Abstract
In the last decade, a new serine protease inhibitor family has been described in arthropods. Eight members of the family were purified from locusts and share a conserved cysteine array (Cys-Xaa9–12-Cys-Asn-Xaa-Cys-Xaa-Cys-Xaa2–3-Gly-Xaa3–6-Cys-Thr-Xaa3-Cys) with nine inhibitory domains of the light chain of the crayfish protease inhibitor, pacifastin (PLDs; pacifastin light chain domains). Using cDNA cloning, several pacifastin-related precursors have been identified, encoding additional PLD-related peptides in different insect species. In the present study, two isoforms of a novel pacifastin-related precursor (SGPP-4) have been identified in the desert locust, predicting the previously identified SGPI-5 (Schistocerca gregaria PLD-related inhibitor-5) peptide and two novel PLD-related peptide sequences. One novel peptide (SGPI-5A) was synthesized chemically, and its inhibitory activity was assessed in vitro. Although proteases from a locust midgut extract were very sensitive to SGPI-5A, the same peptide proved to be a relatively poor inhibitor of bovine trypsin. By an in silico datamining approach, a novel pacifastin-related precursor with seven PLD-related domains was identified in the mosquito, Aedes aegypti. As in other insect pacifastin-related precursors, the Aedes precursor showed a particular domain architecture that is not encountered in other serine protease inhibitor families. Finally, a comparative real-time RT-PCR analysis of SGPP-4 transcripts in different tissues of isolated- (solitarious) and crowded-reared (gregarious) locusts was performed. This showed that SGPP-4 mRNA levels are higher in the brain, testes and fat body of gregarious males than of solitarious males. These results have been compared with data from a similar study on SGPP-1–3 transcripts and discussed with respect to a differential regulation of serine-protease-dependent pathways as a possible mechanism underlying locust phase polymorphism.
Keywords: pacifastin-like peptide precursor, phase polymorphism, protease inhibitor, serine peptidase, Schistocerca gregaria (desert locust), transcript profiling
Abbreviations: AAPP-1, Aedes aegypti pacifastin-related precursor 1; AEBSF, 4-(2-aminoethyl)benzenesulphonyl fluoride; BPVApNA, N-benzoyl-Phe-Val-Arg-p-nitroanilide; EST, expressed sequence tag; Fmoc, fluoren-9-ylmethoxycarbonyl; IA%, inhibitory activity; LMPI, Locusta migratoria PLD (pacifastin light chain)-related inhibitor; ORF, open reading frame; PLD, pacifastin light chain domain; PP, pacifastin-related precursor; RACE, rapid amplification of cDNA ends; RT, reverse transcriptase; SBTI, soybean trypsin inhibitor; SGPI, Schistocerca gregaria PLD-related inhibitor; SGPP, Schistocerca gregaria PP; TFA, trifluoroacetic acid; Tos-Lys-CH2Cl (‘TLCK’), tosyl-lysylchloromethane
INTRODUCTION
Serine peptidases, more commonly known as serine proteases, are widely distributed in the animal kingdom, and their vital role in several physiological processes has been studied intensively for many years. Several of these serine-protease-dependent pathways involve a proteolytic cascade, comprising a set of inactive serine proproteases (zymogens). As in the well-studied blood-clotting reaction, zymogens (e.g. the blood clotting Factor Xa) are activated in a stepwise process (cascade) in response to a specific ‘stimulus’, resulting in the rapid proteolytic activation of an effector protein (e.g. conversion of fibrinogen into fibrin). More recently, it has become apparent that some vertebrate serine-protease-dependent processes are remarkably paralleled in invertebrates (blood clotting in Limulus [1] and the innate immune response [2]), whereas others (moulting and metamorphosis) are restricted to arthropods. Since unwanted activation of these processes is potentially hazardous and even implicated in disease states, such as inflammation and thrombogenesis, most animals produce a repertoire of serine protease inhibitors as part of a complex regulation mechanism. Therefore various protease inhibitors have also been studied for their potential use as therapeutic agents or for their application in insect pest control.
Despite their huge diversity, the majority of all known animal serine protease inhibitors can be divided into two mechanistically different groups: the serpin family and the ‘canonical’ inhibitors [3]. Based on structural characteristics, the latter group can be subdivided further into at least 18 different families [4], among which is the pacifastin family [5], comprising only members from invertebrates. After 10 years since its initial purification from the freshwater crayfish [6], the light chain of the heterodimeric so-called ‘pacifastin’ protease inhibitor was shown to be composed of nine PLDs (pacifastin light chain domains), each of which contains a characteristic pattern of six conserved cysteine residues (Cys1-Xaa9–12-Cys2-Asn-Xaa-Cys3-Xaa-Cys4-Xaa2–3-Gly-Xaa3–6-Cys5-Thr-Xaa3-Cys6) [7]. Interestingly, in eight peptides purified from the locusts Locusta migratoria [LMPI-1, where LMPI is Locusta migratoria PLD-related inhibitor (PMP-D2), LMPI-2 (PMP-C) and HI] [8–10] and Schistocerca gregaria (SGPI-1–5, where SGPI is Schistocerca gregaria PLD-related inhibitor) [11], the same cysteine array is encountered. All these locust peptides were characterized further as serine protease inhibitors and the P1-position (nomenclature according to [12]), defining the inhibitor specificity, was localized between the two final cysteine residues [10,13,14]. In addition, the three-dimensional structures of LMPI-1–2 and HI, as well as of SGPI-1–2, were elucidated, showing a similar overall conformation, which is stabilized by an identical pattern (Cys1–Cys4, Cys2–Cys6, Cys3–Cys5) of disulphide bridges [15–19]. Based on the disulphide topography, as well as the fold, these PLD-related peptides define a novel family, designated the pacifastin family [5], within the group of the canonical serine protease inhibitors.
Using cDNA cloning, it was shown that most of the locust PLD-related peptides are derived from precursor proteins, which contain multiple inhibitory units [20–24]. In addition, these studies led to the molecular identification of 13 novel PLD-related sequences in locusts. More recently, PPs (pacifastin-related precursors) from insects other than locusts have been identified in the endoparasitoid wasp, Pimpla hypochondriaca [25], and the African malaria mosquito, Anopheles gambiae [26]. Furthermore, an in silico data-mining approach revealed the presence of PPs in insects which belong to different insect orders, such as Lepidoptera, Coleoptera and Siphonaptera [26,27], suggesting a broad distribution of the pacifastin family among arthropods. Transcript profiling studies have shown that several PP-encoding transcripts from gregarious desert locusts are expressed in a tissue- and stage-dependent way [21–24]. Intriguingly, a comparative realtime RT (reverse transcriptase)-PCR analysis of desert locusts in the solitarious (isolated-reared) and the swarm-forming gregarious (crowded-reared) phase revealed a differential phase-dependent transcript profile [28].
The present study reports on the molecular identification of two novel PP isoforms in the desert locust, S. gregaria. Both predicted precursors contain the sequences of two PLD-related peptides. One of these peptides (SGPI-5A) was synthesized chemically, and its inhibitory activity towards both mammalian and locust endogenous midgut proteases was assessed. In addition, the tissue- and phase-dependent distribution of the corresponding transcripts was studied using quantitative real-time RT-PCR analysis.
