A.). Anti-GST antibodies were purchased from Chemicon.
Co-immunoprecipitation experiments
A P3 fraction containing approx. 800 μg of total protein was mixed with 100 μg of Protein A-purified IgG from anti-STNB and non-specific sera in a final volume of 400 μl of Hepes buffer as described above. After 2 h of mixing at 4 °C, the mixture was diluted to 2.5 ml in Hepes buffer, and the P3 fraction was re-precipitated by centrifugation at 125000 g for 1 h. The pellet was then re-homogenized in Hepes buffer containing 1% Triton X-100 and 50 μl (bed volume) of Protein A–Sepharose was added. The Protein A–Sepharose was collected by centrifugation, washed 5 times in 1.5 ml of Hepes buffer, resuspended in SDS/PAGE loading buffer and boiled. The supernatants were then loaded on to SDS gels for Western blotting and probed for STNB, DAP-160 and dynamin.
RESULTS
Genetic interaction between the viable stoned mutants and Dap-160
The strain of flies EP(2)2543 has a P element inserted in the 5′ untranslated region of the Dap-160 transcript [24]. Flies homozygous for this P element insertion are quite normal. However, when the levels of DAP-160 protein in these homozygous flies are measured by homogenizing fly heads straight into SDS/PAGE sample buffer, Western blotting and probing with anti-DAP-160 antibodies, it can be seen that the insertion mutation reduces the DAP-160 levels by 50% (Figure 1A). By placing the EP(2)2543 chromosome over a deficiency that uncovers the Dap-160 locus, the level of DAP-160 protein in fly heads is reduced to 25% of the normal level (Figure 1A). We asked if this reduction in the DAP-160 levels in any way compromised the viability of the stnts and stnC mutants. Given that the stnts mutation interacts with mutations at the shibire (dynamin) locus [8], and that DAP-160 is a major binding partner for dynamin, we anticipated that an interaction might occur between this stoned allele and any genotype that reduced the levels of DAP-160. Crosses were performed (Figure 1B) that allowed the effects of the reduced levels of DAP-160 on stnts and stnC flies to be determined. Flies homozygous for the both the stnts or stnC and the EP(2)2543 mutations were no less viable than the original stnts mutation alone, nor did the flies show any gross behavioural abnormalities, beyond those attributable to the stoned mutations. When these doubly homozygous females were then crossed to the deficiency bearing strain, half of the resulting males will carry the stnts or stnC mutation and have only 25% of the normal DAP-160 levels. The other half of the male population will have normal (≥75%) DAP-160 levels (Figure 1B). The stnts males with reduced levels of DAP-160 eclosed in the expected numbers (asterisk in Figure 1C), but were somewhat late emerging and extremely uncoordinated as adults, dying within 48 h.
In all previous cases where a second mutation has been reported to interact with the stoned locus, the interaction has been allele specific, and has been with the stnts allele [8,14,15]. It was surprising, therefore, to find an interaction between the stnC mutation and the mutant combination reducing the level of DAP-160 to 25%. Survival of the doubly mutant males that derive from these crosses is about 35% of the EP(2)2543/+ heterozygous male siblings (asterisk in Figure 1D). The surviving males are sedentary, extremely uncoordinated and exhibit an extreme stress-sensitive phenotype. They die within 3 days of eclosion.
Early termination of translation of the STNB ORF in the stnC mutation
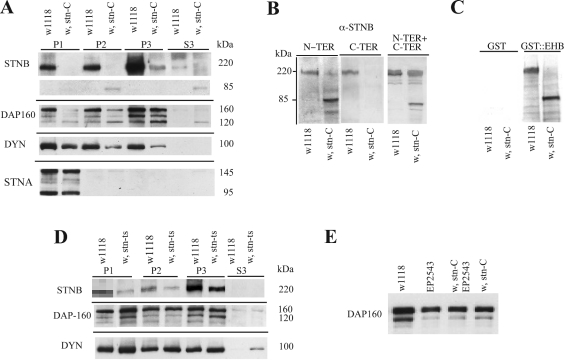
While the sequence alteration in the stnts mutation is known to be a lysine to methionine substitution in the STNA ORF [15], the nature of the stnC mutation had yet to be defined. To understand the nature of the interaction between stnC and the Dap-160 mutant, it became imperative to determine the nature of the stnC mutation. We sequenced PCR-derived stoned genomic DNA from stnC flies, and found no differences between the wild-type and the stnC sequences in the STNA ORF, but found a C→T substitution in the STNB ORF. This results in a UGA nonsense codon in place of arginine (residue 415) in the STNB ORF. This means that the STNB protein would terminate prematurely, after the first five NPF motifs, but before the μ-HD. To confirm this, Western blots of fractionated head homogenates from wild-type and stnC flies were probed with anti-STNB antibodies raised against an N-terminal fragment of STNB (see the Experimental section). As can be seen in Figure 2(A) the STNB protein runs on SDS/PAGE with an estimated molecular mass of 220 kDa. From sequence analysis the expected molecular mass is 135 kDa, suggesting that, like STNA and DAP-160, the STNB protein has an anomalous mobility on SDS/PAGE. More importantly this 220 kDa protein, normally present in extracts from fly heads, is now absent in the P1 and P2 fractions from stnC flies and present at extremely low levels (>10%) in the P3 fraction. A new cross-reacting protein of 85 kDa is present in the stnC extracts, and whereas the 220 kDa protein is not found in the soluble S3 fraction, this 85 kDa species is present in the S3 fraction. The expected molecular mass of the STNB fragment produced in the stnC mutant is 43.5 kDa, however, the new cross-reacting protein has an SDS/PAGE mobility corresponding to twice the expected size of this protein fragment, suggesting that the N-terminal region of the STNB protein may be responsible for the anomalous mobility of STNB on SDS/PAGE. One might have expected that the presence of a UGA nonsense codon would completely remove the full-length STNB protein, so the presence of even low levels of the 220 kDa cross-reacting STNB protein in stnC head extracts was unexpected. It could be that there is a small amount of translational read-through of the UGA codon in stnC flies. Previous reports have indicated that a UGA codon, followed by a uridine, is a poor translation termination signal [25], and there are at least two other examples of read-through of nonsense codons in Drosophila mutants producing full-length proteins [26,27].
