Abstract
Alternative splicing at position 3495 b yields SERCA2 (sarco/endoplasmic reticulum Ca2+ pump 2) RNA species, namely SERCA2a and SERCA2b which differ in 3′-end regions. This results in SERCA2b RNA being less stable. In vitro decay experiments show that, in the presence of protein extracts from nuclei of LVMs (left ventricular myocytes), the rate of decay of both SERCA2b RNA and synthetic RNA from its 3′-region is greater than that of the corresponding SERCA2a RNA. To search for cis-acting instability elements in the 3′-region of SERCA2b, we examined the effects of LVM nuclear protein extracts on the in vitro decay of six short overlapping capped [m7G(5′)ppp(5′)Gm] and polyadenylated (A40) RNA fragments from the 3′-end region (3444–4472) of SERCA2b. The proximal fragment 2B1 (3444–3753) was the most unstable. 2B1 RNA without a cap or a polyadenylated tail was analysed further in electrophoretic mobility-shift assays, and was observed to bind to protein(s) in the nuclear extracts. Based on competition for binding to nuclear proteins between radiolabelled 2B1 RNA and short unlabelled RNA fragments, the cis-acting element involved in this binding was the sequence 2B1-4. 2B1-4 is a 35-base (3521–3555, CCAGUCCUGCUCGUUGUGGGCGUGCACCGAGGGGG) GC-rich region just past the splice site (3495). Nuclear extracts decreased the electrophoretic mobility of the radiolabelled 2B1-4 RNA which bound to two proteins (19 and 21 kDa) in cross-linking experiments. Excess 2B1-4 RNA decreased the decay of the 2B1 RNA by the nuclear protein extract. 2B1-del 4 RNA (2B1 with the 2B1-4 domain deleted) also decayed more slowly than the control 2B1 RNA. Thus SERCA2b contains a novel GC-rich cis-acting element involved in its decay by nuclear proteins.
Keywords: ATPase, calcium pump, nuclear protein, RNA stability, SERCA2b, 3′-untranslated region
Abbreviations: G3PDH, glyceraldehyde-3-phosphate dehydrogenase; LVM, left ventricular myocyte; poly(A+), polyadenylated; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase; UTR, untranslated region
INTRODUCTION
Intracellular calcium ions (Ca2+) are a key second messenger, thus regulation of intracellular Ca2+ concentration is essential in many cellular processes [1]. One means of controlling this concentration is by sequestering Ca2+ into the sarcoplasmic reticulum via SERCA (sarco/endoplasmic reticulum Ca2+ ATPase) pumps [2]. Genes SERCA1, 2 and 3 encode the pump proteins: SERCA1 is expressed mainly in fast-twitch skeletal muscle, SERCA2 in most tissues and SERCA3 in specialized epithelial cells [2–4]. All three SERCA genes encode more than one protein isoform owing to alternative splicing. The human SERCA2 gene, for example, is made up of 25 exons, of which the four terminal exons are alternatively spliced to produce SERCA2a, SERCA2b and two additional low-abundant RNA species [5]. SERCA2a mRNA includes exons 1–21 and 25, and encodes a protein of 997 amino acids, generated by splicing of an optional intron in the 3′-end of the mRNA. SERCA2b includes exons 1–22, which encode a protein of 1042 residues [6,7]. Thus the initial 3495 bases, including the 5′-UTR (untranslated region) and encoding the first 993 amino acids, are common between SERCA2a and 2b. The remaining amino acids of their respective proteins and the 3′-UTR of their mRNA are isoform-specific.
The level of expression of any gene can be modulated at several steps, including mRNA export and decay [8–10]. There appears to be a number of regulatory elements for these steps, including specific sequences within the RNA itself (cis-acting elements) and proteins which bind the RNA (trans-acting factors) [9–14]. Sequences throughout the mRNA and proteins may contribute to the rate of mRNA degradation, but the poly(A+) (polyadenylated) tail and 3′-UTR are probably the most important regions of RNA for regulating its stability [15–19]. The sequences controlling RNA decay may include RNA stability or instability elements. The cis-acting elements do not act alone in controlling the RNA stability: RNA stability is controlled by the interaction between the cis-acting elements and proteins (or trans-acting factors) that bind them. A number of stabilizing and destabilizing proteins have been reported [20–22]. The types of cis-acting elements and trans-acting factors involved and RNA-decay kinetics have been reviewed [10,17,22]. SERCA2b mRNA decays more rapidly than SERCA2a. Protein extracts from nuclei of LVMs (left ventricular myocytes) are more effective in producing the in vitro decay of SERCA2b RNA than those from cytoplasm [23]. This difference in the decay has been observed when tissue RNA or synthetic RNA fragments corresponding to the unique 3′-region of SERCA2 (bases 3444–4472) were used. This difference in the decay between SERCA2a and SERCA2b using nuclear extracts was observed whether one used extracts from LVMs or stomach smooth muscle, but the LVM extracts were more effective. Thus the 3′-end region of SERCA2b RNA contains cis-acting instability elements that are responsible for its decay by nuclear proteins. In the present study, we analysed the 3′-region of SERCA2b for the cis-acting sequences that may play a role in the decay.