MATERIALS AND METHODS
Rearing of animals
Gregarious desert locusts, S. gregaria (Forskål), were reared under crowded conditions with controlled temperature (32±1 °C), light (14 h photoperiod) and relative humidity (40–60%). Depending on the experimental conditions, locusts were synchronized further at the time of ecdysis. Breeding of the solitarious desert locusts (>20 generations) was carried out under isolated conditions, as described previously [28]. Newly hatched solitarious hoppers were taken on the day of emergence and placed in individual containers. Room temperature (21±1 °C), light/dark photoperiod and feeding of the animals were similar for isolated-reared and crowded-reared locusts. For all experiments, sexually mature locusts were taken.
cDNA cloning
cDNA synthesis, PCR and RACE (rapid amplification of cDNA ends)
Locust fat bodies were dissected under a binocular microscope, immediately collected in RNAlater solution (Ambion) to prevent degradation, and stored at −80 °C. RNA extraction and cDNA synthesis were performed as described previously [21]. Based on the amino acid sequence of SGPI-5 [11], a serine protease inhibitor of the pacifastin family, a pair of degenerate PCR primers (Eurogentec, Seraing, Belgium) was designed, resulting in the amplification of a 92-bp fragment, as detailed by Simonet et al. [21]. This specific PCR product was subcloned and sequenced as outlined below. In order to obtain the complete cDNA sequence, a RACE protocol was performed, following the instructions of the Marathon cDNA Amplification kit (Clontech). Adapter primers were included in the kit, whereas an antisense gene-specific primer (see Figure 1, grey arrow) was derived from the original PCR fragment. After sequence analysis (as outlined below) of the 5′-RACE fragment, a second gene-specific primer (sense) was designed (see Figure 1, black arrow): antisense (5′-RACE), 5′-GCTGGCACTCCTACCATTGGAACCGCA-3′ and sense (3′-RACE), 5′-GGATGTGTTACGAAGAGAGAGGTGAACTGC-3′.
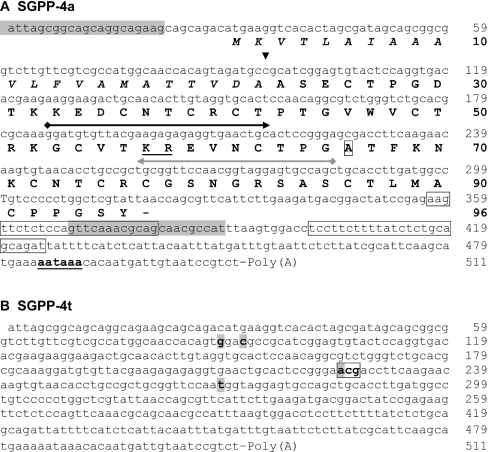
Figure 1. Nucleotide sequences (cDNA) of SGPP-4 isoforms.
(A) Nucleotide sequence (cDNA) and deduced amino acid sequence (in bold) of SGPP-4a. The signal peptide sequence is in italic, and its computed cleavage site [32] is indicated by an arrowhead (▼). The predicted dibasic cleavage site between two PLD-related domains is underlined. 5′- and 3′-RACE primers are indicated as grey and black arrows respectively. Ala66, which differs from the SGPP-4t amino acid sequence, is boxed. A putative polyadenylation signal sequence is indicated in bold and underlined. The nucleotide regions, from which the real-time primers were derived, are boxed. Finally, the nucleotide sequences, corresponding to the primers for the control PCR, are shaded (grey). (B) Nucleotide sequence (cDNA) of SGPP-4t. Nucleotides which differ from the SGPP-4a cDNA sequence are shaded. The codon, encoding Thr66, is boxed.
Finally, the entire cDNA sequences were verified by PCR with primers (see Figure 1), spanning the ORF (open reading frame). Unless mentioned otherwise, all primers were purchased from Invitrogen Life Technologies.
Cloning and sequence analysis
Cloning was performed as described previously [21]. In order to avoid discrepancies in the sequence analysis, for each cloned cDNA fragment, at least three different clones were sequenced in both directions. Nucleotide and amino acid sequence analysis and comparisons were performed by means of AlignX (ClustalW) software (InforMax; Invitrogen Life Technologies). In order to uncover novel PPs, translated (tblastn) BLAST searches were performed against the ‘nr’ database and the recently released Apis mellifera genome (September 2004, Amel1.2; http://www.hgsc.bcm.tmc.edu/projects/honeybee/). In addition, using the Ensembl genome browser (http://www.ensembl.org), the Drosophila melanogaster (Release 23.3a.1), An. gambiae (Release 22.2b.1) and Ap. mellifera (Release 27.1a.1) genomes were searched for annotated PP-encoding sequences.
Chemical synthesis, folding and purification of SGPI-5A
Solid-phase peptide synthesis
The SGPI-5A peptide was synthesized at a 0.1 mmol scale on an HMP [(4-hydroxymethyl)phenoxymethyl polystyrene] resin using amino acids with Fmoc (fluoren-9-ylmethoxycarbonyl)-protected α-amino groups and appropriate side-chain-protecting groups on a 431A solid-phase peptide synthesizer (Applied Biosystems). The C-terminal residue was linked to the resin by symmetrical anhydride binding. After each coupling step, incomplete peptide chains were capped with acetic anhydride. The Fmoc group was removed from the α-amino group of the peptide resin on the instrument, and the side-chain-protecting groups and the resin were removed from the peptide in the following cleavage mixture: 82.5% (v/v) TFA (trifluoroacetic acid), 0.75 g of crystalline phenol, 2.5% 1,2-ethanedithiol, 5% thioanisole and 5% water (100 min at room temperature). The cleaved peptide was separated from the resin by filtration, precipitated and washed into cold methyl t-butyl ether, freeze-dried and stored at 4 °C until further purification.
Peptide purification and folding
Before oxidation, the synthetic peptide was purified on a Resource RPC column (Amersham Biosciences) equilibrated with 0.1% TFA (solvent A). Peptides were eluted using an acetonitrile gradient, and were analysed by electrospray ion-trap MS (Esquire-LC; Bruker Daltonic). Fractions, containing the pre-purified synthetic peptide, were dried and air-oxidized in water adjusted with a Tris/HCl (pH 8.0) buffer. The completion of the oxidation was checked after reversed-phase HPLC on an analytical C18 Symmetry column (4.6 mm×250 mm, 5 μm; Waters). Finally, the authenticity and the purity of the oxidized peptide was confirmed by electrospray MS on a Q-TOF (quadrupole time-of-flight) system (Micromass, Manchester, U.K.) and N-terminal sequencing on a Procise 491 microsequencer (PerkinElmer/Applied Biosystems).