Figure 2. Levels and distribution of STNB, DAP-160 and dynamin in stoned mutants.
(A) Equivalent volumes (see the Experimental section) of P1, P2, P3 and S3 fractions from homogenates of heads of w stnC and w1118 control flies, were subjected to SDS/PAGE (7% gel), Western blotted and probed with anti-STNB antibodies. The expected >220 kDa protein is absent from all but the P3 fraction of w stnC. A smaller 85 kDa protein can be seen in the P2 and S3 from w stnC flies. The same fractions were also probed with anti-DAP-160, anti-dynamin (DYN) and anti-STNA antibodies. A very obvious reduction in the levels of DAP-160 and dynamin is observed, but there is little effect on STNA levels. (B) For each lane, six heads from w stnC and w1118 control flies were homogenized directly into SDS/PAGE sample buffer, boiled and centrifuged (10000 g for 5 min) to remove particulate material, and loaded on an SDS/7% polyacrylamide gel. The resulting blot was then probed with three preparations of anti-STNB antibodies, those described in the present report and raised against the N-terminal fragment of STNB, those raised against the C-terminal region of the protein [10] or raised against a mixture of an N-terminal and C-terminal peptide [15]. (C) A blot the same as that for (B) was prepared and overlayed with either GST alone or a GST–DAP-160(EH2) fusion protein. The overlay blots were developed with anti-GST polyclonal antibodies. This shows that both N-terminal anti-STNB antibodies and the GST–DAP-160(EH2) overlay method both identify a 85 kDa truncated protein in the w, stnC extracts that is absent when using the C-terminal-specific antibodies. (D) P1, P2, P3 and S3 fractions from homogenates of heads from w stnts and w1118 control flies were run on SDS/PAGE (7% gel) as in (A), Western blotted and probed with anti-STNB, anti-DAP-160 and anti-dynamin antibodies. While there is a reduction in the levels of STNB in w stnts heads, both DAP-160 and dynamin are at normal levels and there is little difference in their distribution. (E) Heads (five) from w1118, EP2543, w stnC:EP2543 and w stnC were treated as for (A). The resulting Western blot was then probed with anti-DAP-160 antibodies. All of the mutant flies showed the same level of DAP-160, which corresponds to approx. 50% of the wild-type levels.
We also asked whether this truncation of STNB in any way affected the quantity/distribution of DAP-160 and dynamin in these fractions. Similar blots were probed with anti-DAP-160 and anti-dynamin antibodies. There was a 50% reduction in the levels of DAP-160 observed in the stnC extracts (Figure 2A), and as can be seen, the 120 kDa form of DAP-160 is more prevalent in the stnC extracts. There is also much more of this 120 kDa species in the stnC S3 fraction than in the equivalent wild-type S3 fraction. When the blot was probed for dynamin (Figure 2A) there was also a greater than 50% reduction in the levels of dynamin in all but the P1 fraction of stnC extracts as compared with wild-type, but dynamin remains absent from the S3 fraction in stnC. Finally the blots were probed with anti-STNA antibodies. STNA is found almost exclusively in the P1 fraction. This did not change in the stnC mutant, and the levels of STNA were comparable in the wild-type and stnC extracts (Figure 2A).
We sought further evidence in support of the conclusion that the novel 85 kDa anti-STNB cross-reacting protein in stnC extracts (Figure 2A) is the N-terminal truncated fragment of STNB. A Western blot of whole head fractions from wild-type and stnC mutant flies was probed with three different anti-STNB antibodies (Figure 2B). Whereas the antibody prepared against the N-terminal STNB fragment shows the wild-type and stnC truncated proteins, antibodies raised against a C-terminal fragment of STNB [10] only gave a reactive species at 220 kDa in the wild-type extracts. A third antibody raised against a mixture of N- and C-terminal peptides [15] also showed both the full-length and truncated versions of STNB, but also showed non-specific cross-reactivity with a protein species of greater apparent molecular mass than the full-length STNB. A further indication that the truncated protein is indeed the N-terminal fragment of STNB comes from a blot similar to those in Figure 2(B) used in an overlay assay with the GST–DAP-160(EH2) fusion protein used as a probe. The GST–DAP-160(EH2) fusion interacts with the N-terminal region of STNB (see Figure 3). Figure 2(C) shows that the overlay assay duplicates the pattern generated using anti-STNB antibodies. While the EH2 domain of DAP-160 interacts with the full-length STNB in wild-type fly extracts, it also interacts with the 85 kDa protein in the stnC extracts.
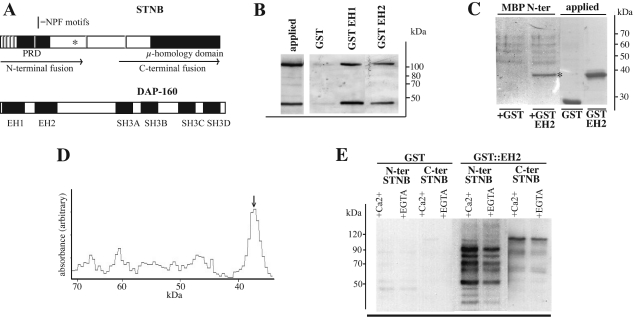
Figure 3. DAP-160 EH domains interact with recombinant STNB.
