Abstract
The catabolism of melatonin, whether naturally occurring or ingested, takes place via two pathways: ∼70% can be accounted for by conjugation (sulpho- and glucurono-conjugation), and ∼30% by oxidation. It is commonly thought that the interferon-induced enzyme indoleamine 2,3-dioxygenase (EC 1.13.11.42), which oxidizes tryptophan, is also responsible for the oxidation of 5-hydroxytryptamine (serotonin) and its derivative, melatonin. Using the recombinant enzyme expressed in Escherichia coli, we show in the present work that indoleamine 2,3-dioxygenase indeed cleaves tryptophan; however, under the same conditions, it is incapable of cleaving the two other indoleamines. By contrast, myeloperoxidase (EC 1.11.1.7) is capable of cleaving the indole moiety of melatonin. However, when using the peroxidase conditions of assay – with H2O2 as co-substrate – indoleamine 2,3-dioxygenase is able to cleave melatonin into its main metabolite, a kynurenine derivative. The present work establishes that the oxidative metabolism of melatonin is due, in the presence of H2O2, to the activities of both myeloperoxidase and indoleamine 2,3-dioxygenase (with lower potency), since both enzymes have Km values for melatonin in the micromolar range. Under these conditions, several indolic compounds can be cleaved by both enzymes, such as tryptamine and 5-hydroxytryptamine. Furthermore, melatonin metabolism results in a kynurenine derivative, the pharmacological action of which remains to be studied, and could amplify the mechanisms of action of melatonin.
Keywords: indoleamine 2,3-dioxygenase; indole ring; kynurenine; melatonin; myeloperoxidase; oxidative catabolism
Abbreviations: AFMK, N1-acetyl-N2-formyl-5-methoxykynurenine; AMK, N1-acetyl-5-methoxykynurenine; IndoDO, indoleamine 2,3-dioxygenase; MyelPO, myeloperoxidase; TFA, trifluoroacetic acid; TrypDO, tryptophan 2,3-dioxygenase
INTRODUCTION
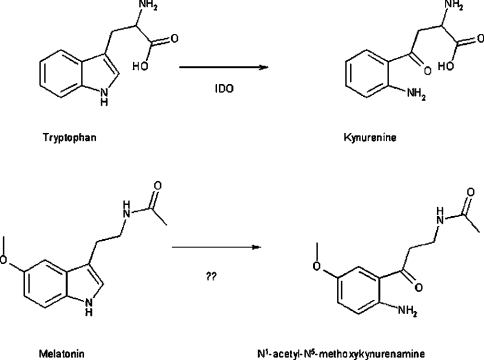
Melatonin is a hormone that regulates multiple physiological functions [1]. It is synthesized in the pineal gland, through a pathway in which the first steps are tryptophan hydroxylation and the subsequent decarboxylation thereof. The final reaction is the methylation of the N-acetyl-5-hydroxytryptamine [3] immediately after the N-acetylation of 5-hydroxytryptamine (serotonin). The rate-limiting enzyme of this cascade is the pineal arylalkylamine N-acetyltransferase (serontonin N-acetyltransferase; EC 2.3.1.87) [4]. The pathway of synthesis of melatonin has been known for 30 years, but its catabolism is less well understood, apart from the conjugation steps that account for approx. 70% of an ingested dose ([5] and references therein). Some melatonin (∼15%) is excreted untransformed. The remaining catabolism involves the oxidative cleavage of the indole moiety, similar to what is reported for tryptophan (Scheme 1).
Scheme 1. Comparative oxidative catabolism of tryptophan and melatonin.
IDO, IndoDO.
Indole rings can be opened by three different enzymatic mechanisms, depending on the co-substrate used and the mechanism of action: (i) TrypDO (tryptophan 2,3-dioxygenase; EC 1.13.11.11; accession number P48775) uses molecular oxygen [6]; (ii) IndoDO (indoleamine 2,3-dioxygenase; EC 1.13.11.42 [7]; accession number P14902) uses a complex catalytic mechanism during which, as a first step, the enzyme iron is oxygenated; the oxygen moiety is then transferred to open the indole ring, with concomitant reduction of Fe3+ to Fe2+; and (iii) peroxidases [MyelPO (myeloperoxidase); EC 1.11.1.7], another prosthetic family of enzymes that are able to detoxify H2O2 into two highly reactive superoxide anions, which are able to react and open the indole moiety in two steps [8].
The specificities of these enzymes have been partly described: (i) TrypDO recognizes only tryptophan, and is strictly a liver enzyme; (ii) IndoDO has a wider specificity, and has been reported to be able to cleave several bioamines, such as 5-hydroxytryptamine, tryptamine and melatonin, as well as the amino acid tryptophan [7,9]; and (iii) MyelPO, which is thought to cleave a very large spectrum of compounds, including melatonin [10,11].
Another source of confusion may arise from the fact that IndoDO catalyses two types of activity: one is the well described dioxygenase reaction, which is H2O2-independent, which converts tryptophan into kynurenamine, while the other (less well studied) is a peroxidase-like reaction, using H2O2 as co-substrate [8]. This feature seems to be quite common in haem-containing enzymes. Both reactions, however, give identical products. The conditions under which one can measure IndoDo activity are quite complex. Indeed, as described in the original report [7], and confirmed in numerous others [12–15], the experimental procedure involves ascorbic acid, Methylene Blue and catalase. Methylene Blue permits the transfer of electrons from ascorbic acid to molecular oxygen, leading to the formation of the superoxide anion. This chemical species attacks the enzyme haem, while catalase protects the enzyme from the overproduction of a side product, H2O2. Obviously, under these conditions, the peroxidase-like activity of IndoDO cannot operate, since H2O2 is destroyed by catalase [6,16]. Nevertheless, the fact that melatonin is cleaved by IndoDO is so widely accepted by the scientific community that some authors have started recent reviews with the statement that IndoDO converts tryptophan, melatonin and 5-hydroxytryptamine into their corresponding kynurenine counterparts (e.g. Grohmann et al. [17]).
Our laboratory group has been involved for several years in understanding the mechanisms of action of melatonin at the molecular level, including the enzymology of its biosynthesis [18–21] and its interaction with melatonin receptors [22–25]. In order to explore further the catabolism of melatonin, which to date is based on only a few reports, we expressed recombinant human IndoDO in bacteria, leading to expression of a protein tagged with a hexahistidine moiety [12]. We then assessed the analytical conditions that allowed us to use HPLC as a tool for determination of the nature of the compounds produced by this catabolic reaction, especially when coupled with MS. After having precisely determined the enzymatic conditions under which IndoDO is able to cleave tryptophan, we undertook structure–activity studies, using other reported substrates such as 5-hydroxytryptamine and melatonin, as well as some of their analogues or derivatives. We also checked the capacity of these compounds to inhibit the IndoDO-mediated catabolism of tryptophan. We found that, under conditions where IndoDO is able to cleave tryptophan and some of its derivatives, it was impossible to record any breakdown of 5-hydroxytryptamine, melatonin or tryptamine. In contrast, when we used conditions under which IndoDO uses H2O2 as co-substrate, melatonin was cleaved. We also tested a hypothesis reported in the literature [10] that melatonin can be cleaved by other enzymes, namely MyelPO. Using optimized enzymatic conditions for MyelPO activity, this was indeed found to be the case, leading to the formation of the kynurenine derivative AFMK (N1-acetyl-N2-formyl-5-methoxykynurenine), which is then rapidly metabolized further to AMK (N1-acetyl-5-methoxy-kynurenine). Finally, we examined some aspects of MyelPO specificity.
EXPERIMENTAL
Materials
Ampicillin, kanamyin, isopropyl β-D-thiogalactoside, haemin, PMSF, EDTA, D- and L-tryptophan, ascorbic acid, Methylene Blue, catalase, L-kynurenine, 1-methyl-D,L-tryptophan, 5-methoxy-L-tryptophan, tryptophol, tryptamine, 5-hydroxytryptamine, 5-methoxytryptamine, 6-methoxytryptamine, 5-methoxy-D,L-tryptophan, melatonin, indole-2-carboxylic acid, indole-3-carboxylic acid, indole-3-acetic acid, indole-3-glyoxylic acid, 3-methylindole, 2-methylindole, N-acetyl-5-hydroxytryptamine, benzimidazole, 4-hydroxyquinazoline, 5-hydroxy-L-tryptophan, 1-methylindole, MyelPO and indole-2-acetic acid were all purchased from Sigma (St Louis, Mo, U.S.A.). AFMK and AMK were synthesized by Professor Gerald Guillaumet (Faculté d'Orléans, France).
Expression and production of His6-tagged human recombinant IndoDO
Human IndoDO expressed in Escherichia coli strain EC538-pQE9 IndoDO-pREP4 was generously provided by Dr S. Tone (Kawasaki Medial School, Okoyama, Japan) and was obtained as described by Littlejohn et al. [12]. Briefly, E. coli strain EC538-pQE9 IndoDO-pREP4 was grown at 37 °C in Luria broth supplemented with ampicillin (100 μg/ml) and kanamyin (50 μg/ml) until it reached an A595 of 0.6. Isopropyl β-D-thiogalactoside, haemin and PMSF were then added to final concentrations of 10 μM, 7 μM and 1 mM respectively, and the culture was maintained at 37 °C for 3 h. EDTA (1 mM) was added at the end of the culture. Bacteria were harvested by centrifugation (5000 g, 4 °C, 10 min), frozen in solid CO2 and stored at −80 °C until further use.
