Abstract
Members of the highly related TEF-1 (transcriptional enhancer factor-1) family (also known as TEAD, for TEF-1, TEC1, ABAA domain) bind to MCAT (muscle C, A and T sites) and A/T-rich sites in promoters active in cardiac, skeletal and smooth muscle, placenta, and neural crest. TEF-1 activity is regulated by interactions with transcriptional co-factors [p160, TONDU (Vgl-1, Vestigial-like protein-1), Vgl-2 and YAP65 (Yes-associated protein 65 kDa)]. The strong transcriptional co-activator YAP65 interacts with all TEF-1 family members, and, since YAP65 is related to TAZ (transcriptional co-activator with PDZ-binding motif), we wanted to determine if TAZ also interacts with members of the TEF-1 family. In the present study, we show by GST (glutathione S-transferase) pull-down assays, by co-immunoprecipitation and by modified mammalian two-hybrid assays that TEF-1 interacts with TAZ in vitro and in vivo. Electrophoretic mobility-shift assays with purified TEF-1 and GST–TAZ fusion protein showed that TAZ interacts with TEF-1 bound to MCAT DNA. TAZ can interact with endogenous TEF-1 proteins, since exogenous TAZ activated MCAT-dependent reporter promoters. Like YAP65, TAZ interacted with all four TEF-1 family members. GST pull-down assays with increasing amounts of [35S]TEF-1 and [35S]RTEF-1 (related TEF-1) showed that TAZ interacts more efficiently with TEF-1 than with RTEF-1. This differential interaction also extended to the interaction of TEF-1 and RTEF-1 with TAZ in vivo, as assayed by a modified mammalian two-hybrid experiment. These data show that differential association of TEF-1 proteins with transcriptional co-activators may regulate the activity of TEF-1 family members.
Keywords: muscle, transcriptional co-activator, transcriptional co-activator with PDZ-binding motif (TAZ), transcriptional enhancer factor-1 (TEF-1), TEF-1, TEC1, ABAA domain (TEAD), Yes-associated protein 65 kDa (YAP65)
Abbreviations: Co-IP, co-immunoprecipitation; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; ETF-1, embryonic TEA-domain-containing factor; GST, glutathione S-transferase; HA, haemagglutinin; HSV, herpes simplex virus; IVT, in vitro transcription/translation; LMC, low-mobility complex; MCAT site, muscle C, A and T sites; MEF2, myocyte enhancer factor 2; βMHC, β-myosin heavy chain; PARP, poly(ADP-ribose) polymerase; RT, reverse transcriptase; SRF, serum-response factor; TAZ, transcriptional co-activator with PDZ-binding motif; TEF-1, transcriptional enhancer factor-1; DTEF-1, divergent TEF-1; RTEF-1, related TEF-1; Tk, thymidine kinase; UAS, upstream activator sequence; Vgl, Vestigial-like protein; WT, wild-type; YAP65, Yes-associated protein 65 kDa
INTRODUCTION
There are four members of the TEF-1 (transcriptional enhancer factor-1) gene family (reviewed in [1]): TEF-1 [also called NTEF-1 (nominal TEF-1) or TEAD1 (TEAD is TEF-1, TEC1, ABAA domain)], RTEF-1 (related TEF-1) (TEFR1, TEF-3 or TEAD4), ETF-1 (embryonic TEA-domain-containing factor) (TEF-4 or TEAD2) and DTEF-1 (divergent TEF-1) (TEF-5, ETFR-1 or TEAD3). TEF-1 binds to MCAT (muscle C, A and T sites), sequence related to CATTCC(A/T), in promoters active in cardiac, skeletal and smooth muscle, placenta, and neural crest. Recently, R. Tsika and co-workers found that TEF-1 binds weakly to A/T-rich binding sites in muscle promoters [2], expanding the promoters that are potentially regulated by TEF-1. The vast majority of cellular promoters that are MCAT-dependent are muscle-specific [1,3–5]. In cardiac muscle, MCAT sites are also required for the full activity of promoters in the presence of hypertrophic signals [6–9]. There may be differences between TEF-1 family members, since TEF-1 and DTEF-1 are implicated in cardiac gene regulation, and RTEF-1 is involved in skeletal muscle differentiation [4,10–13]. TEF-1 is developmentally important, since TEF-1-knockout mice die at ED (embryonic day) 10–11 from an abnormally thin heart ventricular wall [14], an MCAT site is required for PAX3 expression in the neural crest [15] and ETF-1 (TEAD2) is active in the early embryo [16].
TEF-1 family members are broadly expressed [17–19], but function as transcriptional activators only in a subset of their expression domains (cardiac, skeletal and smooth muscle, placenta, and skin; reviewed in [1]). The activity of TEF-1 family members in vertebrates and in Drosophila is regulated by co-factors [20,21]. Known TEF-1 co-factors can be divided into four classes. Class 1: interacting proteins with co-activation function [p160 steroid hormone receptor co-activators and YAP65 (Yes-associated protein 65 kDa)]. These TEF-1 co-factors contain transactivation domains, interact with TEF-1 and activate transcription of MCAT-dependent promoters ([22–24], and see below). Class 2: interacting proteins without co-activation function [TONDU (Vgl-1, Vestigial-like protein-1), Vgl-2 and Vgl-3]. TONDU, Vgl-2 and Vgl-3, vertebrate proteins related to the Drosophila co-activator Vg [20,25,26], interact with TEF-1 proteins, but do not contain a transactivation domain. Expression of these is tissue restricted (placenta, skeletal muscle and heart), supporting the hypothesis that TEF-1 activity is regulated by tissue-specific co-factors. Class 3: DNA co-binders {PARP [poly(ADP-ribose) polymerase]}. PARP forms a tertiary complex with TEF-1 and the sequences flanking MCAT sites in vivo, and may remodel the chromatin of MCAT-dependent promoters [27–30]. Class 4: other transcription factors [MAX, MEF2 (myocyte enhancer factor 2) and SRF (serum-response factor)]. The basic helix–loop–helix factor, MAX, MEF2 and SRF interact with the DNA-binding domain of TEF-1 [31–33], and may therefore co-operate with TEF-1 to activate muscle transcription.
YAP65 was originally identified through its interaction with the Src family tyrosine kinase Yes [34]. YAP65 is approx. 45% identical with the PDZ-binding protein TAZ (transcriptional co-activator with PDZ-binding motif) [23]. YAP65 and TAZ also have similar domain structures; one or two WW domains, a 14-3-3-binding site and a C-terminal PDZ-binding motif [23]. Phosphorylated YAP65 and TAZ are bound to the phospho-serine/threonine-binding protein 14-3-3 in the cytoplasm, and dephosphorylated YAP65 and TAZ are found in the nucleus, where they function as transcriptional co-activators [23,24]. Intracellular localization of YAP65 can be regulated by overexpression of Akt/protein kinase B [35]. However, no endogenous cell signalling system has been shown to influence the intracellular localization of YAP65 or TAZ.
YAP65 and TAZ are transcriptional co-activators for a number of transcription factors: p63/p73 [36], the TEF-1 gene family [24], the ErbB4 nuclear fragment [37], Runx2/Cbfa1 [38] and TTF-1/Nkx2.1 [39]. YAP65/TAZ-interacting transcription factors have diverse functions, including cell-cycle regulation, pro- and antiapoptosis, DNA repair, skeletal muscle and osteoblast differentiation, cardiac trabeculae and conduction system development, and thyroid- and lung-specific gene expression.
TAZ is related to YAP65 in the TEF-1-interaction domain of YAP65, and is also expressed in tissues where the TEF-1 gene family is expressed and active (see below and [23,24]). Owing to this homology with YAP65 and co-expression with TEF-1, we examined whether TAZ interacts with the TEF-1 gene family. We found that TAZ interacts with TEF-1 in vitro, in vivo and with TEF-1 on MCAT–DNA. TAZ interacted with TEF-1 to activate transcription, both as a GAL4-fusion protein and through interaction of TAZ with endogenous TEF-1 proteins on MCAT-dependent promoters. We also found that TAZ interacts with all four TEF-1 family members in vitro, but TAZ interacts more efficiently with TEF-1 than with RTEF-1. In vivo, TAZ was a more efficient co-activator for TEF-1 than for RTEF-1. These data show that TAZ is a transcriptional co-activator of the TEF-1 gene family, and that the activity of TEF-1 family members may be regulated by differential association of TEF-1 proteins with YAP65 and TAZ.