MATERIALS AND METHODS
Template construction and in vitro transcription
Rabbit SERCA2b sequences correspond to a numbering system that starts from the first base in the 5′-UTR as 1, with the translation initiation codon being at 516 (GenBank® accession number X52496). The cDNA sequence from bp 241 to 389 of G3PDH (glyceraldehyde-3-phosphate dehydrogenase) was used (GenBank® accession number L23961.1). The expected size of the fragment is 147 bases. DNA templates for in vitro transcription reactions were constructed by PCR with the appropriate primers from plasmids containing rabbit cDNA [6]. The SP6 RNA polymerase site was included in each template using extended primers during a second round of PCR. The templates used for RNA synthesis in the in vitro decay assays were made by PCR using extended downstream primers containing dT40. A DNA fragment named 2B1-del 4 containing the sequence 3444–3520, contiguous with 3556–3753, was synthesized by PCR without cloning. Thus 2B1-del 4 had a deletion of 35 bp (3521–3555) out of the domain 3444–3753. In vitro transcription was carried out with the SP6 MaxiScript Transcription kit (Ambion, Austin, TX, U.S.A.) using 50 ng of template per 40 μl of reaction volume. For synthesis of radiolabelled RNA, the transcription reactions included [33P]α-UTP (2500 Ci/mmol, Amersham Biosciences, Baie d'Urfe, Canada). The transcripts used for the in vitro decay, but not those used for the mobility-shift experiments contained an m7G(5′)ppp-(5′)Gm cap analogue (added using an m-Message m-Machine kit from Ambion) and a poly(A+) tail. Unlabelled RNA fragments shorter than 40 bases were synthesized commercially (Dharmacon, Lafayette, CO, U.S.A.).
In vitro decay assay
Protein extracts from LVM nuclei were prepared from 2-day-old rabbits, and were stored as described earlier [23]. In vitro decay assays were conducted and analysed by gel electrophoresis as described previously [23]. Typically, 5 ng of 33P-labelled RNA was incubated with 25 μg of the protein extracts for 0–30 min at 37 °C in a reaction buffer containing 10 mM Mops/NaOH (pH 7.2), 200 mM NaCl, 2.5 mM magnesium acetate, 1 mM ATP, 0.1 mM spermine, 2 mM dithiothreitol and 1 unit/μl RNAguard (Amersham Biosciences). Intensity of the bands was determined using the ImageQuant software, and the mean of three background intensities obtained from the same gel was subtracted. Each time point for each RNA was repeated in duplicate on each gel, and the mean intensity of these two bands was calculated. Relative RNA remaining over the time course was determined by assuming the RNA intensity at zero time to be 100%, and expressing the intensities of each band average relative to this. Decay rates were then calculated for each experiment using the Fig.P software (Biosoft) and the equation for mono-exponential (first-order) decay kinetics (A×exp[−kx]+R), assuming A=100 and no residual R=0.
Electrophoretic mobility-shift assay
Electrophoretic mobility-shift assays were performed as described previously [24], with minor modifications. Briefly, for protein concentration curves, 0–25 μg of protein was pre-incubated on ice for 30 min with the binding buffer containing 10 mM Hepes/NaOH (pH 7.6), 40 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 10% (v/v) glycerol and 0.1% (v/v) Nonidet P40, with 13 units of RNasin (Promega, Madison, WI, U.S.A.), 6.5 units of SUPERasin (Ambion) and 9 units of RNAguard (Amersham Biosciences). Yeast tRNA (750 ng) was added as a competitor, and the incubation was continued on ice for another 15 min. Radiolabelled sense RNA (5 ng) was added, and the samples were incubated for an additional 15 min on ice. Samples were then electrophoresed in 4% or 6% non-denaturing acrylamide gels containing 0.4× Tris/borate/EDTA buffer. In competitive mobility-shift experiments, the tRNA was replaced with the specified concentrations of the unlabelled fragments of the same RNA, RNA oligonucleotides, poly(C) RNA, poly(U) RNA or poly(A) RNA. The DNA fragment containing an SP6 polymerase site and the 35-bp fragment 2B1-4 (3521–3555), synthesized by Dharmacon, was end-labelled using [33P]ATP. RNA was precipitated with ammonium acetate and ethanol to remove the bulk of the unincorporated radioactivity.