Purification of human recombinant IndoDO
All procedures were performed at 0 – 4 °C. Approx. 2.5 g of a frozen bacterial pellet was thawed in 10 ml of PBS containing a cocktail of protease inhibitors (EDTA-free, Complete; Boehringher Mannheim; 1 tablet/50 ml). The bacterial suspension was sonicated (VibraCell 72446; Bioblock, Illkirch, France; setting 90/100; 2 mm diameter probe) for 8×1 min, with 1 min intervals between sonications. The preparation was centrifuged immediately (27000 g, 15 min) and the supernatant was slowly (40 ml/h) passed through a phosphocellulose P11 column (2 ml bed volume; Whatman, Maidstone, Kent, U.K.) equilibrated with buffer A (50 mM potassium phosphate, pH 6.5, 1 mM PMSF). The column was initially washed with 20 ml of buffer A. His6–IndoDO was eluted sequentially with 15 ml volumes of 100, 200 and 300 mM potassium phosphate buffer, pH 6.5. Fractions eluted by 100 and 200 mM potassium phosphate buffer were collected (35 ml), 10% (v/v) glycerol and imidazole (10 mM, final concentration) were added, and the fractions were passed through a Ni2+-nitrilotriacetate column (3 ml bed volume; Qiagen, Venlo, The Netherlands) equilibrated with buffer B (250 mM potassium phosphate, pH 6.5, 1 mM PMSF, 10% glycerol) containing 10 mM imidazole. The column was washed with 30 ml of the same buffer. His6–IndoDO was eluted stepwise with 15 ml volumes of buffer B containing 100, 150, 200 and 250 mM imidazole. Fractions were concentrated on Centriprep YM10 membranes (Amicon, Millipore, Saint Quentin en Yvelines, France) and incubation buffer C (50 mM Tris/HCl, pH 7.4, 1 mM EDTA) was then added. The collected fractions were assayed for IndoDO activity, and those containing activity were pooled and stored at −80 °C. The protein concentration was determined using the Bradford assay (Protassay; Bio-Rad, Ivry-sur-Seine, France) with BSA as standard.
IndoDO assay
The assay for IndoDO activity was based upon a reverse-phase HPLC method using absorbance detection of L-kynurenine. In this method, 30 μl aliquots of solution were analysed by reverse-phase HPLC using a Xterra RP18 column (100 mm×4.6 mm; Waters) on a Hewlett Packard 1100 system. The column was eluted with a linear gradient of 0–80% (v/v) acetonitrile in water containing 0.1% TFA (trifluoroacetic acid) at a flow rate of 1 ml/min for 32 min. In this system, kynurenine is eluted at 6 min. Known quantities of this compound were injected into the HPLC system, and the area under the peak at 210 nm was quantified. These measurements permitted us to construct an abascus, from which we were able to systematically measure the actual amount of kynurenine formed during the various experiments. To start with, the reaction mixture contained 50 μl of IndoDO (2.2 μg/ml) in a phosphate buffer (50 mM sodium phosphate, pH 6.5), with 200 μM L-tryptophan as substrate, plus 20 mM ascorbic acid, 10 μM Methylene Blue and 200 μg/ml catalase, in a final volume of 200 μl, as described by Yamamoto and Hayaishi [26] or Shimizu et al. [9] with minor modifications [12]. The role of each component of the catalytic reaction medium was checked, alone or in pairs, to evaluate their importance for the catalytic activity of IndoDO in the presence of two potential substrates, tryptophan and melatonin. The kinetics, linearity as a function of protein concentration, pH dependence and temperature dependence were all studied systematically. After a 15 or 30 min incubation at 37 °C, reactions were stopped by the addition of 30% (v/v) trichloroacetic acid solution (40 μl). Incubations were performed in black Eppendorf® microtubes or in black 96-well plates, away from direct light sources. The assays were analysed by HPLC, as described above.
At a later stage, we performed experiments using L-[3H]tryptophan (26 μCi, 32 Ci/mmol), 5-[3H]hydroxytryptamine (7 μCi, 116 Ci/mmol) or [3H]melatonin (10 μCi, 83 Ci/mmol) (all from Amersham Pharmacia Biotech, Orsay, France) as substrates to increase the sensitivity of the IndoDO system. The HPLC gradient and buffers were the same as described above for the 32 min run. Radioactivity was followed on-line after addition of scintillation cocktail (3 ml/min) using a Berthold detector (EGG, Bad Wildbad, Germany).
For specificity studies, candidate substrates were substituted for the substrate in the standard assay. A first run was performed, in the absence of enzyme, in order to determine the retention times of all of these compounds in the complete system. No interference by any of the components of the assay was recorded, permitting us to evaluate the capacity of the enzyme to accommodate each compound. For inhibitor studies, incubation times and substrate concentrations were chosen in order to be within the linear portion of enzyme activity.
MyelPO assay
To assay the activity of human leucocyte MyelPO (catalog no. M 6908; Sigma), 0.84 μg of enzyme preparation was preincubated with 150 μM H2O2 for 2 min at 37 °C. Melatonin (1 mM) in PBS supplemented with potassium phosphate (55 mM at pH 5.4) was then added (50 μl) and incubated at 37 °C for an additional 20 min. The total volume of the assay was 200 μl. The reaction was stopped by adding 40 μl of trichloroacetic acid (30%, v/v). A 30 μl aliquot of the final solution was injected into the same analytical system as described above, permitting a direct comparison of retention times when needed. Similarly, L-[3H]-tryptophan, 5-[3H]-hydroxytryptamine and [3H]melatonin were also assessed as potential substrates of MyelPO. Again, the incubation times and substrate concentrations for inhibitor studies were chosen in order to be within the linear portion of enzyme activity.
Kinetic studies
Except where noted otherwise, all measurements were obtained using the HPLC assay. Kinetic data were collected using the classical procedures described by Segel [27]. For determination of the apparent Km values of IndoDO and MyelPO, substrate concentrations were varied from 0.005 to 10 mM. At least five concentrations were routinely used, and the experiments were repeated three times. Data were fitted to the Hill equation by linear regression.
Peroxidase-like activity of IndoDO
IndoDO activity was also measured in the presence of oxygen peroxide, which acts as a co-substrate in a potassium phosphate buffer (55 mM, pH 5.4), during 20 min at 37 °C [8]. The incubations were treated as in the IndoDO standard assay, and analysed by HPLC, using the same system as described above.
MS studies
Electrospray ionization mass spectra were recorded on a Q-TOF2 instrument (Waters-Micromass, Manchester, U.K.). LC separations for the LC/MS experiments were performed using an HP 1090 binary pump equipped with a UV detector (Hewlett Packard, Palo Alto, CA, U.S.A.). The samples were passed through a 20 μl injection loop into the Xterra RP18 column (see above) at a flow rate of 1 ml/min, and into the mass spectrophotometer after a split ratio of 1:4, resulting in a flow rate of approx. 250 μl/min into the instrument. The elution solutions were: A, 0.01% TFA in deionized water; B, 0.01% TFA in acetonitrile. The gradient sequence was from 100% to 20% (v/v) A in 30 min.
Analytical electrophoresis
SDS/PAGE was performed according to the method of Laemmli [28], followed by Coomassie Blue staining. Immunoblotting was carried out using standard procedures. Blots were developed using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech) with anti-histidine antibody (Sigma).
RESULTS
Purification of recombinant human IndoDO
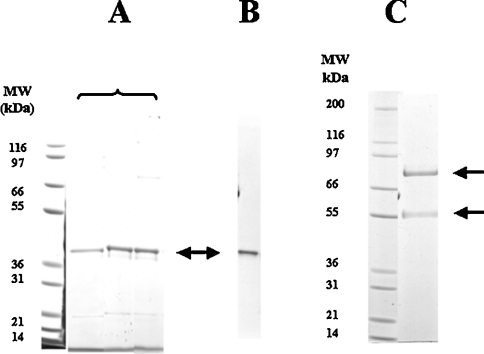
His6-tagged IndoDO was purified using the protocol described by Littlejohn et al. [12]. Figure 1(A) shows the SDS/PAGE characterization of the different steps leading to an apparently unique protein. The gel was Western-blotted with an anti-histidine antibody and showed the presence of a single band at 42.5 kDa (Figure 1B). No ladder (indicating proteolytic degradation of the protein) could be observed. Table 1 summarizes the purification steps and yield. IndoDO was purified only 190-fold compared with the starting material, a poor purification factor, due mainly to the instability of the enzyme over time. The specific activity was diminished by a factor of 35 during the course of the purification, resulting in an overall purification factor of 5.5. Overall, the purification procedure gave an IndoDO enzyme with characteristics similar to those published recently [15]. Furthermore, as IndoDO is a haemoprotein, it is necessary to supplement the bacteria with haemin, to provide the enzyme with sufficient protoporphyric group IX, essential for its activity. The A405/A280 ratio for the fully reconstituted, haem-containing enzyme has been reported as 2.2, while our preparation had an absorbance ratio of 1.35, indicating that approx. 60% [(1.35/2.2)×100%] of the protein was in its active form. Littlejohn et al. [12] reported their own preparation to have an absorbence ratio of 1.7.