EXPERIMENTAL
Plasmids
The following expression plasmids have been described previously [23]: pcDNA3.1/His-TAZ, pEF-TAZ-NFLAG, pEF-TAZ-NFLAG(S89A), pGAL4-TAZ and pGAL4-YAP65. The rat TEF-1 isoforms epitope tagged with the HSV (herpes simplex virus) peptide, XJ40-TEF-1–HSV, were also described previously [40]. Expression vectors for human ETF-1 and DTEF-1 were kindly provided by Dr Irwin Davidson (Institut de Genetique et de Biologie Moleculaire et Cellulaire, Illkirch, France). Expression vectors for human (h) RTEF-1– and hTEF-1–GAL4 fusion proteins {pM, pM-hTEF-1(1-426), equivalent to rat TEF-1ζ (see Figure 2 and [40]), pM-hTEF-1(79-426), and pM-hRTEF-1(79-426)} and a GAL4-UAS (upstream activator sequence)-dependent luciferase reporter (pG5-luc) were kindly provided by Dr Alexandre Stewart (University of Ottawa, Heart Institute, Ottawa, Ontario, Canada) [11,26]. pCMV-FL-TEF-1ε: TEF-1ε [40] was released from XJ40-TEF-1ε–HSV with XhoI and cloned into pCMV-FLAG-5a (Sigma) digested with SalI to create pCMV-FL-TEF-1ε. GST–TAZ-(1–239) (where GST is glutathione S-transferase): a BamHI fragment from pcDNA3.1/His-TAZ (5′ to endogenous site at position 239) was cloned into the BamHI site of pQE30 to yield pQE30-TAZ(1-239) [23]. TAZ-(1–239) was released from pQE30-TAZ(1-239) with BamHI and subcloned into pGEX-4T-1 (Amersham Biosciences). GST–TAZ-(239–395): a BamHI (at position 239) to EcoRI (3′) fragment from pcDNA3.1/His-TAZ was cloned into pGEX-5x-2. pET-TEF-1ε and pCMV-HA-YAP65 (where HA is haemagglutinin): the coding region of rat TEF-1ε and murine YAP65 were amplified by PCR and cloned into pET-Blue-2 (Novagen) and pCMV-HA (Clontech) respectively. All constructs were verified by sequencing.
Figure 2. TEF-1 specifically interacts with the N-terminus of TAZ in vitro.
(A) Interaction between TEF-1 and TAZ was assayed by GST pull-down assays. GST-fusion proteins of the N-terminus (N; amino acids 1–239) and the C-terminus (C; amino acids 239–395) of TAZ were produced and purified. Both empty GST protein (G) and GST–agarose beads alone (−) were used as controls. These proteins (5 μg) were mixed with the following in vitro labelled [35S]methionine TEF-1 isoforms: TEF-1ε, TEF-1η, TEF-1εΔA, TEF-1ζ, TEF-1-D and TEF-1εΔD [see (B) for diagram]. The TEF-1 and TAZ proteins were mixed with interaction buffer and GST–agarose beads overnight at 4 °C. Protein complexes were washed extensively and separated by SDS/PAGE (12.5% gel). Lanes 1–6 represent 7% of the total volume of lysate probed in each reaction. TEF-1εΔD was produced at a lower level, so this portion of the gel is a longer exposure. (B) Schematic representation of TEF-1 isoforms utilized in the GST pull-down assay (left) and summary of the TAZ–TEF-1 interaction assay (right).
RT (reverse transcriptase)-PCR
Total RNA from adult mouse heart and skeletal muscle was obtained from Clontech. RT-PCR was performed with a single-tube RT-PCR system (ProStar HF, Stratagene). PCR primers were TAZ (GenBank® accession number AJ299430): sense, 5′-ATGTGAACCTCCACCCGTCCATCAC-3′ (nt 709–733), and antisense, 5′-GAAGAGAGGGATCAGATCTTCAGACTC-3′ (nt 1349–1323); YAP65 (GenBank® accession number NM 009534): sense, 5′-AAGACGGCCAACGTGCCTCAGA-3′ (nt 379–400), and antisense, 5′-GGAACTCAGCGCTTCCTGCAGACT-3′ (nt 1545–1522).
Cell culture
H1299 cells (a human lung carcinoma cell line) were obtained from Dr France Carrier (University of Maryland, Baltimore, MD, U.S.A.) and maintained in MEM (minimal essential medium) with Earle's salts, supplemented with 10% (v/v) foetal calf serum. C2C12 cells were obtained from the A.T.C.C. (Manassas, VA, U.S.A.) and maintained at low density in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) foetal calf serum. For C2C12 myotubes, cells at 80% confluence were switched from growth media to DMEM supplemented with 2% (v/v) horse serum. Differentiation was allowed to continue for 72 h. Nuclear and cytoplasmic extracts from C2C12 cells were prepared using a commercially available kit (NE-PER; Pierce).
Recombinant protein purification
TAZ constructs in GST expression vectors or full-length TEF-1ε in pET-Blue-2 (Novagen) were expressed in Escherichia coli (BL21-RP; Stratagene), and proteins were purified by glutathione–agarose (Amersham Biosciences) or chelated metal–agarose (Qiagen) affinity chromatography, following the manufacturers' specifications. Coomassie Blue staining (results not shown) confirmed purity of protein expression. Purified proteins were dialysed against 20 mM Hepes, 100 mM NaCl, 0.5 mM EDTA and 1 mM dithiothreitol, pH 7.6, containing protease inhibitors.
GST pull-down assays
35S-labelled TEF-1 isoforms were produced by IVT (in vitro transcription/translation) using a T7 TnT® Quick Coupled system (Promega) following the manufacturer's specifications. Pull-down assays were performed with IVT [35S]TEF-1 proteins (50 μl), 5 μg of purified GST–TAZ-(1–239), GST–TAZ-(239–395), or GST, and glutathione–agarose (25 μl) in 750 μl of interaction buffer (20 mM Tris/HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 10% glycerol and 0.1% Nonidet P40). After 16 h at 4 °C with agitation, the protein complexes bound to beads were centrifuged at 1500 g for 5 min, washed, resuspended in 2× SDS/PAGE sample buffer, and separated by SDS/PAGE (12.5% gel). Gels were fixed, dried, and exposed to a phosphorimaging screen and an autoradiography (Biomax) film. The dose–response curve for interaction of TEF-1 and RTEF-1 was generated using 0, 10, 20, 30, 40 and 50 μl of 35S-labelled TEF-1 or RTEF-1 and unprogrammed lysate (0–50 μl) to a final volume of 50 μl. The amount of input TEF-1 proteins was quantified by trichloroacetic acid precipitation and by phosphorimager analysis of a spotted standard curve. There was good agreement between the methods in the amount of input TEF-1 protein measured.
EMSAs (electrophoretic mobility-shift assays)
EMSAs were performed as described previously [4]. Briefly, purified TEF-1ε (50 ng), GST–TAZ-(1–239) (0.1–2 μg) or GST (5 μg) were mixed with 15000 cpm of 32P 5′-end-labelled probe containing either a single TEF-1-binding site (CATTCCT) or containing tandem TEF-1-binding sites separated by two base pairs (MCAT1 and OGT2-56 [4]). A 0.2 μg amount of anti-GST antibody (Amersham Biosciences) or monoclonal anti-TEF-1 antibody (Signal Transduction Laboratories) was used to super-shift the protein complexes. Complexes were separated on a native 6% gel in TBE (Tris/borate/EDTA). The gel was dried, and exposed to a phosphorimaging screen and an autoradiography (Biomax) film.