(A) Representation of the STNB and DAP-160 proteins, indicating the position of the NPF tripeptide sequences, the proline-rich domain (PRD) and the μ-HD of STNB. The EH and SH3 domains of DAP-160 are shown. Also indicated are the regions included in the two fragments of STNB that are expressed as MBP fusions. The asterisk denotes the position of the stop codon in the stnC mutation. (B) Purified MBP–STNB(N-terminus) fusion protein yielded a protein in excess of 100 kDa, as well as a proteolytic product of 50 kDa, as determined by Coomassie Blue staining of an SDS gel (applied sample). Glutathione resin (25 μl) to which GST (25 μg) or GST–DAP-160(EH1) (40 μg) or GST–DAP-160(EH2) fusion protein (28 μg) was bound, was incubated in TBS with 10 μg of the purified MBP–STNB fusion protein. The bound material was eluted with glutathione, and one fifth of the sample was subjected to Western blot analysis using anti-MBP antibodies as the primary antibody. The relative affinity of each EH domain was seen to be approximately equivalent when normalized for the GST–EH domain protein bound to the resin. (C) Amylose resin (25 μl) to which 8 μg of the MBP–STNB N-terminal fragment is bound was incubated with 25 μg of either GST alone or GST–EH2 fusion protein for 4 h at 4 °C. The MBP was eluted with maltose and the mixture run on SDS/PAGE and the gel stained with Coomassie Blue. Again the MBP–STNB fusion expressed in E. coli was seen to fragment. GST and GST-fusion proteins (12.5 μg of each) were run as controls. The asterisk indicates the GST–EH2 protein eluted with maltose. (D) Densitometric scan of the +GST–EH2 lane from the gel in (C). The arrow indicates the GST–EH2 fusion protein. Integration under these curves along with average estimates of the molecular mass of the MBP–STNB N-terminal fragments suggests an approximately equimolar level of the two protein species. (E) Glutathione resin to which either GST (25 μg) or the molar equivalent amount of the GST–DAP-160(EH2) fusion protein (40 μg) was bound, were incubated with E. coli lysates expressing equivalent amounts (as estimated by the signal generated with anti-STNB antibodies on Western blots) of either the N-terminal or C-terminal fragments of STNB as MBP-fusion proteins. The binding was carried out in TBS with either 100 μM Ca2+ or 1 mM EGTA. The bound material was eluted with glutathione and subjected to Western blot analysis using anti-MBP antibodies as the primary antibody. Exposure times for the N-terminal- and C-terminal-containing lysates were 10 s and 1 min respectively. This indicates the relative binding capacity of the STNB N- and C-terminal fragments.
The STNB, DAP-160 and dynamin levels and their distributions were also determined in the stnts strain. This also acted as a control, as both the stnts and stnC mutants were isolated in the same Oregon-R genetic background. The levels of STNB are reduced in the stnts mutant by 40–50% (Figure 2C), however, in contrast to stnC, the levels of both DAP-160 and dynamin are slightly greater than in the wild-type, and some dynamin is found in the soluble (S3) fraction of stnts heads.
The reduction in the DAP-160 levels in the stnC mutant suggested a possible explanation for the reduced viability of the stnC/Y; EP(2)2543/Df(2)TW65 males. The levels of DAP-160 produced by the EP(2)2543/Df(2)TW65 combination may be further reduced by the presence of the stnC mutation to a critical level. To test this, extracts were made from ten heads of the wild-type (w1118), w, stnC, EP(2)2543 and the w, stnC; EP(2)2543 double mutant, run on SDS/PAGE, blotted and probed with anti-DAP-160 antibody (Figure 2D). As expected, both the stnC mutation and the EP(2)2543 insertion reduced DAP-160 levels by 50%. However, the double mutant combination did not further reduce the levels of DAP-160, indicating that the reduction is not additive, and that the reduced viability of stnC/Y; EP(2)2543/Df(2)TW65 males is not due to a further reduction in DAP-160 levels brought about by the stnC mutation.
EH domains of DAP160 interact with the STNB protein in vitro
The genetic interaction between the Dap-160 mutant and stnC suggested that the DAP-160 EH domains are capable of binding STNB. This was partially confirmed by the overlay assay (Figure 2B), which also indicated that STNB is the major binding-partner in head extracts for the DAP-160 EH2 domain. To determine if the EH domains from DAP-160 show any specificity, a fusion construct was made corresponding to MBP fused to the N-terminal third of STNB (residues 1–481). This fusion protein includes five of the NPF motifs and the proline-rich region of STNB (Figure 3A). This protein is very unstable when expressed in E. coli; however, one purified MBP–N-terminal STNB fusion protein comprised only two forms; one greater than 100 kDa, and a proteolytic product with a molecular mass of 50 kDa. Assuming that the 50 kDa form contained all of the MBP (42 kDa), then this proteolytic fragment would contain only 8 kDa of the N-terminal region of STNB. However, as four of the NPF motifs are present within the first 45 residues of STNB, it might be expected that this smaller fragment would retain binding capacity for the DAP-160 EH domains. Binding assays were performed using this purified MBP–STNB fusion protein with each of the DAP-160 EH domains. The glutathione-eluted material was subjected to SDS/PAGE and probed with anti-MBP serum (Figure 3B). Both of the EH domains of DAP-160 bound to approximately equal amounts of the MBP–STNB fragments. GST alone failed to interact with the MBP–STNB fusion protein. This experiment was repeated in reverse with higher concentrations of both the STNB fusion protein and GST–DAP-160(EH2). The MBP–STNB protein was adsorbed on to amylose resin and mixed with GST–DAP-160(EH2). The complex was then eluted with maltose, run on SDS/PAGE and Coomassie Blue stained (Figure 3C). Although the MBP–STNB fusion had once again broken down, from the densitometric scan (Figure 3D) it was seen that approximately equimolar amounts of the GST– DAP-160(EHB2) had bound to the MBP–STNB fusion protein.