Figure 1. Purity of His6-tagged IndoDO expressed in E. coli EC538, and of commercially available MyelPO.
N-terminally His6-tagged human IndoDO fractions from previous steps were applied on a Ni2+-nitrilotriacetate column, which was washed with 10 mM imidazole buffer and eluted stepwise by imidazole. The fractions were analysed by SDS/PAGE (A) and by Western blot (B) using anti-histidine antibodies. A sample of each chromatographic step was boiled in SDS, applied on a 4–12% gradient SDS/PAGE gel and submitted to electrophoresis. The gel was then fixed, dried and stained with Coomassie Blue. The arrow points at the expected molecular mass of IndoDO. The data presented are a typical example of three independent purifications. MW, molecular mass markers. (C) Commercially available MyelPO (Sigma) was analysed on a 4–12% gradient SDS/PAGE gel and submitted to electrophoresis. The gel was then fixed, dried and stained with Coomassie Blue. The two arrows point to the two subunits of MyelPO.
Table 1. Purification of human recombinant His6-tagged IndoDO expressed in E. coli EC538.
Total activity was determined with tryptophan as substrate using an HPLC assay (see the Experimental section for details).
| Step | Volume (ml) | Total protein (mg) | Specific activity (μmol/h per mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Bacterial pellet | 10 | 152 | 4.2 | 1 | 100 |
| Supernatant | 10 | 104 | 6.1 | 1.5 | 100 |
| Phosphocellulose–agarose | 35 | 3.1 | 27.7 | 6.7 | 14 |
| Ni2+-nitrilotriacetate–agarose | 40 | 0.8 | 22.8 | 5.5 | 3 |
Biochemical characterization of purified human IndoDO
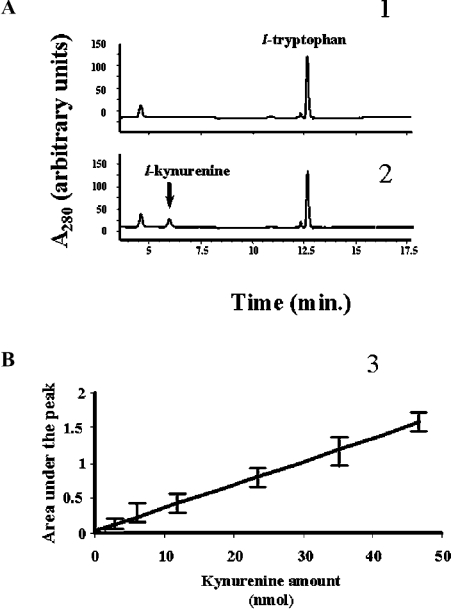
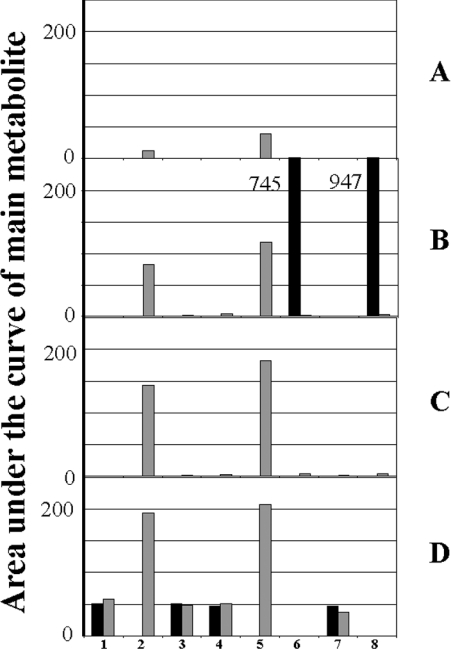
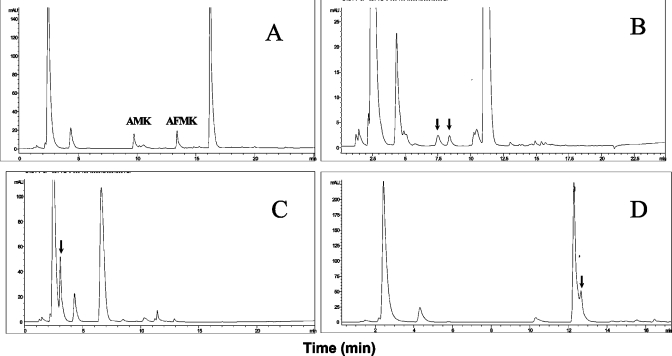
Our HPLC method permitted the analysis of any compound within the chromatographic window. Figure 2(A) shows a typical chromatogram obtained with L-tryptophan, where the product of the reaction, L-kynurenine, can be seen. The identity of kynurenine was confirmed by MS (results not shown). Our main goals were (1) to check the ability of IndoDO to metabolize melatonin, and (2) if not, to determine the conditions under which, and/or other enzyme(s) by which, melatonin can be oxidatively catabolized. A first step was the qualification of the IndoDO assay using tryptophan as substrate. We used a stepwise approach to this system. The experimental conditions used for IndoDO measurement were derived from those described by Yamamoto and Hayaishi [26] and modified slightly by Littlejohn et al. [12]; our conditions were identical to the latter. Briefly, using the HPLC assay with tryptophan and melatonin as substrates, we checked the requirement for the presence of catalase, Methylene Blue and ascorbic acid to obtain cleavage of the substrate. Then all of these components were tested in dual combinations, and finally the complete system was used. We also included a series of experiments where FeCl3 was substituted for the biological source. IndoDO catalysed the oxygenation of tryptophan only with the complete system and in the presence of native IndoDO (Figure 3B); tryptophan oxidative cleavage was not seen with any of the other combinations, or in the absence of enzyme, in the presence of denatured enzyme or in the presence of FeCl3 solution (Figures 3A, 3C and 3D). This last system was used in order to demonstrate that the cleavage is not non-enzymatic. Minute amounts of metabolites were formed in the presence of a 10 mM FeCl3 solution, suggesting that a chemical oxidation of tryptophan was possible under those conditions. When we turned our attention to melatonin, however, the situation was quite different. Indeed, melatonin could be cleaved into AFMK in the presence of ascorbic acid, and more so following the addition of catalase, but this result was independent of the presence of IndoDO (native or denatured) or of FeCl3 (Figure 3). In any case, the amounts of metabolite(s) issued from melatonin were small compared with those obtained following tryptophan cleavage.
Figure 2. Chromatographic profiles for analyses of IndoDO activity as estimated by A280.
(A) Incubations without (panel 1) or with (panel 2) 2.2 μg/ml purified recombinant IndoDO were performed using the complete system described by Hirata et al. [7] containing 200 μM L-tryptophan, 200 μg/ml catalase, 20 mM ascorbic acid and 10 μM Methylene Blue for 30 min at 37 °C. The reaction was stopped by 30% (v/v) trichloroacetic acid and the incubation medium injected into an HPLC system equipped with an Xterra RP18 3.5 μm Waters column (100 mm×4.6 mm). The column was eluted with a linear gradient of 0–80% (v/v) acetonitrile in water (0.1% TFA) at a flow rate of 1 ml/min for 32 min. (B) Using the same conditions, known amounts of the product of the reaction were injected and analysed, providing a way to quantify the compounds from chromatograms. Data are presented as means±S.D. of three determinations. At least one such run was done with daily experiments.
Figure 3. Importance of components of the IndoDO reaction medium.
The four components of the reaction medium were the enzyme, catalase, ascorbic acid and Methylene Blue, as reported by Hirata et al. [7]. The importance of each component was checked by assaying the catalytic activity of the enzyme towards either tryptophan (black bars) or melatonin (grey bars), using one of these components alone, a combination of two components, or the whole system, in the presence of buffer (A), native (B) or denatured (C) IndoDO, or a 10 mM FeCl3 solution (D). Incubations were stopped after 15 min at 37 °C and analysed by HPLC. The area under the curve of the main metabolite – if present – was plotted. Concentrations were as in the standard assay. 1, Buffer alone; 2, ascorbic acid; 3, Methylene Blue; 4, catalase; 5, ascorbic acid plus catalase; 6, ascorbic acid plus Methylene Blue; 7, Methylene Blue plus catalase; 8, complete system. The main product of the reaction was measured at the same retention time as AFMK for melatonin experiments and kynurenine for tryptophan experiments. The whole set of experiments was done twice. A simple mean of the results was calculated; the variation between the two experiments was <10%. Note that in the two conditions where tryptophan was cleaved, the bars are off the scale, with areas under the curve of 745 and 947.