Serine protease inhibitor assay
Mammalian proteases
Bovine trypsin and chymotrypsin, as well as the respective substrates BPVApNA (N-benzoyl-Phe-Val-Arg-p-nitroanilide) and SA2PFpNA (succinyl-Ala-Ala-Pro-Phe-p-nitroanilide) were purchased from Sigma. For both substrates, 10× stock solutions (10 mM) were prepared in DMSO. The reaction buffer used for the chymotrypsin assay was 50 mM Tris/HCl (pH 8.0). For the trypsin assay, this buffer was supplemented with 10 mM CaCl2. Before adding the substrate, 100 μl of enzyme (0.032 μM) and 20 μl of inhibitor, dissolved in reaction buffer (1:5 molar ratio enzyme/inhibitor), were mixed and incubated for 30 min at room temperature. In control reactions, 20 μl of buffer instead of inhibitor was used. Then, 100 μl of the substrate solution (1 mM) was added and mixed during 1 min. The absorbance was monitored at 405 nm (Multiskan RC V1.5 plate reader) every 10 s for an interval of 5 min, corresponding with a linear increase of the reaction products. These data were processed by the Genesis Software (Labsystems) and the ΔA405/min ratio (slope) was determined. The inhibitory activity (IA%) of the tested inhibitors is defined by the following equation:
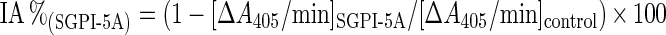 |
Midgut extract
Desert locusts were starved for 12 h. Midguts were dissected, and remnants of the midgut content (undigested food) were removed carefully. Clean preparations were pooled (n=20) and collected in liquid nitrogen. After homogenization in the ‘trypsin reaction buffer’, the homogenate was centrifuged at 7500 g for 10 min at 4 °C, and the protein concentration of the supernatant was quantified using the Bradford method [29]. The final extract was diluted to a working concentration of 0.5 mg/ml. Using BPVApNA as a substrate, the activity, expressed as ΔA405/min, of midgut-derived trypsin-like enzymes was analysed in parallel with different concentrations of bovine trypsin. In all further inhibition experiments, bovine trypsin and locust midgut trypsin-like proteases contained equal activities. In addition to SGPI-5A (0.2 μM), the chemical serine protease inhibitors 1 mM AEBSF [4-(2-aminoethyl)benzenesulphonyl fluoride] and 100 μM Tos-Lys-CH2Cl (‘TLCK’, tosyl-lysylchloromethane) as well as 2.7 μM SBTI (soybean trypsin inhibitor) were tested against bovine trypsin and trypsin-like enzymes from midgut extracts. Stock solutions for both SBTI (0.54 mg/ml) and AEBSF (50 mM) were prepared in water and stored at 4 °C. Fresh 10 mM Tos-Lys-CH2Cl solutions were prepared in methanol. Except for one modification (all reaction volumes were divided by two), the above-mentioned assay conditions were used.
Real-time RT-PCR transcript profiling
Total RNA extraction and cDNA synthesis
After micro-dissection, pooled locust tissues (n≥6), i.e. brain, fat body, gonads and male accessory glands, were homogenized by means of the MagNA Lyser instrument (Roche), according to the manufacturer's instructions. Total RNA was extracted utilizing the RNeasy Lipid tissue mini kit (Qiagen) in combination with a DNase treatment (RNase-free DNase set; Qiagen) to eliminate potential genomic DNA contamination. After spectrophotometric quantification and verification of the RNA quality using the Agilent 2100 Bioanalyser (Agilent Technologies), total RNA was reverse transcribed (Superscript II; Invitrogen Life Technologies) using random hexamers as described in the provided protocol. To minimize variations, all RNA samples were reverse-transcribed simultaneously. Furthermore, several negative-control reactions, i.e. without the RT, were prepared and analysed in parallel with the unknown samples during the PCR assay (see below).
Primer design, PCR amplification and cDNA quantification
PCRs were performed in 35 μl reaction mixture volumes, following the manufacturer's instructions for the iTaq SYBR® Green Supermix (Bio-Rad). In addition to the primers for the endogenous control, β-actin (forward, 5′-AATTACCATTGGTAACGAGCGATT-3′ and reverse, 5′-TGCTTCCATACCCAGGAATGA-3′), a single primer set, based on the 3′-untranslated region of the SGPP-4 (where SGPP is S. gregaria PP) transcripts (see Figure 1), was designed using the Primer Express software (Applied Biosystems). Relative standard curves for the SGPP-4 transcript(s) and the endogenous control were generated by serial (5×) dilutions of a male fat body cDNA sample. Reactions were run in duplicate on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using the following thermal cycling profile: 50 °C (2 min) and 95 °C (10 min), followed by 40 steps of 95 °C for 15 s and 60 °C for 60 s. After 40 cycles, samples were run using the dissociation protocol. This assay was performed twice to minimize variations due to sample handling. In order to compensate for differences in loading and RT efficiency, based on our previous studies, which indicated that the analysed β-actin mRNA levels are quite constant in locust tissues, regardless of the developmental or physiological conditions [22–24,30,31], this β-actin was used as an endogenous control. Thus the reported SGPP-4 transcript levels are normalized relative to β-actin and represent the mean±S.D. for two assays.
RESULTS
cDNA cloning of SGPP-4 isoforms
The initial steps of the cloning procedure overlapped with the identification of the SGPP-5 precursor reported previously [21]. Briefly, an initial RT-PCR with a set of degenerate primers, derived from the SGPI-5 peptide [11], generated a partial 92-bp cDNA sequence. This allowed for the design of an antisense gene-specific primer (Figure 1, grey arrow). However, by performing the RACE protocol, two different 5′-RACE fragments (284 bp and 427 bp) were amplified. Although the latter fragment was previously cloned and shown to code for the N-terminal region of the SGPP-5 precursor [21], the 284-bp fragment was further characterized in this study. After cloning and sequencing of this ‘short’ 5′-RACE fragment, a novel sense gene-specific primer was designed (Figure 1, black arrow). This allowed for the amplification of an overlapping 3′-RACE fragment. However, analysis of different clones, containing the RACE fragments, revealed the occurrence of two homologous cDNAs. To verify these sequences, additional cDNA fragments, spanning the entire ORF, were cloned and analysed. In conclusion, RACE in combination with a control PCR, led to the unambiguous identification of two homologous cDNA sequences, encoding isoforms of a novel PP in S. gregaria (Figure 1). Both cDNA sequences differ at four positions within the ORF, and, from the 96 predicted amino acids, only position 66 differs between each isoform. In accordance with the previously proposed ‘consensus’ terminology [23], the isoform with an alanine residue at position 66 will be designated as SGPP-4a and the isoform with a predicted threonine residue at position 66 will be referred to as SGPP-4t. Each encoded precursor contains an identical predicted signal peptide of 22 amino acids [32], followed by two PLD-related domains, both of which are characterized by a conserved cysteine array and are separated from each other by a pair of basic amino acids. A similar domain organization, i.e. multiple peptide sequences (e.g. SGPI-1 and SGPI-2), which are cleaved from a precursor (SGPP-1) at dibasic cleavage sites, has been encountered in several other reported insect PPs [26]. By analogy with this observation, processing of the SGPP-4 isoforms would result in two individual PLD-related peptides, which are, starting from the N-terminal end, denoted as SGPI-5A and SGPI-5B, (Figures 2A and 3). However, while the SGPP-4a and SGPP-4t isoforms share a similar N-terminal peptide sequence (SGPI-5A), the encoded C-terminal peptide sequences (SGPI-5B) are different. Therefore, to discriminate between these isoform-specific peptides, they will be referred to as SGPI-5Ba and SGPI-5Bt respectively.