Cross-linking experiments
UV cross-linking assays with the radiolabelled fragment 2B1-4 were performed as described previously [24], with minor modifications. Briefly, LVM nuclear protein extract (8 μg of protein/20 μl) was pre-incubated on ice for 15 min with the binding buffer containing 10 mM Hepes/NaOH (pH 7.6), 40 mM NaCl, 3 mM MgCl2, 2 mM dithiothreitol, 10% (v/v) glycerol, 0.1% (v/v) Nonidet P40 and 13 units of RNasin. Each of yeast tRNA, poly(U) RNA and poly(C) RNA was added (750 ng), and the samples were incubated for another 30 min. Radiolabelled sense RNA (2 ng) was added, and the samples were incubated for an additional 15 min. After the incubation, while still on ice, the samples were irradiated with 2.4 mJ/cm2 of UV light using a Hoeffer UVC ultraviolet cross-linker (Amersham Biosciences). The samples were then treated with 25 μg of RNase A1 and 25 units of RNase T1 at 37 °C for 30 min. The samples were then dissolved in a SDS/PAGE sample buffer and electrophoresed in Tris/glycine-based 4–20% acrylamide (Bio-Rad, Missisauga, Canada). The gels were dried and examined using a PhosphorImager. Molecular masses of the bands were estimated using a low-molecular-mass pre-stained rainbow protein marker (Fermentas, Burlington, Canada).
RESULTS
In vitro decay of SERCA2b 3′-region RNA fragments
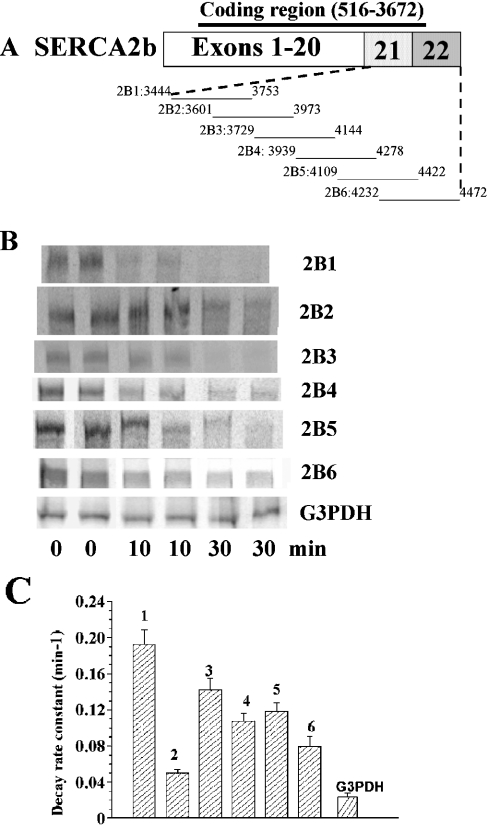
To span the full length of the SERCA2b 3′-region (3444–4472), we constructed six overlapping fragments (2B1–2B6; Figure 1A). Each RNA fragment was capped at the 5′-end with a 5′-m7G(5′)-ppp(5′)Gm cap analogue and extended at the 3′-end with a 40-base poly(A+) tail. Representative images for the decay of 2B1–2B6 RNA fragments in the presence of 25 μg of nuclear extract protein are shown in Figure 1(B). A summary of decay constants of such fragments 2B1–2B6 and G3PDH RNA from three to five experiments is shown in Figure 1(C). Based on a one-way ANOVA of the 2B fragments, 2B1 was the least stable, and 2B2 was more stable than all the others (P<0.05). The overall order of decay constants appeared to be 2B1>2B3>2B5>2B4>2B6>2B2. The segment of G3PDH RNA used as a control was significantly more stable than any of the 2B fragments (P<0.05).
Figure 1. In vitro decay of SERCA2b 3′-region fragments.
(A) Scheme showing SERCA2b mRNA and the overlapping fragments 2B1–2B6. Numbers along the lines are sequence locations. (B) Representative PhosphorImages of gels with RNA fragments after in vitro decay of 5 ng/20 μl of RNA by 25 μg/20 μl of protein from nuclear extracts. (C) Means±S.E.M. of decay constants for SERCA2b fragments 2B1–2B6 and G3PDH in three to five experiments each.