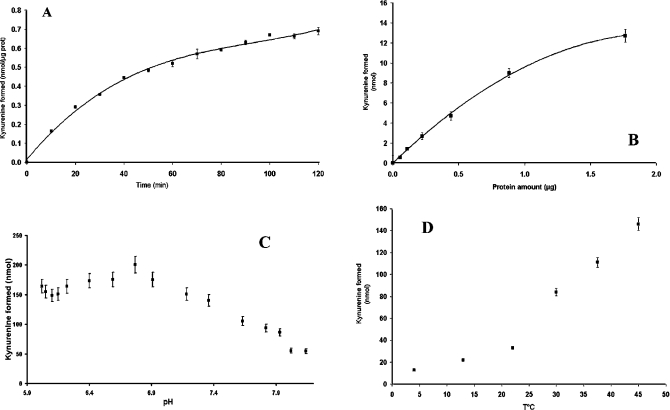
The experimental conditions under which this activity can be measured necessitated the incorporation of three cofactors: ascorbic acid, Methylene Blue and catalase. In order to measure enzyme activity under optimal conditions, we tested the linearity of the reaction in relation to time (Figure 4A), protein concentration (Figure 4B), pH (Figure 4C) and temperature (Figure 4D). Accordingly, the quantity of enzyme was 830 ng per assay (2.2 μg/ml), the reaction was stopped after 15 min, and was carried out at pH 7.0 and at 37 °C (despite the surprising shape of the curve, with apparent enhancement of the catalytic activity with temperature up to 45 °C). The biochemical characterization of purified IndoDO using the HPLC method is reported in Table 2. Characterization was conducted with the natural substrates of the enzyme, i.e. D- and L-tryptophan [12], and also using the inhibitor 1-methyl-D,L-tryptophan [29]. We checked that the colorimetric assay gave similar results to the HPLC analyses. The affinities measured for L-tryptophan and D-tryptophan were 43 μM and 5.2 mM respectively, similar to those reported in the literature [12] and those obtained with the enzyme purified from human placenta [30]. We observed, as did Yamamoto and Hayaishi [26], that high concentrations (>250 μM) of L-tryptophan inhibited enzyme activity (results not shown). These results indicated that addition of the His6 tag did not change the catalytic characteristics of the enzyme, nor its specificity.
Figure 4. Enzymatic assessment of the IndoDO reaction.
The various parameters of the oxidative cleavage of tryptophan by IndoDO were assessed. (A) Incubation time. Experiments were started at a defined time and stopped sequentially after the indicated periods. (B) Linearity with regard to the amount of pure protein in the incubation medium. (C) Variation of the reaction rate as a function of pH of the assay. The experiments were conducted with the complete assay system in a phosphate buffer with a pH of between 6 and 8.1. The pH was measured by a microelectrode inside each test well. (D) Variation of the reaction rate as a function of temperature. Each set of experiments was run at 5, 13, 23, 30, 37 and 45 °C. All assay media contained in 122 μl of 50 mM phosphate buffer, pH 7.09, unless otherwise indicated, 12 μl of catalase (3.3 mg/ml), 8 μl of ascorbic acid (500 mM), 8 μl of Methylene Blue (250 μM), 40 μl of L-tryptophan (1 mM) and 10 μl of purified enzyme. All reactions were stopped by addition of 40 μl of aqueous trichloroacetic acid (30%, v/v). Unless mentioned otherwise the experiments were run for 15 min at 37 °C.
Table 2. Biochemical characterization and sensitivity to inhibitors of recombinant human IndoDO.
Km values were measured as described in the Experimental section, with optimal concentrations of the other reaction components: 200 μg/ml catalase, 20 mM ascorbic acid and 10 μM Methylene Blue. Values are means±S.D. of three independent experiments. For determination of inhibition, the compounds were tested with L-tryptophan as substrate. Products were used as inhibitors in the standard assay at concentrations ranging from 10 mM to 100 nM. Experiments were performed two to four times. Results are given as means ±S.D.
| Substrate/inhibitor | Km (μM) | Vmax (mol/min per mg of protein) | IC50 (nM) | Ki (μM) |
|---|---|---|---|---|
| L-Tryptophan | 43±4 | 47±3 | – | |
| D-Tryptophan | 5200±250 | 47±9 | – | |
| 1-Methyl-D,L-tryptophan | – | – | 134±11 | 23 |
| 5-Hydroxy-L-tryptophan | – | – | 409±32 | 72 |
| Tryptamine | – | – | 1440±300 | 255 |
| 2-Methylindole | – | – | 895±205 | 158 |
| 5-Hydroxytryptamine | – | – | >1000 | |
| N-Acetyl-5-hydroxytryptamine | – | – | >1000 | |
| Melatonin | – | – | >10000 |
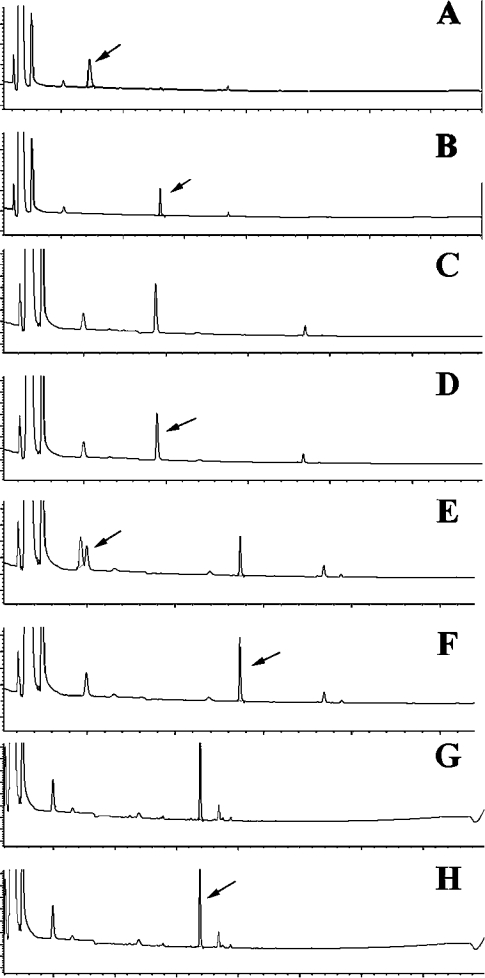
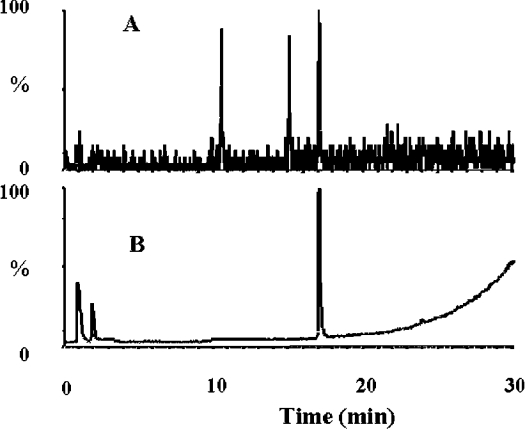
Owing to the lack of results with most of the unconventional candidate substrates, particularly 5-hydroxytryptamine, tryptamine and melatonin, we ‘pushed’ the system by using 8.8 μg of enzyme and an incubation time of 120 min. The detection system for these determinations was set at 210 nm. Other indolic compounds were tested, i.e. 5-hydroxytryptamine, 5′-methoxytryptophan and melatonin. Some raw chromatographic results are presented in Figure 5. Tryptophan (Figures 5A and 5B) and 5′-methoxytryptophan (Figures 5E and 5F) were clearly degraded by IndoDO, but neither 5-hydroxytryptamine nor melatonin was metabolized. We then used some pure tritiated substrates, without isotopic dilution: L-[3H]tryptophan, 5-[3H]hydroxytryptamine and [3H]melatonin. They were tested using eight different isotopic dilutions, from 0.5 μM to 100 μM (final) in the assay, using 26 μCi, 7 μCi and 10 μCi respectively. The enormous amounts of labelled compounds used in these tests resulted in the appearance on the chromatograms of minor contaminants in the profile that were also found in the controls (Figure 6). Tryptophan was again cleaved by IndoDO under these conditions (Figures 6A and 6B), but neither melatonin (Figures 6B and 6C) nor 5-hydroxytryptamine (results not shown) was recognized by this enzyme under these standard conditions.
Figure 5. Survey of indole derivatives as potential substrates of human recombinant IndoDO.
Compounds (200 μM) were incubated in the conventional assay in the presence or absence of 8.8 μg/ml purified IndoDO, plus 200 μg/ml catalase, 20 mM ascorbic acid and 10 μM Methylene Blue for 2 h at 37 °C. The consumption of the products was followed by HPLC. All experiments were performed at least three times independently; identical results were obtained in all tests. (A) L-Tryptophan with enzyme; the arrow points at the product of the reaction. (B) Same as (A), without enzyme; the arrow points at tryptophan. (C) 5-Hydroxytryptamine with enzyme. (D) Same as (C), without enzyme; the arrow points at 5-hydroxytryptamine (serontonin). (E) 5′-Methoxy-L-tryptophan with enzyme; the arrow points at the product of the reaction. (F) Same as (E), without enzyme; the arrow points at 5′-methoxytryptophan. Note the contaminating peak in (E) and (F). (G) Melatonin with enzyme. (H) Same as (G), without enzyme; the arrow points at melatonin.