A second MBP–STNB fusion protein was produced which comprised the C-terminal half of the protein (residues 642–1262) and included a single NPF motif and the μ-HD (Figure 3A). E. coli lysates expressing equivalent amounts of both the N-terminal and C-terminal STNB–MBP fusion proteins were incubated with the GST–DAP-160(EH2) fusion protein bound to glutathione resin. Figure 3(C) shows the results of these pull-down experiments. It should be noted that neither of the MBP–STNB fusion proteins is particularly stable when expressed in E. coli (Figure 3C), however, it is clear that there is a very strong interaction between the EH2 domain of DAP-160 and the N-terminal fragment of STNB in these crude lysates. Although to a lesser extent, the EH2 domain also binds the C-terminal fragment. This probably reflects the single NPF in this fragment as compared with the five NPF motifs present in the N-terminal fragment.
Within the EH domains of DAP-160 there are presumptive EF-hand Ca2+-binding domains. The effects of the Ca2+ chelator EGTA on the DAP-160(EH-2)–STNB interactions were also studied. As compared with the binding in the presence of 100 μM Ca2+, 1 mM EGTA did appear to reduce the binding of the DAP-160 EH2 domain to the STNB-fusion proteins, but only by about 25%. Although Ca2+ may be having an effect on the binding equilibrium of the DAP-160(EH2) to STNB, there is no great effect of Ca2+ on this interaction.
SH3C domain of DAP-160 also interacts with the STNB protein
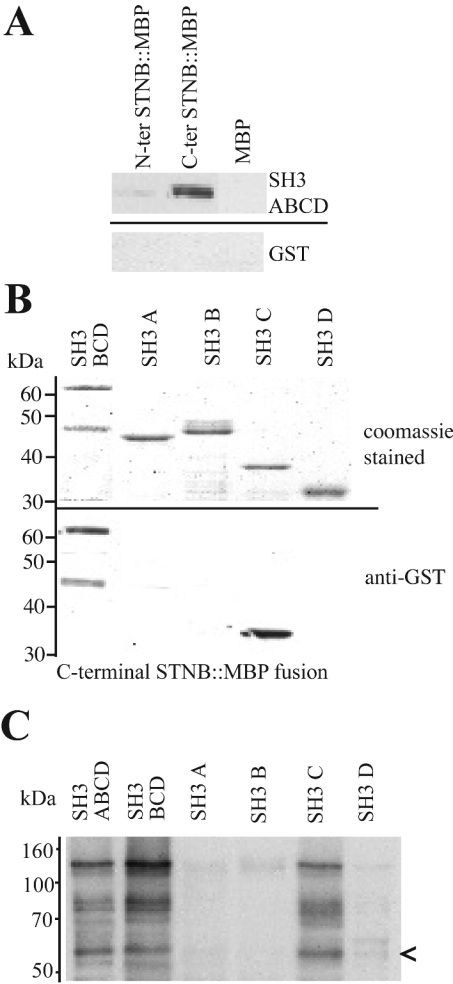
As previously mentioned, the N-terminal region of the STNB protein also contains a large proline-rich domain. Between amino acids 101 and 300, proline constitutes 25% of the residues, and this region contains a number of motifs that might bind to SH3 domains. A GST–DAP-160 fusion protein containing all four SH3 domains was used in pull-down experiments using equivalent amounts of the MBP–STNB N- and C-terminal fusions bound to amylose resin. Surprisingly, although the MBP–STNB(N-terminus) pulled down a small amount of the GST–DAP-160(SH3ABCD) fusion, the major interaction was with the MBP–STNB(C-terminus) fragment (Figure 4A).
Figure 4. DAP-160 SH3 domains interact with recombinant STNB.
(A) Approximately molar equivalent amounts of MBP alone (20 μg) and MBP-fusion protein N- and C-terminal STNB fragments (50 μg) of STNB were bound to amylose resin and mixed with 50 μg of purified GST–DAP-160 SH3 domain protein containing all four SH3 domains. The resulting material, after washing, was eluted, and one tenth of the eluate was run on SDS/PAGE, Western blotted and probed with anti-GST antibodies. The C-terminal fragment of STNB preferentially interacts with the DAP-160 SH3 domains. (B) An experiment, as outlined in (A), using the MBP–C-terminal STNB fragment (50 μg) bound to amylose resin, but with separate GST–DAP-160 fusion proteins consisting of GST–SH3BCD, GST–SH3A, GST–SH3B, GST–SH3C and GST–SH3D (50 μg of each/assay). The upper panel shows the Coomassie Blue stained gel of the applied fusion proteins (10 μg of each), and the lower panel shows the resulting Western blot, one fifth of the total elution from the resin, probed with anti-GST antibodies. The SH3C domain specifically interacts with STNB. (C) The reverse of the experiment shown in (B) where each of the GST–SH3 domain fusion proteins (50 μg of each) are bound to glutathione resin and used to pull down STNB MBP–STNB(C-terminus) from an E. coli lysate. The blot is probed with anti-MBP antibodies. The arrow indicates a proteolytic product of 58 kDa that is still able to bind to the SH3C domain.
It has been shown that the SH3 domains from DAP-160 show differences in their affinity for binding partners [22]. To investigate which of the four SH3 domains of DAP-160 contributed to the binding of this fragment of STNB, equivalent amounts of GST fusions corresponding to the SH3BCD and the individual SH3 A, B, C and D domains were mixed with the MBP–STNB-(C-terminus) fusion, bound to amylose resin, washed, eluted and resolved on SDS/PAGE (10% gel). The blot of this gel was then probed with anti-GST antibodies. As can be seen from Figure 4(B), the SH3C domain appeared to show a specific interaction with the MBP–STNB(C-terminus) fusion protein. This specificity was confirmed in the reverse pull-down experiment where the GST–SH3 domain fusions were bound to resin and used to pull down the MBP–STNB(C-terminus) fusion from an E. coli lysate (Figure 4C).