Defining a sequence element in fragment 2B1 by electrophoretic mobility-shift assay
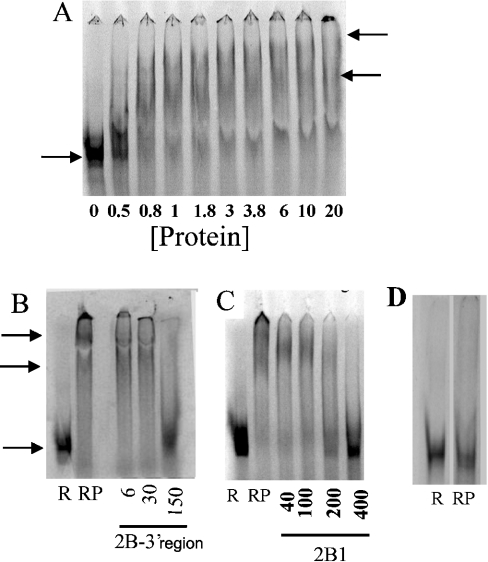
Since the RNA fragment 2B1 was the least stable, we then set out to define a minimal cis-acting sequence that would bind to proteins extracted from LVM nuclei. Since protein binding to the 5′-cap and the 3′-poly(A+) tail would make the results on binding to other domains difficult to interpret, 2B1 RNA without these was synthesized and incubated with proteins extracted from LVM nuclei. This incubation decreased the electrophoretic mobility of the RNA in a protein-concentration-dependent manner (Figure 2A), and resulted in two types of complexes: one with a lower molecular mass and another with a higher one. However, the bands observed for these complexes were somewhat diffuse and hence lower in intensity. To test whether the lower intensity corresponded to RNA degradation, we measured the total intensity of all the RNA in each lane. There was no correlation between the total intensity in each lane and the amount of protein (r2=0.0006, P>0.05), indicating that the lower intensity of the diffuse bands did not represent RNA degradation. Diffuse bands in such experiments may be due to loss of RNA from the RNA–protein complexes, since the binding may not be very tight. Such complexes have been reported previously in the literature [32]. Experiments using radiolabelled 2B1 showed that excess unlabelled 2B1 RNA (Figure 2C) or the 2B 3′-region RNA (3444–4472; Figure 2B) competed for binding. However, excess poly(C), poly(U) or poly(A) RNA did not compete (results not shown). Thus the competition experiments showed that the 2B1 binding to proteins from LVM nuclei was specific.
Figure 2. Electrophoretic mobility-shift assays with SERCA2b fragments and protein extract from LVM nuclei.
(A) Protein-concentration-dependence. Radiolabelled RNA (5 ng/20 μl) was used with protein concentrations (μg/20 μl) given along each lane. The lower arrow indicates the location of the band without any proteins, and the two higher arrows show the mobility-shifted RNA. (B) and (C) Competition of various unlabelled RNA fragments with radiolabelled 2B1 (5 ng/20 μl) for mobility shift by protein from LVM nuclear extracts (2 μg/20 μl). R, radiolabelled RNA alone; RP, radiolabelled RNA+protein, but no unlabelled RNA. Protein was included in all the lanes with radiolabelled RNA plus different amounts of unlabelled RNA (ng/20 μl, shown with each panel). Arrows indicate the position of the radiolabelled RNA in the absence or the presence of the proteins from the nuclear extracts. (D) G3PDH RNA. R, radiolabelled RNA alone; RP, radiolabelled RNA+protein (1 μg/20 μl).
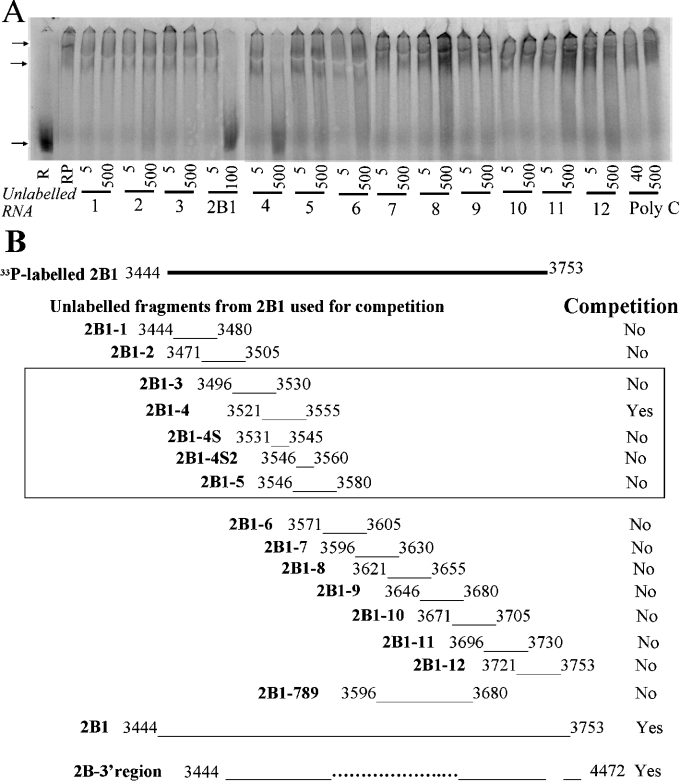
To define a minimal cis-acting sequence that would bind to proteins extracted from LVM nuclei, we examined the competition between radiolabelled 2B1 RNA and several small synthetic RNA fragments. We determined the mobility shift of radiolabelled 2B1 RNA in the presence of 12 overlapping 35 base RNA fragments, 2B1-1 to 2B1-12, together covering the full sequence of 2B1 (see the schematic diagram of Figure 3B). Only the fragment 2B1-4 (3521–3555, CCAGUCCUGCUCGUUGUGGGCGUGCACCGAGGGGG) was able to reverse the shift (Figure 3A). The fragment 2B1-3 contains the initial ten bases of 2B1-4, and 2B1-5 contains the last ten bases. Neither reversed the mobility shift of 2B1 by the LVM nuclear proteins (Figure 3A). Results of these and additional RNA fragments used are summarized in Figure 3(B). These included 2B1-4S that contained the middle 15 bases (3531–3545: UCGUUGUGGGCGUGC) of 2B1-4, and 2B1-4S2 that contained the last ten bases of 2B1-4 and five additional ones (Figure 3B). Neither reversed the mobility shift of 2B-1. Thus the minimal element resides within the 35-base GC rich sequence 3521–3555 and is longer than the middle 15 bases.