Co-IP (co-immunoprecipitation) assay and immunoblotting
H1299 cells were co-transfected with expression vectors (5 μg/100 mm dish) encoding both pXJ40-TEF-1ε-HSV and pEF-TAZ-NFLAG (wild-type or the predominantly nuclear S89A mutant) with FuGENE 6 (Roche) according to the manufacturer's specifications. Whole-cell extracts were prepared [24] by resuspending the cell pellet 2 days after transfection in EBC (50 mM Tris/HCl, pH 8.0, 120 mM NaCl, 0.5% Nonidet P40 and 1 mM EDTA). Extracts were pre-cleared with normal mouse IgG and Protein G–agarose beads for 1 h at room temperature (25 °C). The precleared extracts were incubated overnight at 4 °C with Protein G–agarose beads and either monoclonal anti-HSV antibody (Novagen) or normal mouse IgG (Upstate Biotechnology). The beads were subsequently washed five times with EBC and resuspended in 2× SDS/PAGE sample buffer. Proteins were resolved by SDS/PAGE (12.5% gel) and transferred on to a PVDF membrane. Western blot analysis was performed with affinity-purified polyclonal anti-YAP65/TAZ antibody (Orbigen) [23]. pCMV-FL-TEF-1ε and pCMV-HA-YAP65 were transfected as above and complexes of FLAG–TEF-1ε with HA–YAP65 isolated by anti-FLAG-conjugated agarose (Sigma) or Protein G–agarose as a control. Complexes were eluted from the FLAG–agarose with peptide and analysed by immunoblotting as above or purified further by MCAT DNA-affinity chromatography as described in [3,24]. Association of FLAG–TEF-1 with endogenous YAP65/TAZ was done as described above using the anti-YAP65/TAZ polyclonal antibody in the Western blot. Immunoblotting of C2C12 nuclear extracts (20 μg of protein) was performed with the anti-YAP65/TAZ polyclonal antibody.
Transient transcriptional activation assays
H1299 cells were transfected using FuGENE 6 (Roche). Each six-well dish contained 250 ng of luciferase reporter plasmids, 10 ng of Tk (thymidine kinase)–Renilla (Promega) as a co-transfection control, 5–100 ng of GAL4-fusion proteins as activators, and 100 ng of co-activators (YAP65/TAZ). Empty expression vectors were used to keep the final amount of transfected DNA constant. Expression vectors for GAL4-fusions or YAP65/TAZ did not affect Tk–Renilla activity. At 2 days after transfection, cellular extracts were collected and assayed for promoter activity using the Dual Luciferase Assay system (Promega). Experiments were performed in duplicate and were repeated four to six times. Data are expressed as means±S.E.M. and were analysed by the paired two-tailed Student's t test or ANOVA.
RESULTS
YAP65 and TAZ are related and expressed in muscle tissues
YAP65 is a 65 kDa protein found in the cytoplasm, bound to 14-3-3 proteins, and in the nucleus, bound to a number of transcription factors [24,34,36–39]. YAP65 and the TEF-1 family protein, ETF-1 (TEAD2) interact specifically in the nucleus in stable complexes that were isolated by two rounds of affinity chromatography [24]. Mapping studies showed that the N-terminal portion of YAP65 (amino acids 31–139) interacts with the C-terminal portion (amino acids 115–445) of ETF-1 (Figures 1A and 1B). YAP65 interacts with all four TEF-1 family members, and is a strong transcriptional co-activator for TEF-1. Vassilev et al. [24] proposed that mitogenic (or other) signals cause a dephosphorylation of YAP65 and release from the 14-3-3 complex. YAP65 subsequently translocates to the nucleus where it can interact with TEF-1 proteins (or other factors) and activate transcription.
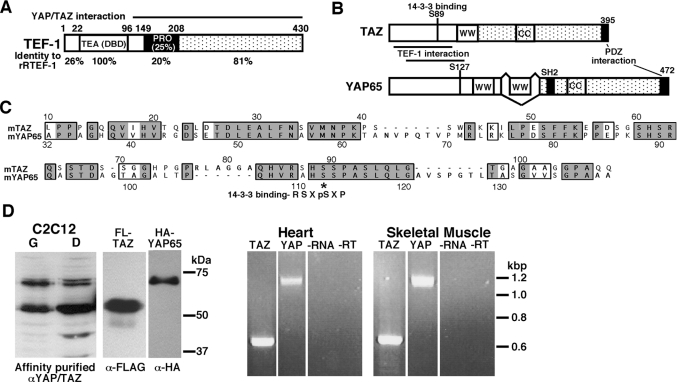
Figure 1. TEF-1, YAP65 and TAZ proteins.
(A) The TEF-1 family of proteins has a number of conserved domains: the DNA-binding TEA domain (DBD), the proline-rich domain (PRO) and the C-terminal half of TEF-1. The region of TEF-1 that interacts with YAP65 is shown. Additionally, the amino acid identity between rTEF-1 and rRTEF-1 is shown for each domain. (B) TAZ is homologous with YAP65. The diagram shows the structure of YAP65 and TAZ. The following domains are highlighted: the WW (two conserved tryptophan residues spaced 20–22 amino acids apart) domains, the CC (coiled-coil) domains, the PDZ-interaction domain, the TEF-1-interaction domain, and the specific amino acid which regulates interaction with 14-3-3 (Ser89 in murine TAZ and Ser112 in mouse YAP65). (C) Homology of YAP65 and TAZ. Sequence alignment of the mouse TAZ and mouse YAP65 proteins within the region of YAP65 that interacts with the TEF-1 family of proteins [24]. Identical amino acids are boxed and shaded, and similar amino acids are boxed. *, 14-3-3-interaction motif: R-S-X-pS-X-P. (D) TAZ and YAP65 are expressed in muscle tissues. Left: Western blots on nuclear extracts (20 μg) from C2C12 myoblasts and myotubes were performed with the anti-YAP65/TAZ antibody (αYAP/TAZ). The immunoreactive bands are specific, as they co-migrate with overexpressed TAZ-N–FLAG and HA–YAP65 and pre-incubation of the antibody (anti-FLAG, α-FLAG, or anti-HA, α-HA) with immunogenic peptide resulted in no immunoreactivity (results not shown). Right: RT-PCR analyses on RNA from adult mouse heart and skeletal muscle (leg) were carried out with primers for YAP65 and TAZ. Both YAP65 and TAZ are expressed. Controls for these reactions, no RNA and no RT (−RNA −RT), resulted in no DNA products. Sizes in kDa (left) or kb (right) are indicated.
TAZ was identified as a 14-3-3-binding protein. TAZ is related to YAP65 in primary sequence and structure (Figure 1B) [23]. Importantly, in the TEF-1 interacting region YAP65 is highly related to TAZ, with 65% identity and 77% similarity (Figures 1B and 1C) [23,24]. Both YAP65 and TAZ are expressed in tissues where TEF-1 proteins regulate transcription. YAP65 and TAZ proteins are expressed in the nuclei of myoblasts and myotubes from the skeletal muscle cell line C2C12 (Figure 1D) and in neonatal rat cardiac myocytes (results not shown). YAP65 and TAZ mRNA were also detected by RT-PCR in adult mouse heart and skeletal muscle (Figure 1D). These results are consistent with the RNA expression studies previously reported [23]. Owing to the co-expression of TEF-1 proteins and YAP65/TAZ, and the homology between YAP65 and TAZ, we examined whether TAZ also interacts with the TEF-1 gene family.