Figure 2. Amino acid sequence alignment of PLD-related peptides.
(A) Peptide sequences were aligned using the ClustalW program. In addition, gaps were introduced between the first and the second, as well as between the fourth and the fifth, cysteine residues, based on the position of particular residues, which have been shown to be crucial for the fold (B) and to reveal (highlighted in black) the cysteine array, which is typical of pacifastin inhibitors. The names of the precursor-encoded PLD-related peptides are in accordance with the proposed ‘consensus’ terminology [23]. Peptide names are depicted at the left-hand side, and the corresponding precursor is denoted at the right-hand side. Peptides that have been identified exclusively by cDNA cloning are in black, while peptides that have been purified and characterized by Edman degradation (frequently used synonyms are within parentheses), are depicted in grey. Additional C-terminal amino acids in the predicted LMPI-3 sequence, which are not encountered in the previously purified peptide, HI, are underlined. Conserved residues are shaded. On top of the Figure, a diagram showing the disulphide bond pairing and an arrowhead (▼), marking the P1 position within the reactive site, are represented. (B) Pairwise amino acid alignment of the SGPP-4-encoded peptide SGPI-5A and three homologous trypsin-like locust inhibitors. The P1–P1′ reactive peptide bond is underlined. Residues that are important for the backbone fold of the peptides (see text) are shaded.
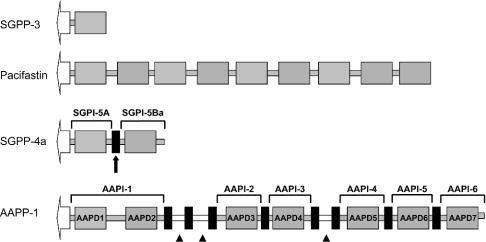
Figure 3. Domain organization of pacifastin-related peptide precursors.
Pacifastin-related precursors contain a predicted signal sequence, indicated as an open arrow, followed by one (SGPP-3) or more PLD-related domains (grey boxes, e.g. AAPD-3). Putative dibasic cleavage sites are depicted as small black boxes. Grey bars correspond with amino acid sequences outside the conserved cysteine array, which are connected directly to the PLD-related domains. White bars correspond to (small) peptide sequences (indicated by ▲), which are flanked by putative dibasic cleavage sites, suggesting a processing into small individual peptides without the conserved cysteine array. The SGPP-4a isoform contains two inhibitory units, SGPI-5A and SGPI-5Ba. The predicted SGPI-5Ba sequence corresponds to the previously purified single-domain peptide, SGPI-5, confirming the effective cleavage (processing) of SGPP-4a at the predicted dibasic cleavage site (see black arrow).
In silico data mining
An in silico data-mining approach revealed two mRNA sequences (AY440036 and AY432935), encoding additional PPs in the mosquitoes Armigeres subalbatus and Aedes aegypti respectively. Both sequences are derived from EST (expressed sequence tag) clusters, which were generated during a recent EST project on cDNA libraries from immune-response-activated mosquito haemocytes [33]. However, while the Ae. aegypti mRNA sequence represents the consensus sequence of 14 ESTs, comprising a complete ORF, the Ar. subalbatus sequence was derived from only two ESTs, and lacks a predicted stop codon. In line with other PPs, the predicted precursor sequence from Ae. aegypti contains multiple inhibitory units, all of which share six conserved cysteine residues according to the ‘pacifastin signature’ (Figures 2A and 3). When searching the Ap. mellifera genome, two putative PLD-related domains were found (gnl|Amel_1.2|Group12.12: 438138-438224 and 438036-438224) but, so far, preliminary annotation of the genomic data showed no corresponding gene product.
Peptide synthesis and protease inhibitor assay
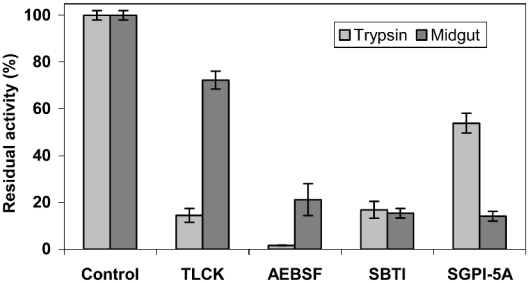
After SGPI-5A synthesis, followed by an initial purification, peptide fractions with a mass of 3651 Da were air-oxidized to realize the formation of three disulphide bridges. The oxidation products were analysed by reversed-phase HPLC on a C18 column, and different peaks were collected as individual fractions (results not shown). These elution fractions were analysed further by MS, revealing a molecular mass of 3645 Da for the fraction with a retention time of 27.6 min. This experimental mass differed by exactly 6 Da from the calculated mass, confirming the effective formation of three disulphide bridges. Subsequently, the N-terminal peptide sequence was verified by Edman degradation, enabling a quantification of the purified peptide. The inhibitory activity of SGPI-5A against bovine chymotrypsin and trypsin (5:1 molar ratio) was assessed, showing a relatively weak inhibition of trypsin, whereas no inhibition of chymotrypsin was observed (Table 1). On the other hand, addition of SGPI-5A to a midgut extract resulted in a strong decrease in the trypsin-like activity (<20%), while, under the same reaction conditions, bovine trypsin retained more than 50% of its activity (Figure 4).
Table 1. Protease-inhibitory activity of SGPI peptides.
The inhibitory activity of SGPI-5A, as defined by the equation IA%(SGPI-X)=(1−[ΔA405/min]SGPI-X/[ΔA405/min]control)×100, was assessed against bovine chymotrypsin and trypsin (1:5 molar ratio of enzyme/inhibitor) and is compared with the inhibitory activities of SGPI-1 and SGPI-2. The P1 residue within the reactive site is indicated in bold. The computed [ΔA405/min] values represent the means±S.D. (n=8). IA% values<5 are indicated by NI (no inhibition). ND, not determined.
Figure 4. Inhibition of bovine trypsin and locust digestive enzymes.
Inhibition of trypsin(-like) activity by SGPI-5A, Tos-Lys-CH2Cl (‘TLCK’), AEBSF and SBTI was analysed using the selective substrate BPVApNA. The (residual) activity of the experimental samples is represented as the percentage of the activity in control reactions, which contained buffer instead of inhibitor. Values represent the mean±S.D. (n=4).