Figure 3. Competitive mobility shift of 2B1 with smaller fragments.
(A) Competition of 35-base RNA fragments with radiolabelled 2B1 (5 ng/20 μl) for mobility shift by protein from LVM nuclear extracts (2 ng/20 μl). R, radiolabelled RNA alone; RP, radiolabelled RNA+protein, but no unlabelled RNA. Protein was included in all the lanes with the radiolabelled RNA plus 5 or 500 ng/20 μl of unlabelled RNA. The identities of the 2B1-1 to 2B1-12 RNA fragment are given as numbers 1–12 at the bottom of the panels. Unlabelled 2B1 RNA (5 or 100 ng/20 μl) was used as a positive control for the competition. The experiment was replicated three times. (B) Summary of results from (A) and other competition experiments using radiolabelled smaller RNA fragments to compete with binding to radiolabelled 2B1 RNA. Yes indicates that the RNA competed for the binding and No indicates that it did not. The approximate location of each base is shown schematically, and the base numbers correspond to the full-length SERCA2b sequence. Arrows indicate the position of the radiolabelled RNA in the absence or the presence of the proteins from the nuclear extracts.
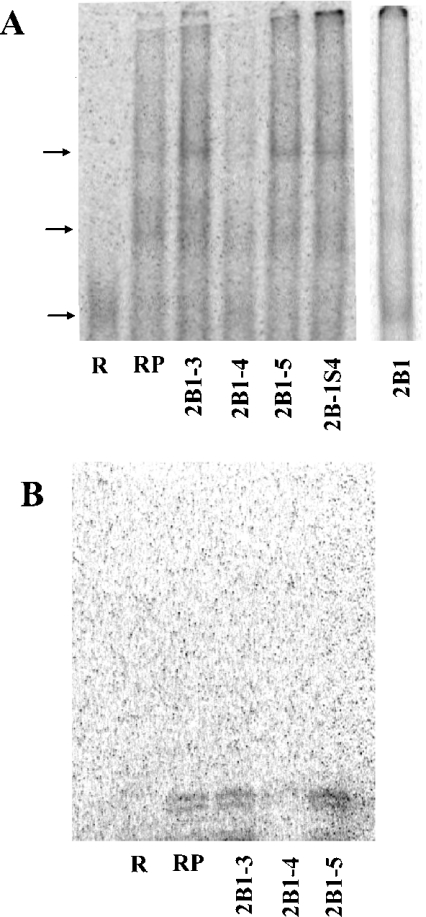
We next radiolabelled the 35-base RNA fragment 2B1-4, and examined if it would bind to proteins extracted from the LVM nuclei (Figure 4A). Protein binding to 2B1-4 resulted in two bands of slower mobility, a pattern not that different from that obtained with the 2B1 RNA. To characterize this binding further, we conducted competition experiments with unlabelled RNA. Excess 2B1-4 decreased the mobility shift as anticipated (Figure 4A). Unlabelled 2B1 RNA that contains this sequence also reversed the shift of 2B1-4. Neither the RNA fragments 2B1-3 and 2B1-5 (Figure 4A) nor poly(U) or poly(C) RNA (results not shown) reversed the mobility shift of 2B1-4. The fragments 2B1-3 (GUAAAGAGUGUGUGCAGCCUGCCCCCCAGUCCUGC) and 2B1-5 (ACCGAGGGGGUCUCCUGGCCGUUUGUGCUGCUCAU) are also GC-rich, like the fragment 2B1-4. Their failure to reverse the mobility shift of 2B1-4 indicates that a specific sequence, rather than a general GC-rich domain, is involved. We confirmed these results by UV cross-linking of 2B1-4 (radiolabelled at bases C and U) with the LVM nuclear protein extracts. Whereas no band was observed in the absence of the LVM protein, incubation with the protein resulted in two bands: one at 19±1 kDa and another at 21±1 kDa. Including excess unlabelled 2B1-4 abolished these bands, indicating that the binding is saturable. Excess unlabelled 2B1-3 or 2B1-5 did not affect the appearance of these bands, confirming the specificity of the binding. Consistent with the mobility-shift experiments, excess unlabelled 2B1-4S also did not affect the appearance of these bands (results not shown).