TAZ interacts with TEF-1 in vitro and in vivo
To determine if TAZ interacts with TEF-1 in vitro, GST pull-down assays were performed. The N- and C-terminal halves of TAZ were fused to GST to yield GST–TAZ-(1–239) and GST–TAZ-(239–395) respectively. GST–TAZ-(1–239), GST–TAZ-(239–395) and GST alone were produced in bacteria, purified, and used to determine if multiple TEF-1 isoforms [40] and deletions interacted with TAZ (Figure 2). Specifically, these proteins are: TEF-1ε, the predominant TEF-1 protein in cardiac myocytes (I.K.G. Farrance and P. C. Simpson, unpublished work); TEF-1ζ, the rat equivalent of hTEF-1 [40,41]; TEF-1η, a rare splicing isoform of TEF-1 lacking the C-terminal portion of the TEA domain (amino acids 90–110); TEF-1ε(ΔA), deletion of amino acids 1–89; TEF-1-D (deletion of amino acids 1–114); and TEF-1εΔD, deletion of amino acids 115–430. GST-fusions were incubated with [35S]TEF-1 proteins, produced by coupled IVT, incubated with glutathione–agarose, washed and analysed by SDS/PAGE. All TEF-1 proteins containing amino acids 115–430 of TEF-1ε interacted with GST–TAZ-(1–239), but not with GST–TAZ-(239–395), GST or beads alone (Figure 2A). TEF-1εΔD did not interact with TAZ. These results, interaction of the C-terminal portion of TEF-1 with the N-terminal half of TAZ, agrees with the results of Vassilev et al. [24], for the interaction of the TEF-1 gene family with YAP65.
Co-IP assays were used to determine whether TEF-1 interacted with TAZ in vivo. Two TAZ proteins were analysed for interaction with TEF-1: wild-type FLAG-tagged TAZ (TAZ-N–FLAG) and a preferentially nuclear 14-3-3-binding site mutant [TAZ-N–FLAG(S89A)] [23]. Expression vectors for HSV-tagged TEF-1ε and TAZ-N–FLAG (WT or S89A) (where WT is wild-type) were introduced into the human lung cell line H1299, and extracts from the cells were subjected to Co-IP with anti-HSV monoclonal antibody at low stringency to precipitate HSV–TEF-1ε and associated proteins (Figure 3A). TEF-1ε interacts specifically with both TAZ-N–FLAG(WT) and TAZ-N–FLAG(S89A). Co-IP assays were also carried out with FLAG–TEF-1ε and HA-tagged YAP65 using anti-FLAG–agarose (M2; Sigma) or Protein G–agarose as control, to isolate FLAG–TEF-1ε and associated proteins (Figure 3B). These experiments show that YAP65 interacts with TEF-1 and directly confirm the results of Vassilev et al. [24]. Exogenous TEF-1 also interacts with endogenous YAP65/TAZ proteins (Figure 3C). Extracts from H1299 cells containing FLAG–TEF-1 were subjected to Co-IP using anti-FLAG–agarose or Protein G–agarose as control. Western blot with an anti-YAP65/TAZ antibody (Orbigen) showed that endogenous YAP65/TAZ interacts with the transfected FLAG–TEF-1.
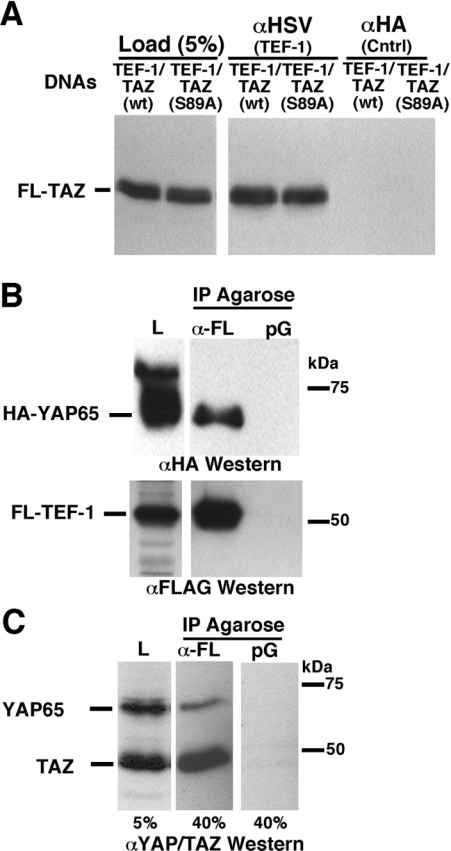
Figure 3. TEF-1 interacts with TAZ in vivo.
(A) The interaction between TEF-1 and TAZ was confirmed by an in vivo Co-IP assay. H1299 cells were co-transfected with expression vectors for both TEF-1ε–HSV (XJ40-TEF-1ε-HSV) and FLAG–TAZ [pEF-TAZ-NFLAG, wild-type (wt) or the predominantly nuclear S89A mutant]. Cell extracts, antibodies [anti-HSV monoclonal antibody (αHSV, TEF-1) or the anti-HA antibody (αHA, Cntrl)] and Protein G–agarose were allowed to interact overnight at 4 °C. Proteins complexed to TEF-1ε–HSV were washed and separated by SDS/PAGE (10% gel). TAZ-N–FLAG (FL-TAZ) was identified by Western blot analysis with a polyclonal anti-YAP65/TAZ antibody. (B) YAP65 interacts with TEF-1. C2C12 cells were co-transfected with expression vectors encoding HA–YAP65 (pCMV-HA-YAP65) and FLAG–TEF-1ε (pCMV-FL-TEF-1ε). Complexes of FLAG–TEF-1 with HA–YAP65 were isolated by anti-FLAG–agarose (α-FL) or Protein G–agarose (pG) as control. Complexes were eluted from the FLAG–agarose with FLAG peptide, resolved by SDS/PAGE and HA–YAP65 (upper panel) and FLAG–TEF-1ε (FL-TEF-1; lower panel) were detected by Western blot with anti-HA (αHA) or anti-FLAG (αFLAG) antibodies. L, load. (C) Exogenous TEF-1 interacts with endogenous YAP65 and TAZ. H1299 cells were transfected with the FLAG–TEF-1 expression vector (pCMV-FL-TEF-1ε) and FLAG–TEF-1ε was isolated using anti-FLAG–agarose (α-FL) or Protein G–agarose (pG) as control. TEF-1 and associated proteins were separated by SDS/PAGE. Western blot analysis was performed with a polyclonal anti-YAP/TAZ antibody (αYAP/TAZ), showing that both endogenous TAZ and YAP65 are complexed to TEF-1 in vivo. Sizes in kDa are indicated in (B) and (C).
TEF-1 and TAZ interact while bound to MCAT DNA
TEF-1 binds to MCAT sequences [related to CATTCC(A/T)] or A/T-rich sites within the promoter region of many genes, including βMHC (β-myosin heavy chain), cardiac troponin T and skeletal α-actin [1,2]. The above results show that TAZ and YAP65 interact with TEF-1, but the significance of this interaction to transcriptional regulation depends on these interactions occurring while TEF-1 is bound to DNA. To address this issue, we performed EMSAs, with bacterially produced and purified TEF-1ε, radio-labelled MCAT DNA probes and increasing amounts of GST–TAZ-(1–239). MCAT probes were (i) a single MCAT site, MCAT1 from the cTNT promoter, and (ii) 2×GTIIC, which contains two MCAT sites separated by 2 bp [3,4]. TEF-1 formed a single complex with MCAT1 and two complexes with 2× GTIIC (Figure 4A, lanes 2 and 13). Addition of GST–TAZ-(1–239) caused the formation of an LMC (low-mobility complex) (Figure 4A, lanes 3–7 and 14–18). The amount of LMC increases as the concentration of TAZ increases. The TAZ–TEF-1–MCAT complexes are specific, as GST–TAZ fails to bind MCAT-dependent DNA when TEF-1 is not present (Figure 4A, lanes 9 and 20). The LMC complex is not due to the GST portion of the fusion, as addition of GST alone does not form the LMC (Figure 4A, compare lanes 8 and 19 with lanes 2 and 13). Addition of anti-GST (Figure 4A, lanes 10 and 21) or anti-TEF-1 antibody (Figure 4A, lanes 11 and 22) supershifts the LMC, indicating that the LMC contains both TAZ and TEF. These results show that TEF-1 and TAZ interact while bound to MCAT DNA.