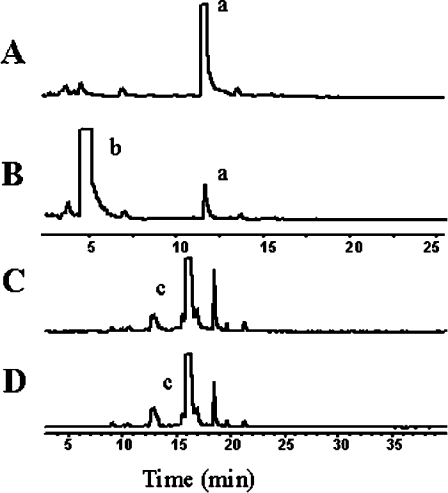
Figure 6. Incubation of human recombinant IndoDO with L-[3H]tryptophan and [3H]melatonin as substrates.
L-[3H]Tryptophan or [3H]melatonin was incubated with human recombinant IndoDO in a standard assay, as described in the Experimental section. Once stopped by acidification, a sample of the reaction medium was injected into an HPLC system equipped with a Xterra RP18 column and online radioactivity detection. Chromatograms were obtained after incubation of L-[3H]tryptophan without (A) or with (B) IndoDO. Peak a, [3H]tryptophan; peak b, [3H]kynurenine. The same experiments were done with [3H]melatonin after incubation without (C) or with (D) IndoDO. Peak c, [3H]melatonin. To reach maximal sensitivity, a large amount of tritiated substrate was used in each experiment.
Substrate specificity of purified human IndoDO
A number of indole derivatives (200 μM) were incubated in the complete enzyme system, i.e. with ascorbic acid, catalase and Methylene Blue [12]. The consumption of the putative substrates was followed by HPLC. Besides L-tryptophan, 5-methoxy-D,L-tryptophan was also found to be a substrate of IndoDO (results not shown), as expected, as was 5-methoxy-L-tryptophan. None of the other candidates were recognized by the enzyme, with the possible exception of 2-methylindole. The compounds tested were D-tryptophan, tryptophol, tryptamine, 5-hydroxytryptamine, 5-methoxytryptamine, 6-methoxytryptamine, melatonin, indole-2-carboxylic acid, indole-3-carboxylic acid, indole-3-acetic acid, indole-3-glyoxylic acid, 3-methylindole, N-acetyl-5-hydroxytryptamine, and two indole analogues, i.e. benzimidazole and 4-hydroxyquinazoline.
Inhibition of recombinant human IndoDO by indole derivatives
In order to complete the characterization of IndoDO, we used the same series of compounds as possible inhibitors in the same assay, with L-tryptophan as substrate (200 μM). The only compounds capable of inhibiting IndoDO activity were 5-hydroxy-L-tryptophan, tryptamine and 2-methylindole. The IC50 values measured under our conditions were 134±11 nM for 1-methyl-D,L-tryptophan (the reference inhibitor), 895±205 nM for 2-methylindole and 1.44±0.3 μM for tryptamine (see also Table 2).
Oxidative metabolism of melatonin
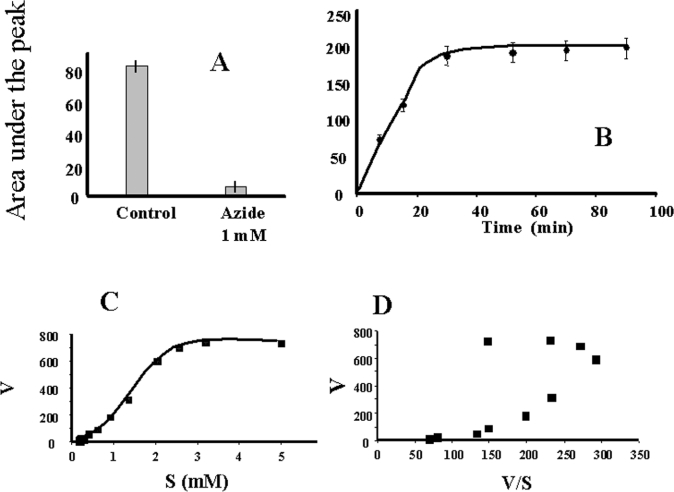
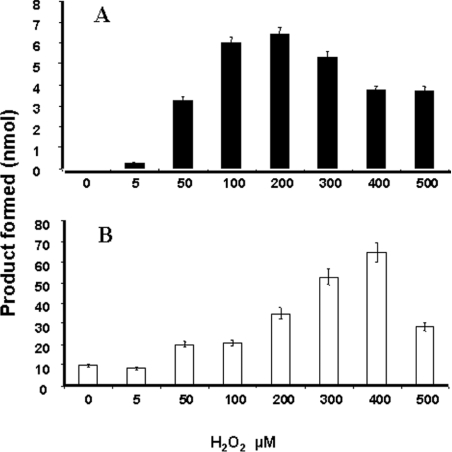
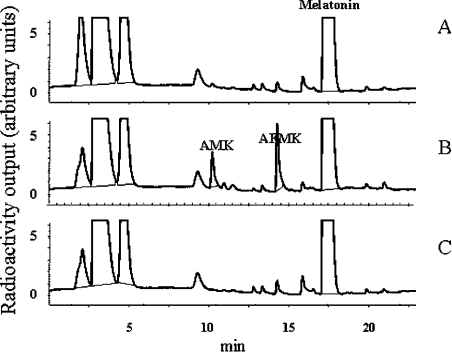
Melatonin is cleaved in vivo either by monocytes [31,32] or by activated macrophages [33]. The identity of the enzyme involved in this process is still unknown. However, our data indicated that it is not IndoDO, which agrees with the results of Rodrigues et al. [34]. The following experiments were designed to identify the enzyme involved in this breakdown. It was anticipated that MyelPO would be a good candidate for catalysing the oxidative catabolism of melatonin. We undertook the characterization of MyelPO with the methodology used to perform the IndoDO experiments. The purity of the commercially available enzyme was checked and found to be satisfactory for further use (Figure 1C). Its incubation with melatonin was performed as described by Andrews and Krinsky [35]. The specific activity of this enzyme was 6.67 nmol/h per mg of protein. We tested the minimal parameters of the enzyme, especially its susceptibility to azide (Figure 7A), and the linearity of the reaction with time (Figure 7B) and protein concentration (results not shown). All of these parameters permitted us to use the conditions described previously in the literature. We found that the optimal concentration of the co-substrate, H2O2, for AFMK production was between 100 and 200 μM (Figure 8A). Using these conditions, we incubated melatonin and other derivatives with MyelPO. The analytical data are presented in Figure 9. Within 30 min, approx. 5% of the melatonin was converted into two products (with retention times of 10.6 and 14.1 min; Figure 9A), which were identified by MS as AMK and AFMK (Figure 10). This experiment clearly demonstrates that melatonin is cleaved by MyelPO, leading to the formation of AFMK. The formyl group is labile under the conditions of the incubation, resulting in the formation of AMK (the kynurenine suspected to be an in vivo melatonin-derived catabolite). No catabolites were produced in the absence of H2O2 or MyelPO. At higher concentrations, H2O2 inhibited the reaction.
Figure 7. Enzymatic characteristics of MyelPO.
(A) Melatonin oxidation was performed in complete reaction mixture (see Experimental section) in the presence or absence of 1 mM azide. (B) The complete enzymatic system was tested over a 90 min time period. At each time point, an aliquot of the reaction mixture was taken and the reaction was stopped by addition of 40 μl of aqueous trichloroacetic acid (30%, v/v), and the amount of reaction product was measured by HPLC. (C) Direct representation of the enzymatic reaction (V) as a function of melatonin concentration in the assay (S). (D) Eadie–Hofstee representation of the data in (C). Experiments in (A) and (B) were performed three times, and data are given as means±S.D. For (C) and (D), data are presented as means±S.D. of triplicate values from a single experiment, representative of three independent experiments. The error bars are within the the experimental points.
Figure 8. Optimization of the concentration of H2O2, the co-substrate of MyelPO or IndoDO, in the presence of melatonin.
The reaction medium for MyelPO (A) and IndoDO (B) contained 0.84 μg of enzyme preparation incubated with increasing concentrations of H2O2 and 1 mM melatonin in PBS supplemented with potassium phosphate (55 mM, pH 5.4) in a volume of 50 μl and incubated at 37 °C for an additional 20 min. The reactions were then stopped by addition of trichloroacetic acid (30%, v/v), and the products were analysed by HPLC. The peak of AFMK was quantified as described in the Experimental section. The experiments were repeated three times, and the values are means±S.D.
Figure 9. Examples of the specificity of MyelPO towards four substrates.
MyelPO (0.88 μg/ml) was incubated with complete reaction medium containing 150 μM H2O2 in phosphate buffer (pH 5.4) at 37 °C for 30 min, plus one of the candidate substrates. The reaction was stopped by adding trichloroacetic acid (30%, v/v) and products were analysed as described in the legend to Figure 6. (A) Melatonin. The chromatogram shows the generation of two new peaks, labelled AMK and AFMK (see Figure 10). (B) Tryptamine. The chromatogram shows the generation of two new peaks (arrows). (C) 5-Hydroxytryptamine. The chromatogram shows the generation of one new peak (arrow). (D) N-Acetyl-5-hydroxytryptamine. The chromatogram shows the generation of one new peak (arrow). The experiments were performed at least three times, and representative chromatograms are shown.