Quantitative real-time RT-PCR analysis
In order to perform a comparative study of the relative abundance of the SGPP-4 transcripts in gregarious and solitarious locusts, a real-time RT-PCR analysis was undertaken on samples from different tissues of sexually mature locusts, reared under crowded and isolated conditions respectively (Figure 5). Since a single primer set for both SGPP-4-encoding transcripts was designed from the identical 3′-untranslated region (Figure 1), both mRNAs were detected with the same efficiency. Therefore the analysed transcript levels correspond with the sum of the relative quantities of both homologous transcripts. Analysis of the dissociation curves of the experimental and the β-actin control samples showed a single melting peak, which indicates a specific signal, corresponding to the SGPP-4 target sequences and the endogenous control respectively. In all negative-control samples, no amplification of the fluorescent signal was detected, proving that the extraction procedure, including the DNase treatment, effectively removed genomic DNA from the RNA samples.
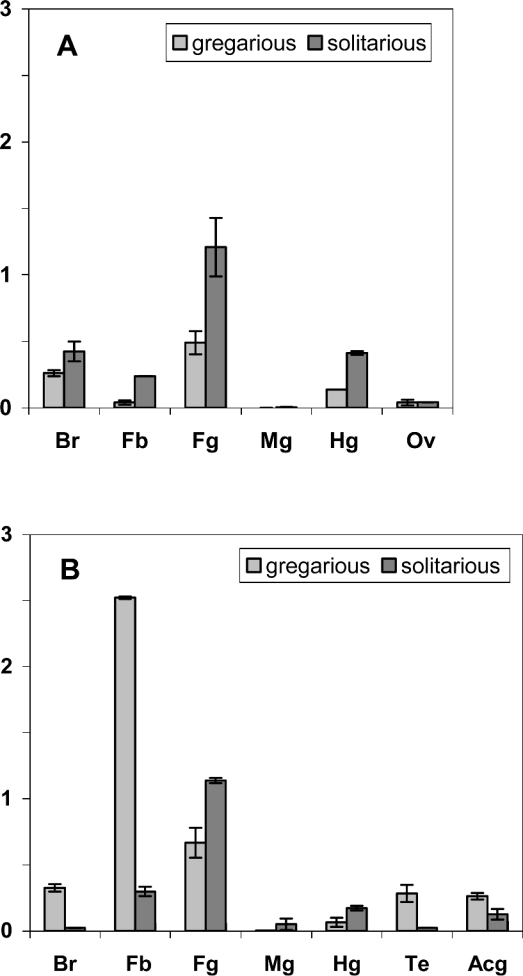
Figure 5. Real-time RT-PCR analysis of SGPP-4 transcripts from female (A) and male (B) locusts.
SGPP-4 transcript levels were measured in different tissues, i.e. brain (Br), fat body (Fb), foregut (Fg), midgut (Mg), hindgut (Hg), ovaries (Ov), testes (Te) and male accessory glands (Acg) from isolated-reared (solitarious) and crowded-reared (gregarious) desert locusts. Samples were analysed in duplicate, and this assay was repeated once. All data are normalized relative to β-actin and represent means±S.D. (of the two assays).
The normalized values (Figure 5) show that the SGPP-4 transcripts are most abundant in the fat body from gregarious males, corresponding to an approx 10-fold higher amount than in fat body extracts from solitarious males. Furthermore, in the fat body from gregarious locusts, a dramatic difference in the SGPP-4 transcript levels between female and male locusts was recorded, whereby the SGPP-4 transcript levels in males exceeded approx. 50 times the fat body mRNA levels of females. On the other hand, fat body transcript levels in solitarious locusts are, regardless of the sex, relatively low as compared with gregarious animals. In addition to male fat body mRNA levels, brain and testes transcripts are also detected more abundantly in gregarious males than in solitarious males.
DISCUSSION
The present paper describes the molecular characterization of two cDNAs, encoding two isoforms, denoted as SGPP-4a and SGPP-4t, of a novel PP in the desert locust, S. gregaria (Figure 1). Each predicted precursor contains two PLD-related domains with the ‘pacifastin signature’, i.e. a conserved array of six cysteine residues (Figure 2A). In contrast with the encoded C-terminal peptides (SGPI-5Ba and SGPI-5Bt), the predicted N-terminal peptide (SGPI-5A) is identical in both isoforms. While SGPI-5A and SGPI-5Bt represent novel members of the pacifastin family, the predicted SGPI-5Ba peptide is identical with the SGPI-5 inhibitor purified previously [11]. At present, 13 complete PPs have been identified [21–26], which predict 38 PLD-related peptide sequences in total (Figure 2A). In line with the continuously growing sequence information on protein-encoding genes in insects, an in silico search revealed two mRNA sequences from the mosquitoes Ae. aegypti and Ar. subalbatus (GenBank accession numbers AY432935 and AY440036 respectively), which are homologous with locust PP-encoding cDNAs. More detailed sequence analysis showed that the latter mRNA sequence constitutes a partial sequence. The Aedes mRNA, on the other hand, predicts a PP with seven PLD-related domains, which will be referred to as Ae. aegypti PP 1 or, briefly, AAPP-1. Moreover, an analysis of the recently released genome of Ap. mellifera suggests the occurrence of at least two pacifastin inhibitor domains in Hymenoptera, providing additional proof for the wide distribution of this serine protease inhibitor family in insects. Intriguingly, although PPs have been predicted in numerous insects, including three different dipteran species, i.e. the mosquitoes An. gambiae, An. aegypti and Ar. subalbatus, no PP-encoding genes have been identified in the genome of another intensively studied dipteran, notably Drosophila melanogaster. Therefore it can be speculated that, whereas a complex variety of PP-encoding genes has evolved in most insects from a conserved ‘ancestral’ pacifastin gene through mechanisms such as gene duplication, the genetic information for such a PP has been ‘lost’ in the genome of fruit flies after the divergence of mosquitoes and flies some 250 million years ago [34]. Evidently, sequencing of additional fly genomes, as is currently in progress for two more species of the Drosophila genus, is needed to verify this hypothesis.
Most insect PPs are composed of multiple inhibitory units, which are preceded by a signal peptide sequence and flanked by putative dibasic cleavage sites (Figure 3). In fact, only one single-domain precursor (SGPP-2) has been reported so far [24]. Interestingly, a very similar domain organization is encountered in many neuronal or endocrine peptide precursors [35], which suggests that PPs are post-translationally modified and secreted into the extracellular space. In line with the effective processing of locust PPs is the observation that all locust inhibitors (SGPI-1-5, LMPI-1-2 and HI [8–11]), which have been purified to date, were identified as monomers. Moreover, all these peptides, except for SGPI-3, are derived from multi-domain precursors, as shown in the present study for the previously purified SGPI-5 peptide, which is encoded by the SGPP-4a transcript. Nevertheless, the biological role and the mechanisms underlying the processing of PPs remain elusive, especially in the light of the recent study of Szenthe et al. [36], showing that the two PLD-related domains of the SGPP-1 precursor have similar inhibition properties as the corresponding monomers, SGPI-1-2. Unlike the insect PPs, the light chain of pacifastin, which represents the only identified crustacean PP at present, was purified as a single multi-domain inhibitor with nine sequential PLDs, conforming to the absence of dibasic cleavage sites [7]. This resembles the domain architecture of ovomucoids, comprising three homologous Kazal domains, each of which can inhibit target enzymes independently, as they are connected by short and flexible linkers [10,37]. Interestingly, based on the presence of eight putative dibasic cleavage sites, processing of the novel AAPP-1 would result in both a ‘bis-headed’ inhibitor, containing two PLD-related domains, as well as in five single-domain peptides (Figures 2A and 3). In addition, three relatively short peptide sequences without the ‘pacifastin signature’ would be generated. Although the occurrence of non-PLD-related peptide sequences on PPs has already been shown in different insect species, none of these peptides has been characterized so far. Since no significant homology with other known peptides is apparent, their biological function remains elusive at present. Both the domain architecture, as well as the post-translational processing of insect PPs is unique among all multi-headed canonical inhibitor families.