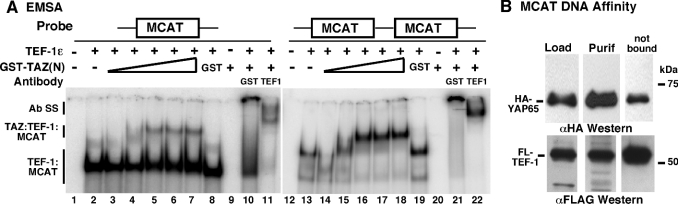
Figure 4. MCAT DNA-bound TEF-1 interacts with TAZ and YAP65.
(A) Interaction of TEF-1 and TAZ on DNA was assayed by EMSAs. Purified TEF-1ε was mixed with [32P]MCAT DNA [MCAT1 from the cTNT promoter, containing a single MCAT site, or 2×GTIIC, containing two tandem MCAT sites separated by two base pairs (lanes 2 and 13)]. One (MCAT1) or two (2×GTIIC) TEF-1–MCAT complexes were formed. Increasing amounts of GST–TAZ-(1–239) were added to the reaction mixtures (lanes 2–7 and 13–18), causing formation of an LMC (TAZ–TEF-1–MCAT). Increasing TAZ concentrations caused the LMC to become more prominent. The LMC contains TEF-1 and TAZ, since both anti-GST (lanes 10 and 21) and anti-TEF-1 (lanes 11 and 22) antibodies supershifted this complex (Ab SS). No complexes were seen between MCAT DNA and GST alone (lanes 9 and 20) or between MCAT DNA and either the anti-GST or anti-TEF-1 antibodies (results not shown). (B) Interaction of TEF-1 and YAP65 on DNA was assayed by DNA-affinity chromatography. C2C12 cells were co-transfected with expression vectors encoding HA–YAP65 (pCMV-HA-YAP65) and FLAG–TEF-1ε (pCMV-FL-TEF-1ε). Complexes of FLAG–TEF-1ε with HA–YAP65 were isolated by and eluted from anti-FLAG–agarose as in Figure 3(B). Released complexes were purified further by MCAT DNA-affinity chromatography using biotinylated MCAT1 DNA [3]. After separation by SDS/PAGE, HA–YAP65 (upper panel) and FLAG–TEF-1 (lower panel) were detected by Western blot with anti-HA (αHA) or anti-FLAG (αFLAG) antibody. A significant portion of FLAG–TEF-1 was not purified by MCAT DNA (not bound). HA–YAP65 was found in fractions eluted from anti-FLAG–agarose (Load) and purified by MCAT DNA (Purif), indicating that YAP65 binds to TEF-1 on DNA. Sizes are indicated in kDa.
The interaction of YAP65 with TEF-1 on DNA was tested in a different type of experiment (Figure 4B). Extracts from differentiated C2C12 cells co-expressing HA–YAP65 and FLAG–TEF-1 were subjected to Co-IP using anti-FLAG–agarose, as shown in Figure 3(B). TEF-1/YAP65 complexes were eluted from the resin with FLAG peptide and were purified further by DNA-affinity chromatography with biotinylated MCAT DNA [3]. Western blot of these fractions showed that HA–YAP65 co-purified with TEF-1 during MCAT DNA-affinity chromatography; HA–YAP65 and FLAG–TEF-1 are found in starting material (Figure 4B, Load) and complexes purified with MCAT DNA (Figure 4B, Purif). These two experiments demonstrate that both YAP65 and TAZ interact with TEF-1 while TEF-1 is bound to DNA.
Interaction between TEF-1 and TAZ activates transcription
To determine if the TEF-1–TAZ interaction regulates transcription in vivo, two sets of experiments were performed; (i) modified mammalian two-hybrid transactivation, and (ii) activation of MCAT-dependent promoters by exogenous YAP65 and TAZ. For the modified mammalian two-hybrid experiments, full-length human TEF-1 (equivalent to rat TEF-1ζ) was fused to the GAL4 DNA-binding domain to yield GAL4–TEF-1-(1–426). Expression vectors for GAL4 or GAL4–TEF-1-(1–426) were co-transfected into H1299 cells with expression vectors for YAP65 or TAZ and a GAL-UAS-dependent luciferase reporter, and extracts were prepared and analysed for luciferase activity. YAP65 and TAZ did not interact with GAL4 (Figure 5A). GAL4–TEF-1-(1–426) alone is a weak transcriptional activator. Both YAP65 and TAZ interact with GAL4–TEF-1-(1–426) and activate transcription (50–60-fold) of the GAL4-dependent reporter.
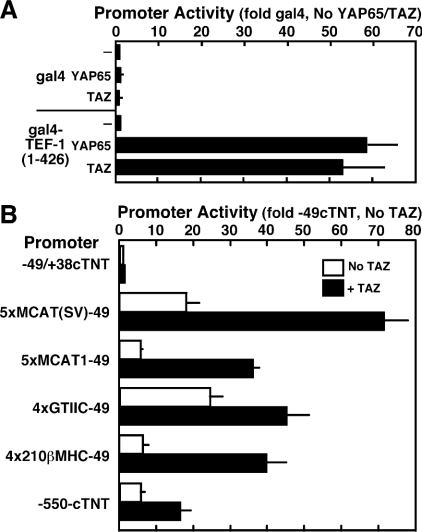
Figure 5. TAZ transactivates TEF-1-dependent transcription.
(A) Mammalian one-hybrid assay. Expression vectors for GAL4 or GAL4–TEF-1-(1–426) (100 ng/six-well dish) and for FLAG–TAZ or HA–YAP65 (pEF-TAZ-NFAG and pCMV-HA-YAP65, 100 ng/well), a GAL4-luciferase reporter (pM5-luc, 250 ng) and Tk-Renilla (10 ng), as a co-transfection control, were co-transfected into H1299 cells using FuGENE 6. Extracts were made, and luciferase assays were performed as described in the Experimental section. Values (promoter activity) are expressed relative to the activity of the pM5-luc reporter in the presence of GAL4 and empty expression vector. TAZ and YAP65 interact with TEF-1, but not with GAL4 alone, to activate transcription. The human TEF-1 used to construct the GAL4 fusions used here is the equivalent of rat TEF-1ζ (426 amino acids, see Figure 2), which lacks a four-amino-acid alternatively spliced exon present in TEF-1ε (430 amino acids). (B) TAZ activates MCAT-dependent promoters. Luciferase assays were performed on H1299 cellular extracts from cells co-transfected with MCAT promoters (250 ng/dish) and TAZ expression vector (pEF-TAZ-NFLAG, 100 ng/dish). Values (fold activity) are presented relative to the −49(cTNT)-luc promoter in the presence of empty expression vector. Experiments were repeated four times, and results are means±S.E.M. All activations by TAZ were significant, P<0.05.
Since the above experiment is a somewhat artificial assay system, we wanted to determine if TAZ could be a transcriptional activator for TEF-1 on MCAT-dependent promoters in vivo. We transfected expression vectors for TAZ with MCAT-dependent luciferase reporters into H1299 cells and assayed for an increase in luciferase activity. TEF-1 expression vectors were not used, since exogenous TEF-1 generally represses MCAT-dependent promoters, presumably by titrating necessary co-factors away from endogenous TEF-1 already bound to DNA [21]. The MCAT-dependent promoters assayed were −550/+38 from the chick cTNT promoter (−550-cTNT) [42], and multimers of MCAT sites cloned upstream of −49/+38-cTNT minimal promoter [5,6]: 5× MCAT1-49, multimers (5×) of the cTNT MCAT1 site; 5× MCAT(SV)-49, multimers (5×) MCAT1 core with flanking sequence from the SV40 (simian virus 40) GTIIC MCAT site; 4×GTIIC-49, two copies of the 2× GTIIC oligomer used in Figure 4; 4×210βMHC-49, multimers (4×) of the −210 MCAT from the rat βMHC promoter. Some of these promoters have higher activity in muscle cells than in non-muscle cells (5×MCAT1-49 and −550-cTNT) [43] or contain multimers of MCATs from a muscle promoter (4× 210βMHC-49). Others promoters are not muscle-specific [5× MCAT(SV)-49 and 4× GTIIC-49]. As shown by others, the non-muscle promoters showed higher basal activity in the non-muscle H1299 cells than the muscle promoters. Exogenous TAZ did not activate the basal cTNT promoter. All of the MCAT-dependent promoters were activated by exogenous TAZ (1.7–4-fold). Vassilev et al. [24] observed similar values for activation of MCAT-dependent promoters by YAP65. The lower levels of activation by TAZ of the MCAT-dependent promoters than GAL-UAS-dependent promoter may indicate that TAZ competes with an endogenous TEF-1-containing transactivation complex on MCAT promoters that is not present on the GAL-UAS promoter. TAZ also activated these MCAT-dependent promoters in the skeletal muscle cell line, C2C12 (results not shown). Taken together, these results show that TAZ interacts with TEF-1 in vivo and can activate TEF-1-dependent transcription.