Figure 4. Competitive mobility shift and UV cross-linking of 35-base fragment 4 with different RNA.
(A) Competition of different RNA (500 ng/20 μl) with radiolabelled fragment 4 (5 ng/20 μl) for mobility shift by protein from LVM nuclear extracts (2 μg/20 μl). R, radiolabelled RNA alone; RP, radiolabelled RNA+protein, but no unlabelled RNA. Protein was included in all the lanes with unlabelled RNA. The amount of unlabelled RNA was 500 ng/20 μl. The identities of the RNA fragment, given as numbers B1-3, B1-4, B1-5 and B1-4S, are as in Figure 3(B). Unlabelled 2B1 RNA (5 or 100 ng/20 μl) was used as a positive control. Arrows indicate the position of the radiolabelled RNA in the absence or the presence of the proteins from the nuclear extracts. (B). UV cross-linking of radiolabelled fragment 4 (2 ng/20 μl) with proteins (8 μg/20 μl) from LVM nuclear extracts. R, radiolabelled RNA alone; RP, radiolabelled RNA+protein alone. Competition with specified unlabelled RNA fragments (2.5 μg/20 μl) is shown in the right-hand three lanes.
The mobility-shift experiments showed that the 2B1 RNA (Figures 2 and 3A) and the fragment 2B1-4 (Figure 4A) bound to two proteins. Molecular masses of these bands cannot be determined because non-denaturing gels were used in which the protein mobility depends on charge/mass ratio, rather than on mass alone. The UV cross-linking experiment with the fragment 2B1-4, where denaturing gels were used, gave two bands: at 19 and 21 kDa (Figure 4B). These data do not allow one to determine if the two bands seen in the mobility-shift experiments are simply oligomers of different sizes or if two proteins with different charge/mass ratio are involved. In any event, these data in the mobility-shift and the cross-linking experiments are consistent with the conclusion that two protein species bind to 2B1 RNA.
Role of 2B1-4 in the decay of 2B1
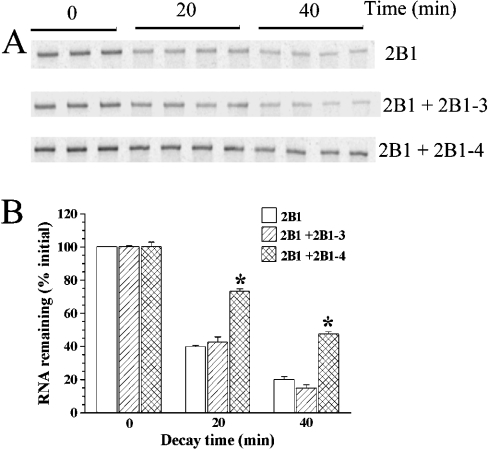
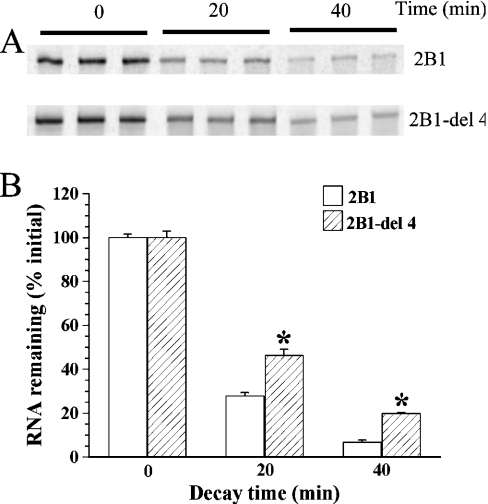
To examine the role of the cis-acting sequence 2B1-4, we conducted two types of experiments. The first experiment was to test whether including excess unlabelled 2B1-4 RNA in the decay reaction would bind the destabilizing protein and thereby protect the radiolabelled 2B1 RNA [capped and poly(A+) as in Figure 1] from decay. As expected, the decay of 2B1 RNA was slower in the presence of excess 2B1-4 RNA (Figure 5). 2B1-3 RNA, which is also GC-rich, was used as a negative control. Consistent with the requirement of a sequence specificity, 2B1-3 RNA did not alter the decay of 2B1. The second experiment tested whether the presence of the 2B1-4 sequence in 2B1 RNA was required for the faster decay. If so, 2B1-del 4, an RNA with the 2B1-4 sequence deleted from 2B1 would decay less rapidly than the 2B1 RNA. The decay of 2B1-del 4 was slower than that of 2B1 (Figure 6), consistent with a destabilizing role for the identified cis-acting element.
Figure 5. Effect of 2B1-4 RNA on in vitro decay of 2B1 RNA.