All purified locust peptides of the pacifastin peptide family have been characterized as serine protease inhibitors, and, based on kinetic and structural data, inhibition was found to conform with the ‘standard’ mechanism [4] of canonical serine protease inhibitors [10,37]. Each inhibitory PLD-related domain contains a single reactive P1–P1′ bond [12], which interacts directly with the catalytic residues of the enzyme in a substrate-like manner. Moreover, the reactive peptide bond is located between the two final cysteine residues (P3–P3′) on an exposed, so-called ‘canonical’, binding loop. Upon interaction with a target enzyme, the P1 residue of canonical inhibitors will fit nicely in the S1 binding pocket of the enzyme and is therefore critical for the inhibitor specificity. While inhibitors of chymotrypsin-like enzymes have usually aromatic or bulky amino acids at the P1 position, trypsin-like enzymes are more sensitive to inhibitors with positively charged P1 residues. However, although LMPI-1, HI and SGPI-1 share a trypsin-specific arginine residue at the P1 position (Figure 2B), they showed only poor activity against mammalian trypsin [10,13]. In agreement with these data, we found that SGPI-5A, which contains the same reactive-site residues as SGPI-1 and LMPI-1, is a rather weak inhibitor of mammalian trypsin (Table 1). On the other hand, the N-terminal SGPP-4-encoded peptide, SGPI-5Ba was reported to be a strong inhibitor of α-chymotrypsin, conforming to a leucine residue at the P1 position [11]. Since no kinetic data on SGPI-5Ba are available, it is worthwhile to consider that the homologous SGPI-2 peptide was shown to be a very potent inhibitor of mammalian chymotrypsin, corresponding to >98% inhibition (same assay as for SGPI-5A) [38] and a Ki value of 6.2×10−12 (Table 1) [13].
Intriguingly, synthetic variants of LMPI-1, SGPI-1 and HI, replacing the arginine residue with leucine, converted these peptides into potent inhibitors of bovine chymotrypsin [10,14]. Moreover, trypsin-like enzymes from midguts of migratory locust or crayfish were shown to be ≥104-fold more sensitive to the locust trypsin inhibitors than mammalian trypsin [14,16]. The unexpected selectivity of these locust peptides was attributed to an additional interaction site, corresponding to residues 20–24 (P6–P10 loop). As a result, the affinity of the inhibitor for mammalian trypsin would be decreased due to a steric ‘clash’ between the inhibitor Pro21 residue (Figure 2B) and Pro173 of bovine trypsin [16,18]. Furthermore, H-NMR studies revealed the importance of the Trp25–Lys10 interaction in SGPI-1, LMPI-1 and HI for the backbone fold, as well as for the positioning of Pro21 [16,17,19]. Interestingly, SGPI-5A shares Trp25 and Lys10, as well as Pro21 in the P6–P10 loop (Figure 2B), with LMPI-1, SGPI-1 and HI. In line with this, trypsin-like enzymes from a midgut extract proved to be more sensitive to SGPI-5A than bovine trypsin (Figure 4). Furthermore, two chemical serine protease inhibitors, Tos-Lys-CH2Cl and AEBSF, were found to be more effective against bovine trypsin, as compared with the arthropod trypsin-like enzymes. Altogether, these data suggest that SGPI-5A has a similar fold (including a second interaction site) as the other locust trypsin inhibitors, and that mammalian and arthropod trypsin-like enzymes have distinct biochemical properties.
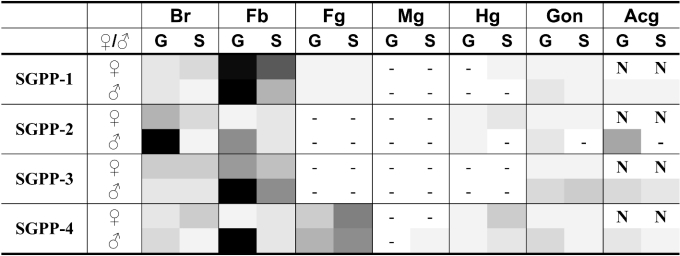
Locusts are characterized by the unique ability to undergo phase transition from the harmless solitarious to the swarm-forming gregarious phase in response to changes in population density. As illustrated by massive invasions of locust swarms in Africa, which have affected approx. 6.5 million ha since October 2003 [FAO (Food and Agriculture Organization) Desert Locust Bulletin January 2005; http://www.fao.org/news/global/locusts/315bull/DL315e.pdf], the economical implications of this phase shift can be enormous. Although this complex process is known to involve drastic changes in behaviour, reproduction and colour, the physiological mechanisms are yet not fully understood. Nevertheless, recent studies have shown phase-dependent profiles of two PLD-related peptides [39,40], as well as a phase-dependent transcriptional regulation of SGPP-1–3 expression [28]. In line with these studies, transcription of SGPP-4 is differentially regulated in a sex- and phase-dependent way, as illustrated by the relatively high SGPP-4 mRNA content in fat body, brain and testes from gregarious males, as compared with solitarious ones (Figure 5). Furthermore, if the SGPP-1–4 transcript profiles are compared (Figure 6), male fat body transcript levels are consistently higher in gregarious than in solitarious desert locusts. In female fat body on the other hand, SGPP mRNAs are less abundant than in males (except for the SGPP-1 transcript), and phase-dependent differences are far less evident than in males. Based on these data, it can be hypothesized that a differential control of serine-protease-dependent pathways, i.e. a stricter regulation (inhibition) in gregarious as compared with solitarious males could be involved in phase transition in male locusts.
Figure 6. Overview of SGPP-1–4 transcript levels in different tissues from isolated- and crowded-reared desert locusts.
SGPP-1–4 transcript levels (data for SGPP-1–3 transcripts from [28]; for SGPP-4, see Figure 3) were measured in different tissues, i.e. brain (Br), fat body (Fb), foregut (Fg), midgut (Mg), hindgut (Hg), gonads (G) and male accessory glands (Acg) from isolated-reared (S) and crowded-reared (G) desert locusts. No data are available on female accessory glands (N). Then, per SGPP transcript, the normalized transcript levels, corresponding to all the different experimental conditions, were expressed as a percentage of the maximal expression level (%MAX). In the schematic colour presentation, the grey-black colour intensity (>0% and ≤100%) corresponds to the %MAX: %MAX<0.5% are indicated as ‘−’, 0.5%≤%MAX≤5% are indicated by a grey tone of 5%, and so on. Thus the highest SGPP-4 transcript level, which is recorded in fat body from gregarious males (%MAX=100%) is represented in black, while the relative mRNA content in fat body from solitarious males (10≤%MAX≤15%) is represented by a 15%-grey tone.