TAZ interacts with all four members of the TEF-1 family in vitro
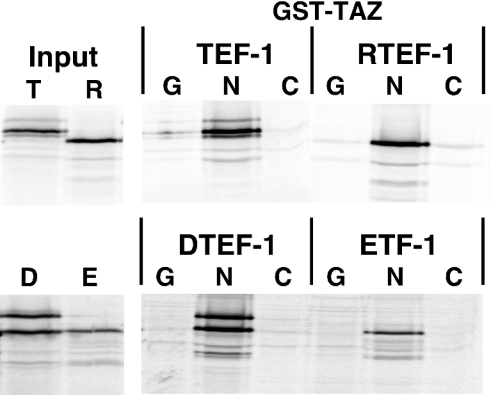
YAP65 interacts with all four members of the TEF-1 family [24]. Since the TEF-1 interaction domain of YAP65 is highly related to TAZ, it is expected that TAZ will also interact with all four members of the TEF-1 family of proteins. To test this, GST pull-down experiments were performed as described above, with 35S-labelled TEF-1, RTEF-1, DTEF-1 or ETF-1, using GST–TAZ-(1–239), GST–TAZ-(239–395) or GST. As shown in Figure 6, the N-terminal portion of TAZ (N) interacts with TEF-1, RTEF-1, DTEF-1 and ETF-1. This interaction between TAZ and TEF-1 is specific, as each TEF-1 protein failed to interact with either the GST protein alone (G) or with the C-terminal portion of TAZ (C).
Figure 6. TAZ interacts with all four members of the TEF-1 family in vitro.
(A) Each TEF-1 family member [TEF-1ε (T), RTEF-1 (R), DTEF-1 (D) and ETF-1 (E)] were produced and 35S-labelled by IVT reactions, and mixed with purified GST–TAZ-(1–239) (N), GST–TAZ-(239–395) (C) or GST (G). GST-fusion proteins and associated TEF-1 proteins were removed from solution with glutathione–agarose and assayed by SDS/PAGE. A total of 10% of each input is shown. Each member of the TEF-1 family specifically interacts with the N-terminal 239 amino acids of TAZ. TAZ interacts more efficiently with TEF-1ε (33% of input) than with RTEF-1 (22%). DTEF-1 and ETF-1 were produced much less efficiently than RTEF-1 and TEF-1ε (10–15%), so the images shown for these TEF-1 proteins are longer exposures, and the efficiency of interaction of DTEF-1 and ETF-1 with TAZ cannot be compared with that of TEF-1 or RTEF-1.
TEF-1 and RTEF-1 interact differentially with TAZ in vivo and in vitro
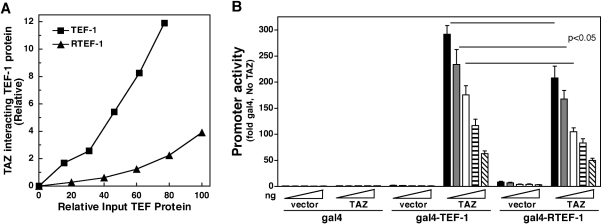
Quantification of the assay shown in Figure 6 showed that a higher percentage of TEF-1 interacted with GST–TAZ-(1–239) than RTEF-1 interacted with GST–TAZ-(1–239); compare the relative band intensity of input TEF-1 and RTEF-1 to GST–TAZ-(1–239)-bound TEF-1 and RTEF-1 (see legend to Figure 6). Since TEF-1 and RTEF-1 were produced at slightly different levels, a dose–response curve was constructed with purified GST–TAZ-(1–239) and increasing amounts of IVT 35S-labelled TEF-1 and RTEF-1 (Figure 7A). This experiment showed that, at all concentrations tested, TEF-1 binds TAZ more efficiently than RTEF-1 binds TAZ. Absolute binding-affinity experiments could not be performed as TEF-1 and RTEF-1 were produced by IVT, and were not purified further.
Figure 7. TAZ interacts more efficiently with TEF-1 than with RTEF-1.
(A) TAZ binds TEF-1 more efficiently than RTEF-1 in vitro. GST pull-down assays were performed with GST–TAZ-(1–239) and 0–50 μl of IVT [35S]TEF-1ε or [35S]RTEF-1. The levels of input TEF-1s were quantified by trichloroacetic acid precipitation and phosphorimager analysis (see the Experimental section). The TAZ-bound TEF-1 and RTEF-1 were quantified by phosphorimager analysis and are expressed relative to the highest level of input 35S-labelled protein (approx. 1.3 pmol RTEF-1 at 50 μl of lysate). (B) Expression vectors for GAL4-fusion proteins [GAL4 DNA-binding domain alone, GAL4–TEF-1-(79–426), GAL4–RTEF-1-(79–426); 5, 10, 25, 50 and 100 ng], transcriptional co-activators (empty expression vector or pEF-TAZ-NFLAG, 100 ng), a GAL4–luciferase reporter (pM5-luc, 250 ng) and Tk-Renilla (10 ng), as a co-transfection control, were co-transfected into H1299 cells using FuGENE 6. Extracts were made, and luciferase assays were performed as described in the Experimental section. Promoter activity is expressed relative to the activity of the pM5-luc reporter in the presence of 5 ng of GAL4 and empty expression vector. TAZ was a more efficient co-activator for TEF-1 than for RTEF-1 at 5, 10 and 25 ng (P<0.05). If data are normalized to the basal activity of GAL4–TEF-1 and GAL4–RTEF-1, TAZ interacts more efficiently with TEF-1 than with RTEF-1 at all concentrations. Western blots to verify equal amounts of RTEF-1 were performed with plates transfected with the equivalent of 100 ng of GAL4 DNAs. Therefore equal production at low concentrations presumes a linear relationship between input DNA and protein production. Even if this assumption is not absolutely correct, since the activation levels of the reporter at 25 ng of GAL4–TEF-1 is approximately the same as for 10 ng of GAL4–RTEF-1, the conclusions of the experiment still hold, since it is unlikely that there is a 2.5-fold difference in protein levels.