Figure 10. Identification of AMK and AFMK produced from melatonin by MyelPO.
Melatonin (1 mM) was incubated with 0.84 μg of MyelPO and 50 μM H2O2. LC separations for the LC/MS experiments were performed using an HP 1090 binary pump equipped with an Xterra RP18 column. The samples were passed through a 20 μl injection loop at 1 ml/min and the flow was split to 250 μl/min into the mass spectrometer. The eluents were: A, 0.01% TFA in deionized water; B, 0.01% TFA in acetonitrile; the gradient sequence was from 0% to 80% A in 32 min. Electrospray ionization mass spectra were recorded on a Q-TOF2 instrument. (A) Reconstructed ionic current for m/z 178 theoretical, the most abundant fragment ion in the mass spectrum of AMK (retention time 10.6 min), and AFMK (retention time 15.1 min). (B) Total ionic current (m/z 20 to 900 theoretical) showing mainly melatonin at a retention time of 17.1 min.
Apparent affinity of MyelPO for melatonin
In order to measure the apparent Km of MyelPO for melatonin, the concentration of the hormone was varied from 0.005 to 5 mM. A plot of the direct data (Figure 7C) shows a sigmoïdal curve typical of positive allosteric behaviour, which was confirmed by the Eadie–Hofstee representation (Figure 7D). MyelPO apparent affinity was determined from a Hill plot (results not shown), and the apparent Km was 2.3±0.8 mM, with a Hill coefficient of 2.2±0.2. A Hill coefficient of >1 confirms positive allostery which is greater than, or equal to, the number of the interaction sites of MyelPO with melatonin. This result was not surprising, since MyelPO is a tetrameric enzyme that has two catalytic sites (see review by Ghibaudi and Laurenti [36]). The Vmax was 661±81 nmol of AFMK formed/min per mg of MyelPO.
MyelPO specificity
Once it was established that melatonin could be cleaved by MyelPO, we wanted to determine the specificity of this enzyme for other potential substrates. After a standard incubation in the presence of 150 μM H2O2, the reaction was stopped and analysed by HPLC. In all cases when a compound was substrate, a new product with a different retention time was visualized by HPLC (see examples in Figure 9). None of the products were characterized further. The compounds cleaved by the enzyme were 5-hydoxy-L-tryptophan (12±2%), tryptophol (14±0.5%), 5-hydroxytryptamine (22±1%) (Figure 9C) and N-acetyl-5-hydroxytryptamine (14±0.1%) (Figure 9D), as well as tryptamine (16±1%) (Figure 9B), 5-methoxytryptamine (11±0.2%) and 6-methoxytryptamine (13±0.1%). Furthermore, 2-methylindole (9±3%) also seemed to be cleaved to some extent, although with poor reproducibility. The results are given as the percentage disappearance of the main peak (100% was the area under the peak measured after incubation and analysis without enzyme). Among the compounds not recognized by MyelPO were 5-methoxy-D,L-tryptophan, 1-methylindole, 3-methylindole, benzimidazole, 4-hydroxyquinazoline, indole-2-carboxylic acid, indole-3-carboxylic acid, indole-2-acetic acid, indole-3-acetic acid and indole-3-glyoxylic acid. Depending on the compounds, two (for inactive) or three (for active) independent measurements were performed.
Characteristics of the peroxidase-like activity catalysed by IndoDO
The fact that IndoDO can show peroxidase-like activity has been largely disregarded in the literature, despite the fact that peroxidase activities are often associated with haem-containing enzymes. Assaying IndoDO under the same conditions as MyelPO, in the presence of H2O2, strictly as described by Takikawa et al. [8], it was possible to measure an activity towards melatonin. It was determined that the optimal H2O2 concentration for the production of AFMK from melatonin by IndoDO was ∼400 μM (Figure 8B). The relative amounts of AFMK and AMK formed varied due to the instability of the first metabolite under the experimental conditions, with either MyelPO or IndoDO. No AFMK was produced in the absence of enzyme or in the presence of denatured IndoDO (Figure 11). We found that the enzyme was Michaelian under these conditions (in contrast with the dioxygenase conditions of activity, where it was allosteric as shown above), and a Km of ∼0.6 mM was recorded (results not shown).
Figure 11. Evidence of the catalytic nature of the peroxidase-like activity of IndoDO towards melatonin.
The peroxidase-like activity of IndoDO was measured by incubating 400 μM H2O2 with melatonin. The reaction was stopped and analysed by HPLC. (A) IndoDO was replaced by water; (B) native recombinant IndoDO; (C) IndoDO was denatured at 95 °C for 15 min. The experiments were performed at least three times; chromatograms are representative of the results obtained.
DISCUSSION
Melatonin is a natural hormone which also has potent antioxidant capacities, albeit at high concentrations (e.g. above 10 μM). This compound is now available ‘over the counter’ in capsule form in doses as high as 20 mg. Melatonin normally circulates in mammals at low concentrations (between 10 and 400 pM), and only at night. Therefore high concentrations of this neurohormone could have unexpected and toxic side-effects. Indeed, adverse effects such as headache, gastrointestinal discomfort and hypo- or hyper-tension have been reported in clinical trials, all of short duration (less than 1 month) (www.iom.edu/includes/dbfile.asp?id=12948). Melatonin is removed from the body primarily through sulpho- and/or glucurono-conjugation, and the conjugates are then eliminated in the urine [38,39]. This classical pathway for drug metabolism accounts for approx. 60–70% of ingested melatonin. The fate of the remaining ∼30% seems to be divided into two categories: unchanged melatonin, present in several tissues, and production of an unknown metabolite that seemed to be analogous to kynurenines [7].
In the present study, we investigated two questions: (i) which enzyme(s) is responsible for the oxidative metabolism of melatonin, and (ii) what are the Km values of this particular compound for its metabolizing enzyme(s)? Several reports [14,40,41] had proposed IndoDO as a candidate for such an oxidative process, probably because of the reportedly wide substrate specificity of IndoDO and the structural similarity between melatonin and tryptophan. It has been demonstrated (and confirmed by us) that tryptophan is indeed cleaved by IndoDO. Previous experiments using melatonin as substrate have been conducted using tissue homogenates [7]. However, using melatonin and recombinant IndoDO, we were unable (under standard dioxygenase conditions) to show cleavage of this compound. One possible explanation for this apparent disparity is that the previous enzyme preparations had not been purified to homogeneity [7], and thus could have contained various amounts of MyelPO (e.g. from blood cells contaminating the tissue preparations). This is important for other reasons. Since IndoDO is regulated by interferon [42,43], it was tempting to link melatonin catabolism with fever and infections. Particularly surprising, but in line with these observations, was the enhanced capacity of IndoDO to cleave tryptophan at 40 °C as compared with 37 °C. We can reasonably rule out the possibility that, after interferon induction of IndoDO, the enzyme would become capable of oxidizing melatonin. Indeed, interferon induces the enzyme, and therefore only enhances its amount, as the increase was inhibited or diminished by actinomycin D and cycloheximide [44]. This induction would not change the enzyme specificity.
IndoDO is also capable of peroxidase-like activity [8,13]. Indeed, IndoDO exists in an oxygenated form that is still active to degrade substrates. Under our experimental (i.e. dioxygenase) conditions, oxygenated species (H2O2 or O2−) could not be produced, and therefore could not oxygenate the enzyme directly (which, in turn, would have oxidized melatonin). We show in the present paper that melatonin can be cleaved by a peroxidase-like activity catalysed by IndoDO under the conditions described by Takikawa et al. [8]. Under the same conditions, tryptophan is not a substrate of IndoDO. Nevertheless, the original report [7] clearly showed that with a complete system (Methylene Blue, catalase and ascorbic acid), melatonin was a substrate of a partially purified preparation of rabbit intestine IndoDO, a result that we were not able to confirm. Under the same conditions used in that paper, no peroxidase-like activity of IndoDO could be observed, ruling out the possibility that IndoDO would act as a peroxidase to cleave melatonin, as it does with aniline [8]. The other report establishing a link between IndoDO and melatonin is a paper by Fujiwara et al. [45], in which a supernatant from pineal glands was used as enzymatic source with an assay containing the same concentrations of the same components (catalase, Methylene Blue and ascorbic acid). It is therefore reasonable to assume (i) that the dioxygenase activity of IndoDO cannot destroy melatonin, and (ii) that the peroxidase-like activity of IndoDO under these conditions is responsible for the oxidative catabolism of melatonin reported in these publications [7,8]. The Km of IndoDO for melatonin is ∼0.6 mM, similar to the affinities of IndoDO for its other substrates, aniline and benzamphetamine [8]. Such a melatonin concentration would be impossible to reach under physiological conditions or after melatonin tablet ingestion (i.e. 2 mg could lead to a mere 2 μM concentration in blood). MyelPO has a Km for melatonin of ∼2 mM. Under these conditions the Michaelis–Menten relationship indicates that IndoDO and MyelPO would be operating at approx. 0.33% and 0.1% respectively of their maximum velocities. Theoretically, this could correspond for MyelPO to ∼0.57 nmol of AMFK produced/min per mg of enzyme, and for IndoDO to ∼1 nmol of AFMK/min per mg.