It should be noted, however, that the molecular processes, mediating phase transition, are most likely a complex interplay of signalling pathways, involving (neuro-)endocrine factors and several effector proteins, some of which depend on proteolytic cascades. Furthermore, since PLD-related peptides are widely distributed among insects, it is evident that their functions are not restricted to locust phase-polymorphism-related processes. In fact, pacifastin has already been shown to inhibit the proteolytic activation of phenoloxidase in crayfish [41], and a similar effect has been described for LMPI-1–2 in locusts [8]. As a consequence, taking into account that (i) the serine protease cascade, activating phenoloxidase, is a crucial element in the innate immune system of arthropods, and (ii) the AAPP-1-encoding transcript is derived from an EST project on cDNA libraries from immune-response-activated mosquito haemocytes [33], a similar role for AAPI-1 as for pacifastin in innate immunity is suggested.
In conclusion, based on the rapidly accumulating data on insect PPs and pacifastin-related peptides, we believe that the study of this family is a promising approach to unravel yet unknown serine-protease-dependent processes, leading to a better insight in different aspects of insect physiology.
Acknowledgments
We are indebted to S. Van Soest for the real-time RT-PCR studies. In addition, J. Gijbels, M. Van Der Eeken, J. Puttemans, M. Christiaens, L. Vanden Bosch, J. Huybrechts and R. Jonckers are thanked for technical or administrative assistance. We gratefully acknowledge the Belgian Interuniversity Attraction Poles Programme (IUAP/PAI P5/30, Belgian Science Policy), the Catholic University of Leuven Research Foundation (GOA/2000/04) and the FWO-Vlaanderen for financial support. J.Vd.B. was Senior Research Associate of the FWO, and G.S. obtained a Postdoctoral fellowship from the Catholic University of Leuven Research Foundation. I.C. obtained a Ph.D. fellowship from the IWT.
References
- 1.Iwanaga S. The molecular basis of innate immunity in the horseshoe crab. Curr. Opin. Immunol. 2002;14:87–95. doi: 10.1016/s0952-7915(01)00302-8. [DOI] [PubMed] [Google Scholar]
- 2.Leclerc V., Reichhart J. M. The immune response of Drosophila melanogaster. Immunol. Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- 3.Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 4.Laskowski M., Qasim M. A. What can the structures of enzyme–inhibitor complexes tell us about the structures of enzyme substrate complexes? Biochim. Biophys. Acta. 2000;1477:324–337. doi: 10.1016/s0167-4838(99)00284-8. [DOI] [PubMed] [Google Scholar]
- 5.Simonet G., Claeys I., Vanden Broeck J. Structural and functional properties of a novel serine protease inhibiting peptide family in arthropods. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002;132:247–255. doi: 10.1016/s1096-4959(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 6.Hergenhahn H. G., Aspan A., Soderhall K. Purification and characterization of a high-Mr proteinase inhibitor of pro-phenol oxidase activation from crayfish plasma. Biochem. J. 1987;248:223–228. doi: 10.1042/bj2480223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Z., Sottrup-Jensen L., Aspan A., Hall M., Soderhall K. Pacifastin, a novel 155-kDa heterodimeric proteinase inhibitor containing a unique transferrin chain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6682–6687. doi: 10.1073/pnas.94.13.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boigegrain R. A., Mattras H., Brehelin M., Paroutaud P., Coletti-Previero M. A. Insect immunity: two proteinase inhibitors from hemolymph of Locusta migratoria. Biochem. Biophys. Res. Commun. 1992;189:790–793. doi: 10.1016/0006-291x(92)92271-x. [DOI] [PubMed] [Google Scholar]
- 9.Nakakura N., Hietter H., Van Dorsselaer A., Luu B. Isolation and structural determination of three peptides from the insect Locusta migratoria: identification of a deoxyhexose-linked peptide. Eur. J. Biochem. 1992;204:147–153. doi: 10.1111/j.1432-1033.1992.tb16617.x. [DOI] [PubMed] [Google Scholar]
- 10.Kellenberger C., Boudier C., Bermudez I., Bieth J. G., Luu B., Hietter H. Serine protease inhibition by insect peptides containing a cysteine knot and a triple-stranded β-sheet. J. Biol. Chem. 1995;270:25514–25519. doi: 10.1074/jbc.270.43.25514. [DOI] [PubMed] [Google Scholar]
- 11.Hamdaoui A., Wataleb S., Devreese B., Chiou S. J., Vanden Broeck J., Van Beeumen J., De Loof A., Schoofs L. Purification and characterization of a group of five novel peptide serine protease inhibitors from ovaries of the desert locust, Schistocerca gregaria. FEBS Lett. 1998;422:74–78. doi: 10.1016/s0014-5793(97)01585-8. [DOI] [PubMed] [Google Scholar]
- 12.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 13.Malik Z., Amir S., Pal G., Buzas Z., Varallyay E., Antal J., Szilagyi Z., Vekey K., Asboth B., Patthy A., Graf L. Proteinase inhibitors from desert locust, Schistocerca gregaria: engineering of both P(1) and P(1)' residues converts a potent chymotrypsin inhibitor to a potent trypsin inhibitor. Biochim. Biophys. Acta. 1999;1434:143–150. doi: 10.1016/s0167-4838(99)00167-3. [DOI] [PubMed] [Google Scholar]
- 14.Patthy A., Amir S., Malik Z., Bodi A., Kardos J., Asboth B., Graf L. Remarkable phylum selectivity of a Schistocerca gregaria trypsin inhibitor: the possible role of enzyme-inhibitor flexibility. Arch. Biochem. Biophys. 2002;398:179–187. doi: 10.1006/abbi.2001.2686. [DOI] [PubMed] [Google Scholar]
- 15.Mer G., Hietter H., Kellenberger C., Renatus M., Luu B., Lefevre J. F. Solution structure of PMP-C: a new fold in the group of small serine proteinase inhibitors. J. Mol. Biol. 1996;258:158–171. doi: 10.1006/jmbi.1996.0240. [DOI] [PubMed] [Google Scholar]
- 16.Kellenberger C., Ferrat G., Leone P., Darbon H., Roussel A. Selective inhibition of trypsins by insect peptides: role of P6-P10 loop. Biochemistry. 2003;42:13605–13612. doi: 10.1021/bi035318t. [DOI] [PubMed] [Google Scholar]
- 17.Mer G., Kellenberger C., Koehl P., Stote R., Sorokine O., Van Dorsselaer A., Luu B., Hietter H., Lefevre J. F. Solution structure of PMP-D2, a 35-residue peptide isolated from the insect Locusta migratoria. Biochemistry. 1994;33:15397–15407. doi: 10.1021/bi00255a021. [DOI] [PubMed] [Google Scholar]