These results imply that there may be differences in the ability of TAZ to be a co-activator for TEF-1 and RTEF-1 in vivo. To test this, modified mammalian two-hybrid experiments were performed as above. Amino acids 79–426 (C-terminus) of human TEF-1 and human RTEF-1 were fused to the GAL4 DNA-binding domain to yield GAL4–TEF-1-(79–426) and GAL4–RTEF-1-(79–426) (kindly provided by Dr Alexandre F. R. Stewart, University of Ottawa Heart Institute, Ottawa, Ontario, Canada) [11,26]. Increasing amounts (5–100 ng) of expression vectors for GAL4–TEF-1-(79–426) and GAL4–RTEF-1-(79–426) were co-transfected into H1299 cells, with a constant amount of expression vector for TAZ and a GAL-UAS-dependent luciferase reporter, and extracts were prepared and analysed for luciferase activity (Figure 7B). YAP65 and TAZ did not interact with GAL4. Both GAL4–TEF-1-(79–426) and GAL4–RTEF-1-(79–426) were relatively weak transcriptional activators, with RTEF-1 being slightly stronger: 1–2-fold compared with 3–8-fold [26]. At all concentrations of input DNA, TAZ interacted with both GAL4–TEF-1-(79–426) and GAL4–RTEF-1-(79–426) and strongly activated transcription. At low concentrations (5, 10 and 25 ng) of GAL4–TEF-1(RTEF-1) plasmids, TAZ was a significantly stronger (P<0.05) co-activator for TEF-1 than for RTEF-1. For example, at 5 ng of GAL4 plasmids, TAZ activated transcription approx. 300-fold for GAL4–TEF-1-(79–426), relative to GAL4 alone with no TAZ, and 200-fold for GAL4–RTEF-1-(79–426). At higher levels of input of GAL4 plasmids, differences between TEF-1 and RTEF-1 were not significant, presumably due to saturation of the system. These results could be influenced by levels of GAL4 proteins in the nucleus, owing to different levels of protein production, stability or nuclear targeting. However, Western blots with polyclonal GAL4-(1–147) antisera (Covance) verified that GAL4–TEF-1-(79–426) and GAL4–RTEF-1-(79–426) were present at approximately the same level in nuclear extracts from transfected cells (results not shown). Taken together, the GST pull-down and transient transfection experiments show that TAZ differentially interacts with TEF-1 family members.
DISCUSSION
TEF-1 family members are broadly expressed, but function as transcriptional activators in only a few tissues, including cardiac muscle, skeletal muscle and placenta [1]. It is proposed that activity of TEF-1 proteins is controlled by interactions with specific co-factors with limited expression patterns. Known vertebrate TEF-1 co-factors include PARP [27], the Vgl family of proteins [25,26,43], YAP65 [24] and p160 [22]. These co-factors are broadly expressed (PARP and p160), expressed in the same tissues as TEF-1 proteins (YAP65) or restricted in their expression to one or two tissues (Vgl proteins).
YAP65 was cloned as a co-factor for ETF-1 (TEAD2) in 3T3 cells, but binds to all TEF-1 family members [24]. The current model is that phosphorylated YAP65 (at Ser112 in mYAP65) is associated with 14-3-3 proteins in the cytoplasm. Upon dephosphorylation, YAP65 accumulates in the nucleus, interacts with target transcription factors and activates transcription through interactions of C-terminal transactivation and PDZ-interaction domains. Phosphorylation in the nucleus of YAP65, perhaps by AKT, results in 14-3-3 binding and nuclear export [35,44]. Since YAP65 is related to TAZ and is expressed in muscle tissues where TEF-1 is known to regulate transcription, we determined whether TAZ interacted with the TEF-1 gene family to activate transcription. We found that TAZ interacts with TEF-1 in vitro, in vivo and with TEF-1 while bound to MCAT DNA. TAZ interacted with TEF-1 to activate transcription, both as a GAL4-fusion protein and through interaction of TAZ with endogenous TEF-1 proteins on MCAT-dependent promoters. Our results, showing the interaction of the C-terminal portion of TEF-1 with the N-terminal half of TAZ, agree with the results for the interaction of the TEF-1 gene family with YAP65 [24]. The WW domains of TAZ and YAP65, required for the interaction of YAP65/TAZ with (AP)PXY motifs in other proteins [23,24,36,38,39], are not required for the interaction of YAP65 and TAZ with TEF-1. This raises the possibility that YAP65/TAZ bound to TEF-1 could also interact with other proteins via the free WW domains and recruit other factors to DNA to activate transcription synergistically.
We also found that TAZ interacts with all four TEF-1 family members in vitro, but that TAZ interacts more efficiently with TEF-1 than with RTEF-1. Also, TAZ was a more efficient co-activator for TEF-1 than for RTEF-1 in vivo. Since YAP65 and TAZ are highly related in their TEF-1 interaction domain, it is likely that YAP65 also differentially interacts with TEF-1 family members. The data presented here are the first example of TEF-1 co-factors showing preferential interactions with different TEF-1 family members.
TEF-1 proteins show a high degree of sequence conservation throughout most of the molecule: overall, rat (r) TEF-1 and rat RTEF-1 are 67% identical and 76% similar. Regions of high homology between rTEF-1ε and rRTEF-1 include the DNA-binding domain (100% identical) and the C-terminus (amino acids 209–430 of rTEF1ε, 81% identical and 91% similar). There are regions of TEF-1 proteins that are less conserved between family members. The N-terminus and the proline-rich region of rat TEF-1 is only 26% identical (39% similar) and 20% identical (32% similar) with rat RTEF-1 respectively. Although the proline-rich region is not conserved at the primary sequence level, it is proline-rich (16–25%) in all TEF-1 family members. Also, the proline-rich region of TEF-1 proteins is required for full interaction with YAP65 and TAZ [24]. Therefore the proline-rich region of TEF-1 proteins probably mediates the differential interaction of TEF-1 and RTEF-1 with YAP65/TAZ.
Although TEF-1 family members are co-expressed in many tissues, family members may have different cellular roles. For example, TEF-1 and DTEF-1 are co-expressed in neonatal rat cardiac myocytes, with TEF-1 representing most of the TEF-1 protein in the cell [4,13]. However, DTEF-1, but not TEF-1, is implicated in the response of skeletal α-actin, but not the βMHC promoter, to hypertrophic signals [6,11–13]. Similarly, TEF-1 and RTEF-1 are expressed in skeletal muscle, but RTEF-1 is up-regulated during C2C12 cell differentiation, and is a component of a skeletal-muscle-specific mobility-shift complex [4,10]. Finally, although cellular targets for TEF-1 have only been identified in a few tissues (heart, muscle, placenta and skin), TEF-1 is broadly expressed and probably regulates transcription of unidentified targets in many cell types.
The data in this paper imply that differential association of TEF-1 proteins with transcriptional co-activators may regulate the activity of MCAT-dependent promoters and suggest a mechanism for cell signalling systems to generate the different cellular roles for TEF-1 family members. First, the TEF-1 proteins contain highly conserved regions and some divergent regions (see Figure 1); the activity of TEF-1 proteins is probably regulated by co-factors that interact preferentially with one TEF-1 family member and co-factors that interact equally with all TEF-1 family proteins. Secondly, endogenous cell signalling pathways, activated by skeletal muscle differentiation and α1-adrenergic treatment of cardiac myocytes, regulate the intracellular localization of the TEF-1 co-factors Vgl-2 and -4 [26,43]. If TEF-1 co-factors that differentially interact with TEF-1 proteins are targeted by cell signalling systems, promoter response would be influenced by which TEF-1 family member is occupying a given promoter or is present in a particular cell type. For example, phosphorylation of YAP65 and TAZ can cause their nuclear export. Therefore TEF-1 and YAP65/TAZ may be the predominant regulators of TEF-1-dependent transcription under conditions where YAP65/TAZ are nuclear. Upon activation of cell signalling pathways (currently unknown) that cause cytoplasmic localization of YAP65/TAZ, RTEF-1 and its preferentially associated co-factors could become the predominant transcriptional regulators.
Acknowledgments
This work was supported by grants from the NHLBI (National Heart, Lung, and Blood Institute), HL27867 (to Dr Giuseppe Inesi, University of Maryland, who was the overall Principal Investigator) and HL071894 to I.K.G.F., and from the NIGMS (National Institute of General Medical Studies), GM60954, to M.B.Y. W.M.M. was supported by the University of Maryland Interdisciplinary Program in Muscle Biology, an NIH (National Institutes of Health)-funded training grant (T32 AR07592). We thank Dr Irwin Davidson and Dr Alexandre Stewart for generously providing plasmids for these studies.