We have shown experimentally that the enzyme responsible for the generation of kynurenine-like compounds from melatonin is most probably MyelPO, as also suggested by others [10,11,46]. The next challenge will be to try to understand the actions of AFMK and its deformylated metabolite, AMK. AFMK has potent antioxidant capacity [47]. It has not been technically possible to measure these compounds in circulating fluids, such as cerebrospinal fluid. However, Rozov et al. [48] reported the absence of AMFK and AMK from serum, except after melatonin treatment. They showed the presence of AFMK at surprisingly high (7 nM) concentrations in microdialysis samples from rat brain lateral ventricle and from retina (∼50 pg per retina). A recent study also revealed that no AFMK could be detected in human plasma using an assay with a detection limit of 65 pmol/l [49]. However, after melatonin administration (100 μg, subcutaneously) in rat, AFMK was found in the plasma. One reason to explain the absence of AMK from serum or cerebrospinal fluid is the extremely low quantities likely to be present. Indeed if we consider that 5% of melatonin is metabolized to AFMK and AMK, and that the night-time levels of melatonin are approx. 400 pmol/l, the resulting AFMK/AMK levels would be below the limit of detection. Nevertheless, significant AFMK/AMK levels could be induced by exogenous melatonin: if we consider that administration of 100 μg of melatonin to rats resulted in a plasma concentration of the two metabolites of 3–4 nmol/l [49], melatonin treatment at a dose of 10 mg would be expected to give 100-fold higher AFMK/AMK concentrations. This is still far from the in vitro pharmacological concentration at which AFMK/AMK could be active [50].
IndoDO cleaves both D- and L-tryptophan, but under identical conditions it does not recognize melatonin, the decarboxylated and 5-methoxylated counterpart of this amino acid. Since IndoDO activity is not inhibited by melatonin, the carboxyl moiety (one of the two structural differences between the two compounds) appears to have an important role in substrate recognition at the IndoDO catalytic site [14]. Interestingly, however, TrypDO, the liver tryptophan-cleaving enzyme, has been reported to be inhibited by melatonin [51], pointing to major differences in enzyme structure between IndoDO and TrypDO. Furthermore, the plasticity of the catalytic site seems to be limited, since close analogues of tryptophan are poorly recognized by the enzyme, if at all. By contrast, MyelPO, showed activity towards melatonin as well as 5-hydroxytryptamine, but not tryptophan.
The relative importance of the two routes for melatonin catabolism is hard to determine. One can hypothesize that, being present in circulating cells, such as macrophages, MyelPO is likely to be exposed more frequently to melatonin than is the tissue enzyme IndoDO. On the other hand, melatonin travels freely across plasma membranes and into cells. Furthermore, these two enzymes are also dependent for their activity on the presence of their co-substrate, H2O2. More exploration will be necessary to detail their relative contributions to melatonin catabolism and subsequent kynurenine production.
Acknowledgments
We are indebted to Dr Gérald Guillaumet (Orléans, France) for the synthesis of AMK and AFMK, to Dr Benoît Malpaux (INRA, Nouzilly, France) for providing sheep cephalo-rachidian liquid samples, and to Mrs Nicole Taimiot for help and support. J. A. B. thanks Dr Emmanuel Canet and Mr Vincent Minvielle for their help in supporting the last part of this project.
References
- 1.Delagrange P., Atkinson J., Boutin J. A., Casteilla L., Lesieur D., Misslin R., Pellissier S., Pénicaud L., Renard P. Therapeutic perspectives for melatonin agonists and antagonists. J. Neuroendocrinol. 2003;15:442–448. doi: 10.1046/j.1365-2826.2003.01016.x. [DOI] [PubMed] [Google Scholar]
- 2. Reference deleted.
- 3.Axelrod J. The pineal gland: a neurochemical transducer. Science. 1974;184:1341–1348. doi: 10.1126/science.184.4144.1341. [DOI] [PubMed] [Google Scholar]
- 4.Klein D. C., Coon S. L., Roseboom P. H., Weller J. L., Bernard M., Gastel J. A., Zatz M., Iuvone P. M., Rodriguez I. R., Begay V., et al. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog. Horm. Res. 1997;52:482–510. [PubMed] [Google Scholar]
- 5.Leone A. M., Francis P. L., Silma R. E. The isolation, purification, and characterisation of the principal urinary metabolites of melatonin. J. Pineal Res. 1987;4:253–266. doi: 10.1111/j.1600-079x.1987.tb00863.x. [DOI] [PubMed] [Google Scholar]
- 6.Ishimura Y., Nozaki M., Jhayaishi O. The oxygenated form of L-tryptophan-2,3-dioxygenase as reaction intermediate. J. Biol. Chem. 1970;245:3593–3602. [PubMed] [Google Scholar]
- 7.Hirata F., Hayaishi O., Tokuyama T., Senoh S. In vitro and in vivo formation of two new metabolites of melatonin. J. Biol. Chem. 1974;249:1311–1313. [PubMed] [Google Scholar]
- 8.Takikawa O., Yoshida R., Hayaishi O. Monooxygenase activities of dioxygenases. Benzphetamine demethylation and aniline hydroxylation reactions catalyzed by indoleamine 2,3-dioxygenase. J. Biol. Chem. 1983;258:6808–6815. [PubMed] [Google Scholar]
- 9.Shimizu T., Nomiyama S., Hirata F., Hayaishi O. Indoleamine 2,3-dioxygenase. Purification and some properties. J. Biol. Chem. 1978;253:4700–4706. [PubMed] [Google Scholar]
- 10.Silva S. O., Ximenes V. F., Catalani L. H., Campa A. Myeloperoxidase-catalyzed oxidation of melatonin by activated neutrophils. Biochem. Biophys. Res. Commun. 2000;279:657–662. doi: 10.1006/bbrc.2000.3993. [DOI] [PubMed] [Google Scholar]
- 11.Allegra M., Furtmüller P. G., Regelsberger G., Turco-Liveri M. L., Tesoriere L., Perretti M., Livrea M. A., Obinger C. Mechanism of reaction of melatonin with human myeloperoxidase. Biochem. Biophys. Res. Commun. 2001;282:380–386. doi: 10.1006/bbrc.2001.4582. [DOI] [PubMed] [Google Scholar]
- 12.Littlejohn T. K., Takikawa O., Skylas D., Jamie J. F., Walker M. J., Truscott R. J. W. Expression and purification of recombinant human indoleamine 2,3-dioxygenase. Protein Expr. Purif. 2000;19:22–29. doi: 10.1006/prep.2000.1214. [DOI] [PubMed] [Google Scholar]
- 13.Terentis A. C., Thomas S. R., Takikawa O., Littlejohn T. K., Truscott R. J. W., Armstrong R. S., Yeh S. R., Stocker R. The heme environment of recombinant human indoleamine 2,3-dioxygenase. Structural properties and substrate-ligand interactions. J. Biol. Chem. 2002;277:15788–15794. doi: 10.1074/jbc.M200457200. [DOI] [PubMed] [Google Scholar]
- 14.Southan M. D., Truscott R. J. W., Jamie J. F., Pelosi L., Walker M. J., Maeda H., Iwamoto Y., Toné S. Structural requirements of the competitive binding site of recombinant human indoleamine 2,3-dioxygenase. Med. Chem. Res. 1996;6:343–352. [Google Scholar]
- 15.Austin C. J., Mizdrak J., Matin A., Sirijovski N., Kosim-Satyaputra P., Willows R. D., Robert T. H., Truscott R. J., Polekina G., Parker M. W., Jamie J. F. Optimised expression and purification of recombinant human indoleamine-2,3-dioxygenase. Protein Exp. Purif. 2004;37:392–398. doi: 10.1016/j.pep.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi T., Hirata F., Hayaishi O. Indoleamine-2,3-dioxygenase. Potassium superoxide as substrate. J. Biol. Chem. 1977;252:4643–4647. [PubMed] [Google Scholar]
- 17.Grohmann U., Fallarino F., Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 18.Ferry G., Loynet A., Kucharczyk N., Bertin S., Rodriguez M., Delagrange P., Galizzi J. P., Jacoby E., Volland J. P., Lesieur D., et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem. 2000;275:8794–8805. doi: 10.1074/jbc.275.12.8794. [DOI] [PubMed] [Google Scholar]