- 18.Roussel A., Mathieu M., Dobbs A., Luu B., Cambillau C., Kellenberger C. Complexation of two proteic insect inhibitors to the active site of chymotrypsin suggests decoupled roles for binding and selectivity. J. Biol. Chem. 2001;276:38893–38898. doi: 10.1074/jbc.M105707200. [DOI] [PubMed] [Google Scholar]
- 19.Gaspari Z., Patthy A., Graf L., Perczel A. Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria. Eur. J. Biochem. 2002;269:527–537. doi: 10.1046/j.0014-2956.2001.02685.x. [DOI] [PubMed] [Google Scholar]
- 20.Kromer E., Nakakura N., Lagueux M. Cloning of a Locusta cDNA encoding a precursor peptide for two structurally related proteinase inhibitors. Insect Biochem. Mol. Biol. 1994;24:329–331. doi: 10.1016/0965-1748(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 21.Simonet G., Claeys I., Van Soest S., Breugelmans B., Franssens V., De Loof A., Vanden Broeck J. Molecular identification of SGPP-5, a novel pacifastin-like peptide precursor in the desert locust. Peptides. 2004;25:941–950. doi: 10.1016/j.peptides.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Simonet G., Claeys I., November T., Wataleb S., Janssen T., Maes R., De Loof A., Vanden Broeck J. Cloning of two cDNAs encoding isoforms of a pacifastin-related precursor polypeptide in the desert locust, Schistocerca gregaria: analysis of stage- and tissue-dependent expression. Insect Mol. Biol. 2002;11:353–360. doi: 10.1046/j.1365-2583.2002.00345.x. [DOI] [PubMed] [Google Scholar]
- 23.Simonet G., Claeys I., Vanderperren H., November T., De Loof A., Vanden Broeck J. cDNA cloning of two different serine protease inhibitor precursors in the migratory locust, Locusta migratoria. Insect Mol. Biol. 2002;11:249–256. doi: 10.1046/j.1365-2583.2002.00331.x. [DOI] [PubMed] [Google Scholar]
- 24.Vanden Broeck J., Chiou S. J., Schoofs L., Hamdaoui A., Vandenbussche F., Simonet G., Wataleb S., De Loof A. Cloning of two cDNAs encoding three small serine protease inhibiting peptides from the desert locust Schistocerca gregaria and analysis of tissue-dependent and stage-dependent expression. Eur. J. Biochem. 1998;254:90–95. doi: 10.1046/j.1432-1327.1998.2540090.x. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson N. M., Conyers C., Keen J., MacNicoll A., Smith I., Audsley N., Weaver R. Towards a comprehensive view of the primary structure of venom proteins from the parasitoid wasp Pimpla hypochondriaca. Insect Biochem. Mol. Biol. 2004;34:565–571. doi: 10.1016/j.ibmb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Simonet G., Claeys I., Franssens V., De Loof A., Vanden Broeck J. Genomics, evolution and biological functions of the pacifastin peptide family: a conserved serine protease inhibitor family in arthropods. Peptides. 2003;24:1633–1644. doi: 10.1016/j.peptides.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Gaspari Z., Ortutay C., Perczel A. A simple fold with variations: the pacifastin inhibitor family. Bioinformatics. 2004;20:448–451. doi: 10.1093/bioinformatics/btg451. [DOI] [PubMed] [Google Scholar]
- 28.Simonet G., Claeys I., Breugelmans B., Van Soest S., De Loof A., Vanden Broeck J. Transcript profiling of pacifastin-like peptide precursors in crowd- and isolated-reared desert locusts. Biochem. Biophys. Res. Comm. 2004;317:565–569. doi: 10.1016/j.bbrc.2004.03.078. [DOI] [PubMed] [Google Scholar]
- 29.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 30.Claeys I., Simonet G., Van Loy T., De Loof A., Vanden Broeck J. cDNA cloning and transcript distribution of two novel members of the neuroparsin family in the desert locust, Schistocerca gregaria. Insect Mol Biol. 2003;12:473–481. doi: 10.1046/j.1365-2583.2003.00431.x. [DOI] [PubMed] [Google Scholar]
- 31.Janssen T., Claeys I., Simonet G., De Loof A., Girardie J., Vanden Broeck J. V. cDNA cloning and transcript distribution of two different neuroparsin precursors in the desert locust, Schistocerca gregaria. Insect Mol. Biol. 2001;10:183–189. doi: 10.1046/j.1365-2583.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen H., Engelbrecht J., Brunak S., vonHeijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Bartholomay L. C., Cho W. L., Rocheleau T. A., Boyle J. P., Beck E. T., Fuchs J. F., Liss P., Rusch M., Butler K. M., Wu R. C., et al. Description of the transcriptomes of immune response-activated hemocytes from the mosquito vectors Aedes aegypti and Armigeres subalbatus. Infect. Immun. 2004;72:4114–4126. doi: 10.1128/IAI.72.7.4114-4126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolshakov V. N., Topalis P., Blass C., Kokoza E., della Torre A., Kafatos F. C., Louis C. A comparative genomic analysis of two distant diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 2002;12:57–66. doi: 10.1101/gr.196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. 2001;22:241–254. doi: 10.1016/s0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]
- 36.Szenthe B., Gaspari Z., Nagy A., Perczel A., Graf L. Same fold with different mobility: backbone dynamics of small protease inhibitors from the desert locust, Schistocerca gregaria. Biochemistry. 2004;43:3376–3384. doi: 10.1021/bi035689+. [DOI] [PubMed] [Google Scholar]
- 37.Krowarsch D., Cierpicki T., Jelen F., Otlewski J. Canonical protein inhibitors of serine proteases. Cell. Mol. Life Sci. 2003;60:2427–2444. doi: 10.1007/s00018-003-3120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonet G., Claeys I., Huybrechts J., De Loof A., Vanden Broeck J. Bacterial production and purification of SGPI-1 and SGPI-2, two peptidic serine protease inhibitors from the desert locust, Schistocerca gregaria. Protein Expression Purif. 2003;31:188–196. doi: 10.1016/s1046-5928(03)00170-0. [DOI] [PubMed] [Google Scholar]
- 39.Rahman M. M., Vanden Bosch L., Baggerman G., Clynen E., Hens K., Hoste B., Meylaers K., Vercammen T., Schoofs L., De Loof A., Breuer M. Search for peptidic molecular markers in hemolymph of crowd- (gregarious) and isolated-reared (solitary) desert locusts, Schistocerca gregaria. Peptides. 2002;23:1907–1914. doi: 10.1016/s0196-9781(02)00175-4. [DOI] [PubMed] [Google Scholar]
- 40.Clynen E., Stubbe D., De Loof A., Schoofs L. Peptide differential display: a novel approach for phase transition in locusts. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002;132:107–115. doi: 10.1016/s1096-4959(01)00538-3. [DOI] [PubMed] [Google Scholar]
- 41.Aspan A., Hall M., Soderhall K. The effect of endogenous proteinase inhibitors on the prophenoloxidase activating enzyme, a serine proteinase from crayfish haemocytes. Insect Biochem. 1990;20:485–492. [Google Scholar]