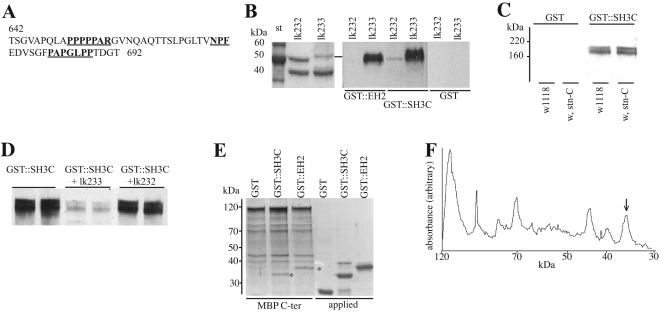
The MBP–STNB(C-terminus) fusion is unstable in lysates and showed a 58 kDa breakdown product that can be pulled out of the lysate by the DAP-160 SH3 domains (arrow in Figure 4C). The fragment is also purified by amylose affinity chromatography and therefore contains MBP. This suggests that the SH3 domain-binding site in STNB lies in the sequence between residues 642 and 750. Analysis of this sequence indicates the presence of a Class 1 SH3 domain-binding site (PPPPAR), corresponding to residues 651–657 (Figure 5A). The fragment of STNB corresponding to residues 642 to 692, containing one NPF motif as well as the putative SH3-binding site, was cloned into pMAL (clone lk233) to produce a MBP fusion protein of 51 kDa. Fusion proteins from this and a control clone (clone lk222), producing a ‘nonsense’ peptide of approximately the same size, were purified and the overlay method was used to determine if the EH2 and SH3C domains of DAP-160 were able to bind. Both the SH3C and EH2 domains bound to this STNB-fusion protein but not to the ‘nonsense’ peptide fusion (Figure 5B).
Figure 5. Further delineation of the interaction of the DAP-160 SH3C domain with STNB.
(A) Sequence of part of the C-terminal fragment of STNB indicating the possible SH3 binding sites (underlined). The MBP–STNB fusion construct starts at amino acid residue 642 of STNB. (B) Two MBP fusion constructs lk232 and lk233 contained an out-of-frame nonsense amino acid sequence or residues 642–692 respectively. These fusion proteins were purified, and both showed the expected fusion protein (upper band), as well as free MBP (lower band). The panel on the left-hand side is the Coomassie Blue stained gel. The blots containing these two fusion proteins were overlayed with GST–DAP-160(EH2) fusion protein, with GST–DAP-160(SH3C) fusion protein or with GST alone, and then developed with anti-GST antibodies. The fragment from lk233 binds both GST fusions, but not GST. (C) Overlay of the GST–SH3C fusion on Drosophila head fractions. The experimental method is identical to that described for (B), but the blot contains crude extracts from the heads of wild-type and stnC flies showing that the overlay is not identifying STNB on these blots. These proteins have been tentatively identified as clathrin [22]. (D) Blots of head fractions as described in (C) were overlayed either with GST–SH3 alone or with GST–SH3 to which had been added 10-fold molar excess of either the lk232 or lk233 fusion proteins. This shows that the region of STNB encoded by the clone lk233 can competitively inhibit the binding of the SH3C domain to the presumptive clathrin. (E) Amylose resin (25 μl) to which 28 μg of MBP–STNB C-terminal fragment is bound was incubated with 50 μg of GST, GST–SH3C or GST–EH2 at 4 °C for 4 h, eluted with maltose and then loaded on to SDS/PAGE along with 12.5 μg of GST and each of the GST fusion proteins as standards. The gel was stained using Coomassie Blue. The asterisks indicate the GST-fusion proteins bound to the MBP-fusion protein. (F) Densitometric scan of the GST–EH2 lane from (E). The arrow indicates the peak corresponding to the GST–EH2 protein. Integration under this curve, and making allowances for the change in molecular mass associated with the proteolytic products of the MBP–STNB fusion, gives an approximate molar ratio of 1:4 for GST–EH2/MBP–STNB. A similar value is calculated for the binding of the GST–SH3C domain.
Using this overlay method, the SH3C domain has been shown to bind to proteins in Drosophila head extracts that have been tentatively identified as clathrin [22]. To determine if this protein could possibly be STNB, we assessed binding of the SH3C domain to extracts from both the wild-type and stnC mutant flies. No difference was found between these strains (Figure 5C). This suggests that the SH3C domain is unable to bind to the complete STNB protein under the overlay conditions. However, addition of the fragment of STNB to the GST–SH3C domain fusion very effectively inhibits the binding of the SH3C domain to the presumptive clathrin (Figure 5D).
We have also attempted to compare the relative binding capacity of the C-terminal domains of STNB to bind the DAP-160 EH2 and SH3C domains. The C-terminal STNB–MBP fusion was bound to amylose resin and incubated with GST, GST–SH3C or GST–EH2. The maltose-eluted material was run on SDS/PAGE and Coomassie Blue stained (Figure 5E). In contrast to the STNB N-terminal fragment (Figure 3C), the C-terminal STNB fragment bound considerably less of the GST–EH2 fragment, with an approximate molar ratio of 1:4 of EH2 to STNB (Figure 5F). However, as was the case with the overlays, both the SH3C domain and the EH2 domain of DAP-160 were bound to the same extent. This is considerably less than the N-terminal STNB fusion binding to the EH2 domain where the ratio was approximately equal (Figure 3D). As might have been expected, this indicates that the overall binding capacity of STNB for the DAP-160 EH2 domains is much greater than for the DAP-160 SH3C domain.