References
- 1.Larkin S. B., Ordahl C. P. Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family. In: Harvey R. P., Rosenthal N., editors. Heart Development. San Diego: Academic Press; 1999. pp. 307–329. [Google Scholar]
- 2.Karasseva N., Tsika G., Ji J., Zhang A., Mao X., Tsika R. Transcription enhancer factor-1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol. Cell. Biol. 2003;23:5143–5164. doi: 10.1128/MCB.23.15.5143-5164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrance I. K., Mar J. H., Ordahl C. P. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J. Biol. Chem. 1992;267:17234–17240. [PubMed] [Google Scholar]
- 4.Farrance I. K., Ordahl C. P. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J. Biol. Chem. 1996;271:8266–8274. doi: 10.1074/jbc.271.14.8266. [DOI] [PubMed] [Google Scholar]
- 5.Larkin S. B., Farrance I. K., Ordahl C. P. Flanking sequences modulate the cell specificity of M-CAT elements. Mol. Cell. Biol. 1996;16:3742–3755. doi: 10.1128/mcb.16.7.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLean B. G., Lee K. S., Simpson P. C., Farrance I. K. Basal and α1-adrenergic-induced activity of minimal rat βMHC promoters in cardiac myocytes requires multiple TEF-1 but not NFAT binding sites. J. Mol. Cell. Cardiol. 2003;35:461–471. doi: 10.1016/s0022-2828(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 7.Kariya K., Karns L. R., Simpson P. C. An enhancer core element mediates stimulation of the rat β-myosin heavy chain promoter by an α1-adrenergic agonist and activated β-protein kinase C in hypertrophy of cardiac myocytes. J. Biol. Chem. 1994;269:3775–3782. [PubMed] [Google Scholar]
- 8.Karns L. R., Kariya K., Simpson P. C. M-CAT, CArG, and Sp1 elements are required for α1-adrenergic induction of the skeletal α-actin promoter during cardiac myocyte hypertrophy: transcriptional enhancer factor-1 and protein kinase C as conserved transducers of the fetal program in cardiac growth. J. Biol. Chem. 1995;270:410–417. doi: 10.1074/jbc.270.1.410. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa K., Lee S. J., Jobe S. M., Markham B. E., Kitsis R. N. cis-Acting sequences that mediate induction of β-myosin heavy chain gene expression during left ventricular hypertrophy due to aortic constriction. Circulation. 1997;96:3943–3953. doi: 10.1161/01.cir.96.11.3943. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu N., Smith G., Izumo S. Both a ubiquitous factor mTEF-1 and a distinct muscle-specific factor bind to the M-CAT motif of the myosin heavy chain β gene. Nucleic Acids Res. 1993;21:4103–4110. doi: 10.1093/nar/21.17.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart A. F., Suzow J., Kubota T., Ueyama T., Chen H. H. Transcription factor RTEF-1 mediates α1-adrenergic reactivation of the fetal gene program in cardiac myocytes. Circ. Res. 1998;83:43–49. doi: 10.1161/01.res.83.1.43. [DOI] [PubMed] [Google Scholar]
- 12.Ueyama T., Zhu C., Valenzuela Y. M., Suzow J. G., Stewart A. F. Identification of the functional domain in the transcription factor RTEF-1 that mediates α1-adrenergic signaling in hypertrophied cardiac myocytes. J. Biol. Chem. 2000;275:17476–17480. doi: 10.1074/jbc.M001970200. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T., Mazzulli J. R., Farrance I. K., Stewart A. F. Mouse DTEF-1 (ETFR-1, TEF-5) is a transcriptional activator in α1-adrenergic agonist-stimulated cardiac myocytes. J. Biol. Chem. 2002;277:24346–24352. doi: 10.1074/jbc.M201171200. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z., Friedrich G. A., Soriano P. Transcriptional enhancer factor-1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 15.Milewski R. C., Chi N. C., Li J., Brown C., Lu M. M., Epstein J. A. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–837. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko K. J., Cullinan E. B., Latham K. E., DePamphilis M. L. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 1997;124:1963–1973. doi: 10.1242/dev.124.10.1963. [DOI] [PubMed] [Google Scholar]
- 17.Jacquemin P., Hwang J. J., Martial J. A., Dolle P., Davidson I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J. Biol. Chem. 1996;271:21775–21785. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- 18.Stewart A. F., Larkin S. B., Farrance I. K., Mar J. H., Hall D. E., Ordahl C. P. Muscle-enriched TEF-1 isoforms bind M-CAT elements from muscle-specific promoters and differentially activate transcription. J. Biol. Chem. 1994;269:3147–3150. [PubMed] [Google Scholar]
- 19.Azakie A., Larkin S. B., Farrance I. K., Grenningloh G., Ordahl C. P. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J. Biol. Chem. 1996;271:8260–8265. doi: 10.1074/jbc.271.14.8260. [DOI] [PubMed] [Google Scholar]
- 20.Simmonds A. J., Liu X., Soanes K. H., Krause H. M., Irvine K. D., Bell J. B. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 22.Belandia B., Parker M. G. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 2000;275:30801–30805. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- 23.Kanai F., Marignani P. A., Sarbassova D., Yagi R., Hall R. A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L. C., Yaffe M. B. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaudin P., Delanoue R., Davidson I., Silber J., Zider A. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF-1 factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–4816. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- 26.Maeda T., Chapman D. L., Stewart A. F. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 2002;277:48889–48898. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 27.Butler A. J., Ordahl C. P. Poly(ADP-ribose) polymerase binds with transcription enhancer factor-1 to MCAT1 elements to regulate muscle-specific transcription. Mol. Cell. Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kun E., Kirsten E., Ordahl C. P. Coenzymatic activity of randomly broken or intact double-stranded DNAs in auto and histone H1 trans-poly(ADP-ribosylation), catalysed by poly(ADP-ribose) polymerase (PARP I) J. Biol. Chem. 2002;277:39066–39069. doi: 10.1074/jbc.C200410200. [DOI] [PubMed] [Google Scholar]
- 29.Tulin A., Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 30.Huang K., Tidyman W. E., Le K. U., Kirsten E., Kun E., Ordahl C. P. Analysis of nucleotide sequence-dependent DNA binding of poly(ADP-ribose) polymerase in a purified system. Biochemistry. 2004;43:217–223. doi: 10.1021/bi0301800. [DOI] [PubMed] [Google Scholar]
- 31.Gupta M. P., Amin C. S., Gupta M., Hay N., Zak R. Transcription enhancer factor-1 interacts with a basic helix–loop–helix zipper protein, Max, for positive regulation of cardiac α-myosin heavy-chain gene expression. Mol. Cell. Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta M., Kogut P., Davis F. J., Belaguli N. S., Schwartz R. J., Gupta M. P. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J. Biol. Chem. 2001;276:10413–10422. doi: 10.1074/jbc.M008625200. [DOI] [PubMed] [Google Scholar]
- 33.Maeda T., Gupta M. P., Stewart A. F. TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem. Biophys. Res. Commun. 2002;294:791–797. doi: 10.1016/S0006-291X(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 34.Sudol M., Bork P., Einbond A., Kastury K., Druck T., Negrini M., Huebner K., Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 35.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 36.Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., Oren M., Sudol M., Cesareni G., Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 37.Komuro A., Nagai M., Navin N. E., Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 38.Cui C. B., Cooper L. F., Yang X., Karsenty G., Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol. Cell. Biol. 2003;23:1004–1013. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park K. S., Whitsett J. A., Di Palma T., Hong J. H., Yaffe M. B., Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- 40.Zuzarte P. C., Farrance I. K., Simpson P. C., Wildeman A. G. Tumor cell splice variants of the transcription factor TEF-1 induced by SV40 T-antigen transformation. Biochim. Biophys. Acta. 2000;1517:82–90. doi: 10.1016/s0167-4781(00)00261-x. [DOI] [PubMed] [Google Scholar]
- 41.Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 42.Mar J. H., Antin P. B., Cooper T. A., Ordahl C. P. Analysis of the upstream regions governing expression of the chicken cardiac troponin T gene in embryonic cardiac and skeletal muscle cells. J. Cell Biol. 1988;107:573–585. doi: 10.1083/jcb.107.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H. H., Mullett S. J., Stewart A. F. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates α1-adrenergic activation of gene expression in cardiac myocytes. J. Biol. Chem. 2004;279:30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 44.Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J. V., Dalal S. N., DeCaprio J. A., Greenberg M. E., Yaffe M. B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]