In vitro decay of capped and poly(A+) radiolabelled 2B1 RNA (2.5 ng/20 μl) was examined in the presence of LVM nuclear protein extract (10 μg/20 μl) without any added unlabelled RNA, or with (500 ng/20 μl) 2B1-3 and 2B1-4 RNA (RNA domains are defined in Figure 3B). The experiment was carried out with three to four replicates. (A) PhosphorImages of triplicate gels. (B) Histogram showing means±S.E.M. of relative intensities of the bands, taking the mean intensity at 0 min as 100%. Asterisks (*) indicate that the relative intensity differed significantly from the control without any added unlabelled RNA. The entire experiment was replicated three times.
Figure 6. In vitro decay of 2B1-del 4 and 2B1 RNA.
In vitro decay of capped and poly(A+) radiolabelled 2B1 or 2B1-del 4 RNA (2.5 ng/20 μl) was examined in the presence of LVM nuclear protein extract (10 μg/20 μl) (RNA domains are defined in Figure 3B). The experiment was carried out with three replicates for 0 min, and four replicates for 20 and 40 min. (A) PhosphorImages of triplicate gels. (B) Histogram showing means±S.E.M. of relative intensities of the bands, taking the mean intensity at 0 min as 100%. Asterisks (*) indicate that the relative intensity differed significantly from the control without any added unlabelled RNA. The entire experiment was replicated three times.
Thus, in the presence of nuclear protein extracts from LVM, there may be several unstable regions in the 3′-region of the SERCA2b mRNA, with the region 2B1 being the least stable. We showed 2B1-4, a 35-base GC-rich cis-acting element to be involved in binding to the proteins from the nuclear extracts, and demonstrated its role as a destabilizing sequence.
DISCUSSION
The Discussion will focus on the validity of the methods used, future directions and possible implications of these results.
Several sequences that influence mRNA stability have already been discovered. The AU-rich pentamer, AUUUA, and nonamer, UUAUUUA(U/A)(U/A), act when clustered or overlapping in the 3′-UTR to destabilize mRNA for many different messages [16–19,22,25,26]. Some theories of their physiological function include attracting the poly(A)-binding protein away from its protective binding at the poly(A+) tail, or increasing RNA degradation by binding to specific AU-binding proteins, such as AUF1 [AU-rich element/poly(U)-binding/degradation factor] or HuR [27,28]. Analysis of the 3′-region sequences of both SERCA2a and 2b RNA show the occurrence of several of the known cis-acting elements. The 3′-region in both SERCA2a (57.5%) and SERCA2b (60.5%) is slightly AU-rich. SERCA2b contains the sequence AUUUA at 4060 and 4342; however, these motifs are not clustered or overlapping. The SERCA2b sequence also contains an unusually large number of novel repeats. For example, the 308-base sequence of 2B1 alone contains GCACCGA at 3472 and 3535, UAACUU at 3545 and 3597, GAUGUG at 3521 and 3591, GGGUCU at 3481 and 3526, UGUGGG at 3463 and 3523, CCUGGU at 3420 and 3515, and several smaller repeats. The 35-base sequence that we have identified for protein binding in 2B1 (CCAGUCCUGCUCGUUGUGGGCGUGCACCGAGGGGG) does not contain any AU repeats, but is, in fact, GC- rich. This sequence is part of the coding region past the alternative splice site, making it unique to SERCA2b. The present study focused on decay by nuclear proteins from LVM, primarily because we found the greatest difference in decay rates of SERCA2a and SERCA2b in these extracts [23]. Much of the literature that is focused on mRNA decay deals with cytoplasmic decay, but more recently the importance of nuclear decay is being recognized [29]. The unique sequence observed here might be novel for this reason.
The fragment 2B2 was stable, but it also gave a mobility shift with the nuclear protein extracts (results not shown). Whether this binding occurred to the same protein as the 2B1 is not known. Although we examined fragment 2B1 in greater detail, the decay and binding data of the remaining fragments also play a role in the RNA decay, particularly the fragment 2B3, which may involve a different cis-acting element. Fragment 2B2 was relatively stable, but downstream fragments showed faster decay, thus an interaction between the 2B1-4 sequence in the coding domain and those further downstream in the 3′-UTR may control the rate of decay of SERCA2b mRNA. A similar relationship between coding region elements and protein binding in the 3′-UTR has already been demonstrated in c-myc and c-fos [30,31]. In addition, the present study of SERCA2b stability in extracts from non-native tissue allows for many other interesting cis- and trans-acting elements to be studied in the future.
We have identified a novel sequence unique to SERCA2b that is able to bind a protein in LVM nuclear extracts. Since rates of transcription of SERCA are similar in tissues that express SERCA2a and SERCA2b mRNA, and it is the RNA degradation by nuclear factors that leads to the difference in their SERCA2 RNA abundance, we conjecture that these cis-acting elements, along with the tissue-specific protein factors may form a developmental control of level of expression of the RNA. This supports our hypothesis that alternative splicing may influence mRNA stability and, subsequently, the level of protein expression. In conjunction with our previous work [23], this also confirms the importance of both isoform-specific sequences and tissue-specific protein factors in the control of SERCA2 stability via the 3′- region.