Co-fractionation and co-immunoprecipitation of STNB and DAP-160
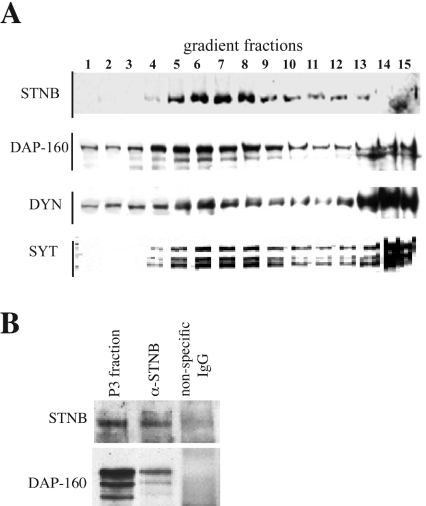
We have previously shown that immunoprecipitation of STNB co-precipitates intact synaptic vesicles and that on Triton X-100 treatment, only SYT-I remains associated with STNB [15]. We also showed that STNB and SYT-I co-sediment with a synaptic vesicle fraction on glycerol gradients [15]. It has also been shown that DAP-160 and dynamin form a high-molecular-mass complex and can be co-immunoprecipitated from a cytosolic fraction [22]. We therefore asked whether DAP-160 and dynamin might co-fractionate with the STNB/SYT-I-associated vesicle fraction. Figure 6(A) shows that both DAP-160 and dynamin are spread throughout the gradient when an S1 fraction from a Drosophila head homogenate is centrifuged on a 5–25% glycerol gradient, whereas STNB and SYT-I form a peak in the middle of the gradient corresponding to free synaptic vesicles. However, unlike dynamin, DAP-160 is also enriched in the STNB/SYT peak in the middle of the gradient. This data places DAP-160, at least partially, within the same biochemical compartment as STNB/SYT, and so mirrors their presynaptic subcellular co-localization. We have used a Triton X-100-treated P3 fraction, which contains synaptic vesicles and is enriched for both STNB and DAP-160, to immunoprecipitate STNB. When Western blotted and probed using anti-DAP-160 antibodies we observed that DAP-160 could be co-precipitated with the STNB protein (Figure 6B). However, we were unable to observe dynamin in these immunoprecipitated fractions.
Figure 6. Co-sedimentation of and co-immunoprecipitation of STNB and DAP-160.
(A) A 5–25% glycerol gradient on which was loaded S1 (1000 g supernatant) from homogenized wild-type fly heads. Fifteen fractions were collected, and each fraction run on SDS/PAGE (7% gel), Western blotted and probed separately with antibodies against STNB, DAP-160, dynamin (DYN) and SYT-I. Fraction 1 represents the top and fraction 15 the bottom of the gradient. The peak of SYT-I in fractions 5 through 8 represents free synaptic vesicles [15]. STNB co-sediments with this fraction, whereas DAP-160, and to a lesser extent dynamin, is enriched in these fractions. (B) A Triton X-100-solubilized P3 fraction (see the Experimental section) contains both STNB and DAP-160. Addition of anti-STNB antibodies, followed by Protein A–Sepharose to this fraction, precipitates both STNB and DAP-160. The amount of the immunoprecipitated fraction loaded on the gel is equivalent in volume to that shown in the P3 fraction. Whereas most of the STNB protein is immunopreciptated from the P3 fraction, only a small amount of the total DAP-160 is co-precipitated.
DISCUSSION
The impetus behind this study was the observation of a genetic interaction between stoned and shibire (dynamin) mutants [8,14]. Those results suggested a molecular interaction between the stoned and dynamin proteins. The identification of DAP-160 as a major binding partner for dynamin, the presence of seven NPF motifs in the STNB protein and two EH domains in DAP-160, made DAP-160 an ideal candidate as the link between the stoned and dynamin proteins. Not surprisingly, we have found that STNB is a high-affinity target for the DAP-160 EH domains in vitro. Moreover, the results of the overlay using the DAP-160 EH domains on fly head extracts indicated that the STNB protein represents the major binding partner for these EH domains. Curiously, the STNB protein also interacts in vitro with the SH3C domain of DAP-160. This domain has previously been shown to interact with a protein putatively identified as clathrin [22], and more recently with synaptojanin [28].
The in vivo genetic interactions that we have observed between the viable stoned mutants and the combination of alleles that reduces DAP-160, suggests that the observed in vitro interactions have some in vivo validity. This is further reinforced by the co-immunoprecipitation of STNB and DAP-160 from the synaptic vesicle (P3) fraction, as well as the observation that the levels of both DAP-160 and dynamin are markedly reduced in the stnC mutant. It is not clear whether this reduction in DAP-160 and dynamin reflects an alteration in the stability of these two proteins or in their regulation. Interestingly, the stnC mutant has also been shown to reduce the levels of SYT-I in head extracts by 50% [29]. It seems likely, therefore, that SYT-I, STNB and DAP-160 function as a complex.
A further similarity exists between the phenotype of the recently described Dap-160 mutants and some of the stoned mutants. At the larval NMJ of presumptive null and hypomorphic Dap-160 mutants numerous satellite boutons are observed [30,31]. This is a phenotype that had previously been reported for stoned mutants [9], although it is not seen in other synaptic vesicle recycling mutants, such as those affecting endophilin and synaptojanin [7,28]. These results suggest that the two proteins may be working together to regulate synaptic morphology, but whether the phenotype is due to a specific second role for these proteins, or merely a result of their failure to correctly recycle synaptic vesicles is unknown. It is also possible that the truncated fragment of STNB in the stnC mutant acts to titrate DAP-160 from its other functions.