- 19.Ferry G., Mozo J., Ubeaud C., Berger S., Bertrand M., Try A., Beauverger P., Mesangeau C., Delagrange P., Boutin J. A. Characterization and regulation of a CHO cell line stably expressing the human serotonin N-acetyltransferase (EC 2.3.1.87) Cell. Mol. Life Sci. 2002;59:1395–1405. doi: 10.1007/s00018-002-8517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferry G., Ubeaud C., Mozo J., Péan C., Hennig P., Rodriguez M., Scoul C., Bonnaud A., Nosjean O., Galizzi J. P., et al. New substrate analogues of human serotonin N-acetyltransferase produce in situ specific and potent inhibitors. Eur. J. Biochem. 2004;271:418–428. doi: 10.1046/j.1432-1033.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferry G., Ubeaud C., Dauly C., Mozo J., Guillard S., Berger S., Jimenez S., Scoul C., Leclerc G., Delagrange P., Boutin J. A. Further attempts to purify human serotonin N-acetyltransferase (EC 2.3.1.87). Comparison with other species and implication for its enzymatic behavior. Protein Expr. Purif. 2004;38:84–98. doi: 10.1016/j.pep.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Nosjean O., Ferro M., Coge F., Beauverger P., Henlin J. M., Lefoulon F., Fauchere J. L., Delagrange P., Canet E., Boutin J. A. Identification of the melatonin binding site MT3 as the quinone reductase 2. J. Biol. Chem. 2000;275:31311–31317. doi: 10.1074/jbc.M005141200. [DOI] [PubMed] [Google Scholar]
- 23.Nosjean O., Nicolas J. P., Klupsch F., Delagrange P., Canet E., Boutin J. A. Comparative pharmacological studies at the melatonin receptors MT1, MT2 and MT3/QR2. Biochem. Pharmacol. 2001;61:1369–1379. doi: 10.1016/s0006-2952(01)00615-3. [DOI] [PubMed] [Google Scholar]
- 24.Audinot V., Mailliet F., Lahaye-Brasseur C., Bonnaud A., Le Gall A., Amosse C., Dromaint S., Rodriguez M., Nagel N., Galizzi J. P., et al. New selective ligands of human cloned melatonin MT1 and MT2 receptors. Naunyn-Schmiedeberg's Arch. Pharmacol. 2003;367:553–561. doi: 10.1007/s00210-003-0751-2. [DOI] [PubMed] [Google Scholar]
- 25.Mailliet F., Audinot V., Malpaux B., Bonnaud A., Delagrange P., Migaud M., Barrett P., Viaud-Massuard M. C., Lesieur D., Lefoulon F., et al. Molecular pharmacology of ovine melatonin receptor. Comparison with recombinant human MT1 and MT2. Biochem. Pharmacol. 2004;67:667–677. doi: 10.1016/j.bcp.2003.09.037. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto S., Hayaishi O. Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes. J. Biol. Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- 27.Segel I. H. New York: John Wiley & Sons; 1975. Enzyme Kinetics. [Google Scholar]
- 28.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Cady S., Sono M. 1-Methyl-DL-tryptophan, beta-(3-benzofuranyl)-DL-alanine (the oxygen analog of tryptophan), and beta-[3-benzo(b)thienyl]-DL-alanine (the sulfur analog of tryptophan) are competitive inhibitors for indoleamine 2,3-dioxygenase. Arch. Biochem. Biophys. 1991;291:326–333. doi: 10.1016/0003-9861(91)90142-6. [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki F., Kuroiwa T., Takikawa O., Kido R. Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem. J. 1985;230:635–638. doi: 10.1042/bj2300635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz J., Corlin R., Oddi F., Kaminker K., Jones W. Myeloperoxidase of the leucocyte of normal human blood. 3. Isolation of the peroxidase granule. Arch. Biochem. Biophys. 1965;111:73–79. doi: 10.1016/0003-9861(65)90324-3. [DOI] [PubMed] [Google Scholar]
- 32.Bos A., Wever R., Roos D. Characterization and quantification of the peroxidase in human monocytes. Biochim. Biophys. Acta. 1978;525:37–44. doi: 10.1016/0005-2744(78)90197-3. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues M. R., Rodriguez D., Catalani L. H., Russo M., Campa A. Macrophage activation includes high intracellular myeloperoxidase activity. Biochem. Biophys. Res. Commun. 2002;292:869–873. doi: 10.1006/bbrc.2002.6724. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues M. R., Rodriguez D., Catalani L. H., Russo M., Campa A. Interferon-gamma independent oxidation of melatonin by macrophages. J. Pineal Res. 2003;34:69–74. doi: 10.1034/j.1600-079x.2003.02944.x. [DOI] [PubMed] [Google Scholar]
- 35.Andrews P. C., Krinsky N. I. Human myeloperoxidase and hemi-myeloperoxidase. Methods Enzymol. 1986;132:369–378. doi: 10.1016/s0076-6879(86)32022-6. [DOI] [PubMed] [Google Scholar]
- 36.Ghibaudi E., Laurenti E. Unraveling the catalytic mechanism of lactoperoxidase and myeloperoxidase. Eur. J. Biochem. 2003;270:4403–4412. doi: 10.1046/j.1432-1033.2003.03849.x. [DOI] [PubMed] [Google Scholar]
- 37. Reference deleted.
- 38.Kopin I. J., Pare C. M. B., Axelrod J., Weissbach H. 6-Hydroxylation, the major metabolic pathway for melatonin. Biochim. Biophys. Acta. 1960;40:377–378. doi: 10.1016/0006-3002(60)91374-3. [DOI] [PubMed] [Google Scholar]
- 39.Kopin I. J., Pare C. M. B., Axelrod J., Weissbach H. The fate of melatonin in animals. J. Biol. Chem. 1961;236:3072–3075. [PubMed] [Google Scholar]
- 40.Hardeland R., Fuhrberg B. Ubiquitous melatonin – Presence and effects in unicells, plants and animals. Trends Comp. Biochem. Physiol. 1996;2:25–45. [Google Scholar]
- 41.Hardeland R., Coto-Montes A., Fuhrberg B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol. Int. 2003;20:921–962. doi: 10.1081/cbi-120025245. [DOI] [PubMed] [Google Scholar]
- 42.Carlin J. M., Ozaki Y., Byrne G. I., Brown R. R., Borden E. C. Interferons and indoleamine 2,3-dioxygenase: role in antimicrobial and antitumor effects. Experientia. 1989;45:533–541. doi: 10.1007/BF01990503. [DOI] [PubMed] [Google Scholar]
- 43.Tan D. X., Manchester L. C., Reiter R. J., Qi W., Zhang M., Weintraub S. T., Cabrera J., Sainz R. M., Mayo J. C. Identification of highly elevated levels of melatonin in bone marrow: its origin and significance. Biochim. Biophys. Acta. 1999;1472:206–214. doi: 10.1016/s0304-4165(99)00125-7. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida R., Imanishi J., Out T., Kishida T., Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by interferon. Proc. Natl. Acad. Sci. U.S.A. 1981;78:129–132. doi: 10.1073/pnas.78.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara M., Shibata M., Watanabe Y., Nukiwa T., Hirata F., Mizuno M., Hayaishi O. Indoleamine 2,3-dioxygenase. Formation of L-kynurenine from L-tryptophan in cultured rabbit fineal gland. J. Biol. Chem. 1978;253:6081–6085. [PubMed] [Google Scholar]
- 46.Ximenes V. F., Catalani L. H., Campa A. Oxidation of melatonin and tryptophan by an HRP cycle involving compound III. Biochem. Biophys. Res. Commun. 2001;287:130–134. doi: 10.1006/bbrc.2001.5557. [DOI] [PubMed] [Google Scholar]
- 47.Tan D. X., Manchester L. C., Burkhardt S., Sainz R. M., Mayo J. C., Kohen R., Shohami E., Huo Y. S., Hardeland R., Reiter R. J. N1-acetyl-N2-formyl-5-methoxykynuramine, a biogenic amine and melatonin metabolite, functions as a potent antioxidant. FASEB J. 2001;15:2294–2296. doi: 10.1096/fj.01-0309fje. [DOI] [PubMed] [Google Scholar]
- 48.Rozov S. V., Filatova E. V., Orlov A. A., Volkova A. V., Zhloba A. R. A., Blashko E. L., Pozdeyevn N. V. N1-acetyl-N2-formyl-5-methoxykynuramine is a product of melatonin oxidation in rats. J. Pineal Res. 2003;35:245–250. doi: 10.1034/j.1600-079x.2003.00081.x. [DOI] [PubMed] [Google Scholar]
- 49.Harthe C., Claudy D., Dechaud H., Vivien-Roels B., Pevet P., Claustrat B. Radioimmunoassay of N-acetyl-N-formyl-5-methoxykynuramine (AFMK): a melatonin oxidative metabolite. Life Sci. 2003;73:1587–1597. doi: 10.1016/s0024-3205(03)00483-1. [DOI] [PubMed] [Google Scholar]
- 50.Stone T. W. Kynurenines in the CNS: from endogenous obscurity to therapeutic importance. Prog. Neurobiol. 2001;64:185–218. doi: 10.1016/s0301-0082(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 51.Walsh H. A., Daya S. Inhibition of hepatic tryptophan-2,3-dioxygenase: superior potency of melatonin over serotonin. J. Pineal Res. 1997;23:20–23. doi: 10.1111/j.1600-079x.1997.tb00330.x. [DOI] [PubMed] [Google Scholar]