What then is the relevance of the STNB–DAP-160 interactions? It has been shown that the STNB protein can be found bound to a subset of synaptic vesicles prior to exocytosis, via the interaction of the μ-HD of STNB and the C2B domain of SYT-I [15], and that stoned mutants alter the rate of vesicle endocytosis [13,14]. SYT-I is found to associate with syntaxin and SNAP-25, and is believed to regulate the dynamics of the fusion pore [32]. The interaction of DAP-160 with STNB, already bound to SYT-I in these ‘primed’ vesicles, may therefore be a mechanism by which dynamin, along perhaps with other proteins, is recruited to the pore region. This could lead to the closure and rapid recycling of this group of synaptic vesicles. In contrast, it has been shown that disruption of endophilin binding to dynamin inhibits vesicle fission and causes the accumulation of clathrin-coated pits [33]. This, along with the evidence from the Drosophila endophilin null mutants, suggests that the endophilin-dependent recruitment of dynamin mediates clathrin-dependent endocytosis [33,34]. The binding of DAP-160 to STNB and synaptic vesicles via synaptotagmin might then serve a function independent of clathrin-coated vesicle recycling. This is in apparent contrast with the presumptive mammalian counterpart of STNB, stonin-2, which has been shown to interact with the AP2 complex via a number of WXXF motifs present in the N-terminal region of stonin-2 [35] and has been implicated in the uncoating of synaptic vesicles that are recycling through the clathrin-dependent pathway [20]. The Drosophila STNB protein lacks these WXXF motifs and we have been unable to show any interaction between the Drosophila STNB protein and AP2, or more specifically α-adaptin. Furthermore, when recycling is blocked using the shibire mutant, STNB is still found associated with synaptic vesicles [15], indicating that the STNB interaction with vesicles is not dependent on their being recently recycled or their interaction with AP2. In the Drosophila endophilin mutants a number of synaptic vesicles remain that appear to recycle independently of endophilin/clathrin. It is presumably this residual recycling capability that allows the mutant larvae to hatch and survive to mid second instar. It is possible that the STNB protein is associated with these vesicles and its interaction with DAP-160 allows dynamin to be recruited in order to recycle this endophilin-independent vesicle pool. The ability of both STNB (present work) and synaptojanin [28] to bind to the same SH3 domain of DAP-160 may be a mechanism whereby DAP-160 bound to STNB excludes any interaction with synaptojanin. Such a mutually exclusive interaction could identify whether a vesicle is destined for recycling through the clathrin-dependent pathway or through an alternative pathway.
The premature termination of the STNB protein in the stnC mutant would mean that the μ-HD of STNB that normally interacts with SYTI and hence synaptic vesicles [15], is no longer present. In fact one might expect the stnC mutation to act as a functional null mutant. Yet these flies survive and have a relatively mild phenotype in that they are non-phototactic and respond to mechanical stimulus by being briefly incapacitated. The stnC mutation also reduces the quantal output from the larval NMJ by some 50%, but does not totally block synaptic transmission [12]. However, some of the other alleles of stoned are embryonic lethal and have a much greater effect on neuromuscular transmission [13]. This may mean that a small amount of translational read-through of the UGA termination codon in stnC flies is sufficient to greatly reduce the severity of the phenotype. That stnC is not a complete null mutation is reinforced by the recent observation that transgenic expression of STNB alone can rescue not only the stnC mutant phenotypes, but also the lethality of the more severe alleles [9], indicating that the transgenic expression of the STNB protein alone is sufficient to overcome embryonic lethality. This embryonic lethal phenotype of some of the stoned alleles [23] is in contrast with the phenotype of endophilin [33,34] or synaptojanin [36] null mutants which do not die until the second instar larval stage, and suggests that the stoned locus is involved in more than just the clathrin-dependent recycling of synaptic vesicles.
Although we cannot exclude the possibility that STNB is also involved in clathrin-dependent synaptic vesicle recycling, as has been suggested for mammalian stonin-2 [20,36], we believe that accumulating evidence points to STNB in Drosophila being associated with a population of synaptic vesicles that may recycle independently of the clathrin-mediated mechanism. Future investigations of the stoned proteins and their interacting partners may help elucidate the specific functions of STNB and STNA, and may identify the molecular processes that distinguish between the modes of synaptic vesicle recycling.
Acknowledgments
This work was supported by a grant from the NHMRC (National Health and Medical Research Council) (to L.E.K.). We thank S. Sweeney for the generation of the DAP-160 fly strains, and M. Ramaswami for discussion on read-through of nonsense codons. L.E.K. would also like to thank L. Clift O'Grady specifically, and other members of the Davis and Kelly labs at UCSF, for their hospitality during the course of some of these experiments.
References
- 1.Ceccarelli B., Hurlbut W. P. Ca2+-dependent recycling of synaptic vesicles at the frog neuromuscular junction. J. Cell Biol. 1980;87:297–303. doi: 10.1083/jcb.87.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palfrey H. C., Artalejo C. R. Vesicle recycling revisited: rapid endocytosis may be the first step. Neuroscience. 1998;83:969–989. doi: 10.1016/s0306-4522(97)00453-3. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi S. P., Stevens C. F. Three modes of synaptic vesicular recycling revealed by single-vesicle imaging. Nature (London) 2003;423:607–613. doi: 10.1038/nature01677. [DOI] [PubMed] [Google Scholar]
- 4.Aravanis A. M., Pyle J. L., Tsien R. W. Single synaptic vesicles fusing transiently and successively without loss of identity. Nature (London) 2003;423:643–647. doi: 10.1038/nature01686. [DOI] [PubMed] [Google Scholar]
- 5.Kuromi H., Kidokoro Y. Two distinct pools of synaptic vesicles in single presynaptic boutons in a temperature-sensitive Drosophila mutant, shibire. Neuron. 1998;20:917–925. doi: 10.1016/s0896-6273(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 6.Koenig J. H., Ikeda K. Synaptic vesicles have two distinct recycling pathways. J. Cell Biol. 1996;135:797–808. doi: 10.1083/jcb.135.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verstreken P., Kjaerulff O., Lloyd T. E., Atkinson R., Zhou Y., Meinertzhagen I. A., Bellen H. J. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell. 2002;109:101–112. doi: 10.1016/s0092-8674(02)00688-8. [DOI] [PubMed] [Google Scholar]
- 8.Petrovich T. Z., Merakovsky J., Kelly L. E. A genetic analysis of the stoned locus and its interaction with dunce, shibire, and Suppressor of stoned variants of Drosophila melanogaster. Genetics. 2001;133:955–965. doi: 10.1093/genetics/133.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estes P. S., Jackson T. C., Stimson D. T., Sanyal S., Kelly L. E., Ramaswami M. Functional dissection of a eukaryotic dicistronic gene: transgenic stonedB, but not stonedA, restores normal synaptic properties to Drosophila stoned mutants. Genetics. 2003;165:185–196. doi: 10.1093/genetics/165.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews J., Smith M., Merakovsky J., Coulson M., Hannan F., Kelly L. E. The stoned locus of Drosophila melanogaster produces a dicistronic transcript and encodes two distinct polypeptides. Genetics. 1996;143:1699–1711. doi: 10.1093/genetics/143.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly L. E. Altered electroretinogram transient associated with an unusual jump response in a mutant of Drosophila. Cell. Mol. Neurobiol. 1983;3:143–149. doi: 10.1007/BF00735278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stimson D. T., Estes P. S., Smith M., Kelly L. E., Ramaswami M. A product of the Drosophila stoned locus regulates neurotransmitter release. J. Neurosci. 1998;18:9638–9649. doi: 10.1523/JNEUROSCI.18-23-09638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fergestad T., Davis W. S., Broadie K. The stoned proteins regulate synaptic vesicle recycling in the presynaptic terminal. J. Neurosci. 1999;19:5847–5860. doi: 10.1523/JNEUROSCI.19-14-05847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stimson D. T., Estes P. S., Rao S., Krishnan K. S., Kelly L. E., Ramaswami M. Drosophila stoned proteins regulate the rate and fidelity of synaptic vesicle internalization. J. Neurosci. 2001;21:3034–3044. doi: 10.1523/JNEUROSCI.21-09-03034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips A. M., Smith M., Ramaswami M., Kelly L. E. The products of the Drosophila stoned locus interact with synaptic vesicles via synaptotagmin. J. Neurosci. 2000;20:8254–8261. doi: 10.1523/JNEUROSCI.20-22-08254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. Binding specificity and in vivo targets of the EH domain, a novel protein–protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owen D. J., Vallis Y., Noble M. E., Hunter J. B., Daffron T. R., Evans P. R., McMahon H. T. A structural explanation for the binding of multiple ligands by the α-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- 18.Scheele U., Alves J., Frank R., Duwel M., Kalthoff C., Ungewickell E. Molecular characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin. J. Biol. Chem. 2003;278:25357–25368. doi: 10.1074/jbc.M303738200. [DOI] [PubMed] [Google Scholar]
- 19.Martina J. A., Bonangelino C. J., Aguilar R. C., Bonifacino J. S. Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J. Cell Biol. 2001;153:1111–1120. doi: 10.1083/jcb.153.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther K., Krauss M., Diril M. K., Lemke S., Ricotta D., Honing S., Kaiser S., Haucke V. Human stoned B interacts with AP-2 and synaptotagmin and facilitates clathrin-coated vesicle uncoating. EMBO Rep. 2001;2:634–640. doi: 10.1093/embo-reports/kve134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M. S., Bonifacino J. S. Adaptor-related proteins. Curr. Opin. Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 22.Roos J., Kelly R. B. Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J. Biol. Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]
- 23.Miklos G. L. G., Kelly L. E., Coombe P. E., Leeds C., Lefevre G. Localization of the genes shaking-B, small optic lobes, sluggish-A, stoned and stress-sensitive-C to a well defined region on the X chromosome of Drosophila melanogaster. J. Neurogenet. 1987;4:1–19. doi: 10.3109/01677068709102329. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd T. E., Verstreken P., Ostrin E. J., Phillippi A., Lichtarge O., Bellen H. J. A genome-wide search for synaptic vesicle cycle proteins in Drosophila. Neuron. 2000;26:45–50. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 25.McCaughan K. K., Brown C. M., Dalphion M. E., Berry M. L., Tate W. P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samson M. L., Lisbin M. J., White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141:1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Washburn T., O'Tousa J. E. Nonsense suppression of the major rhodopsin gene of Drosophila. Genetics. 1992;130:585–595. doi: 10.1093/genetics/130.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verstreken P., Koh Y.-W., Schulze K. L., Zhai R. G., Heisinger P. R., Zhou Y., Mehta S. Q., Cao Y., Roos J., Bellen H. J. Synaptojanin is recruited by endophilin to promote synaptic vesicle unloading. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 29.Fergestad T., Broadie K. Interaction of stoned and synaptotagmin in synaptic vesicle endocytosis. J. Neurosci. 2001;15:1218–1227. doi: 10.1523/JNEUROSCI.21-04-01218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh T.-W., Verstreken P., Bellen H. J. Dap160/Intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 31.Marie B., Sweeney S. T., Poskanzer K. E., Roos J., Kelly R. B., Davis G. W. Dap160/Intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Bai J., Wang C. T., Richards D. A., Jackson M. B., Chapman E. R. Fusion pore dynamics are regulated by synaptotagmin/t-SNARE interactions. Neuron. 2004;41:929–942. doi: 10.1016/s0896-6273(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 33.Gad H., Ringstad N., Low P., Kjaerulff O., Gustafsson J., Wenk M., Ellisman M. H., De Camilli P., Shupliakov O., Brodin L. Fission and uncoating of synaptic clathrin-coated vesicles are perturbed by disruption of interactions with the SH3 domain of endophilin. Neuron. 2000;27:301–312. doi: 10.1016/s0896-6273(00)00038-6. [DOI] [PubMed] [Google Scholar]
- 34.Guichet A., Wucherpfennig T., Dudu V., Etter S., Wilsch-Brauniger M., Hellwig A., Gonzalez-Gaitan M., Huttner W. B., Schmidt A. A. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 2002;21:1661–1672. doi: 10.1093/emboj/21.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walther K., Diril M. K., Jung N., Haucke V. Functional dissection of the interactions of stonin 2 with the adaptor complex AP-2 and synaptotagmin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:964–969. doi: 10.1073/pnas.0307862100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verstreken P., Koh T. W., Schulze K. L., Zhai R. G., Hiesinger P. R., Zhou Y., Mehta S. Q., Cao Y., Roos J., Bellen H. J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]