Acknowledgments
We thank Dr Lin Nie for carrying out one of the PCRs. This work was funded by an operating grant from the Canadian Institute of Health Research (MOP53256). C. M. M. received a Doctoral Scholarship from the Canadian Institute of Health Research and A. K. G. received a Career Investigator Award from the Heart and Stroke Foundation of Ontario.
References
- 1.Clapham D. E. Calcium signaling. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 2.East J. M. Sarco(endo)plasmic reticulum calcium pumps: recent advances in our understanding of structure/function and biology. Mol. Membr. Biol. 2000;17:189–200. doi: 10.1080/09687680010009646. [DOI] [PubMed] [Google Scholar]
- 3.Dode L., Wuytack F., Kools P. F., Baba-Aissa F., Raeymaekers L., Brike F., van de Ven W. J., Casteels R., Brik F. cDNA cloning, expression and chromosomal localization of the human sarco/endoplasmic reticulum Ca2+-ATPase 3 gene. Biochem. J. 1996;318:689–699. doi: 10.1042/bj3180689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misquitta C. M., Mack D. P., Grover A. K. Sarco/endoplasmic reticulum Ca2+ (SERCA)-pumps: link to heart beats and calcium waves. Cell Calcium. 1999;25:277–290. doi: 10.1054/ceca.1999.0032. [DOI] [PubMed] [Google Scholar]
- 5.Eggermont J. A., Wuytack F., Casteels R. Characterization of the 3′ end of the pig sarcoplasmic/endoplasmic-reticulum Ca2+ pump gene 2. Biochim. Biophys. Acta. 1991;1088:448–451. doi: 10.1016/0167-4781(91)90143-a. [DOI] [PubMed] [Google Scholar]
- 6.Khan I., Spencer G. G., Samson S. E., Crine P., Boileau G., Grover A. K. Abundance of sarcoplasmic reticulum calcium pump isoforms in stomach and cardiac muscles. Biochem. J. 1990;268:415–419. doi: 10.1042/bj2680415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lytton J., Zarain-Herzberg A., Periasamy M., MacLennan D. H. Molecular cloning of the mammalian smooth muscle sarco(endo)plasmic reticulum Ca2+-ATPase. J. Biol. Chem. 1989;264:7059–7065. [PubMed] [Google Scholar]
- 8.Pennisi E. The nucleus's revolving door. Science. 1998;279:1129–1131. doi: 10.1126/science.279.5354.1129. [DOI] [PubMed] [Google Scholar]
- 9.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 11.Ross J. A hypothesis to explain why translation inhibitors stabilize mRNAs in mammalian cells: mRNA stability and mitosis. BioEssays. 1997;19:527–529. doi: 10.1002/bies.950190612. [DOI] [PubMed] [Google Scholar]
- 12.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 14.Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 15.Brawerman G. mRNA decay: finding the right targets. Cell. 1989;57:9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- 16.Buzby J. S., Brewer G., Nugent D. J. Developmental regulation of RNA transcript destabilization by A+U-rich elements is AUF1-dependent. J. Biol. Chem. 1999;274:33973–33978. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- 17.Chen C. Y., Shyu A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 18.Fan X. C., Myer V. E., Steitz J. A. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misquitta C. M., Iyer V. R., Werstiuk E. S., Grover A. K. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol. Cell. Biochem. 2001;224:53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- 20.Brewer G., Ross J. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 1990;181:202–209. doi: 10.1016/0076-6879(90)81122-b. [DOI] [PubMed] [Google Scholar]
- 21.Wilson G. M., Brewer G. Identification and characterization of proteins binding A+U-rich elements. Methods. 1999;17:74–83. doi: 10.1006/meth.1998.0709. [DOI] [PubMed] [Google Scholar]
- 22.Wilson G. M., Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 23.Misquitta C. M., Mwanjewe J., Nie L., Grover A. K. Sarcoplasmic reticulum Ca2+ pump mRNA stability in cardiac and smooth muscle: role of the 3′-untranslated region. Am. J. Physiol. Cell Physiol. 2002;283:C560–C568. doi: 10.1152/ajpcell.00527.2001. [DOI] [PubMed] [Google Scholar]
- 24.Wilson G. M., Sun Y., Lu H., Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 25.Aharon T., Schneider R. J. Selective destabilization of short-lived mRNAs with the granulocyte–macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell. Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pende A., Tremmel K. D., DeMaria C. T., Blaxall B. C., Minobe W. A., Sherman J. A., Bisognano J. D., Bristow M. R., Brewer G., Port J. Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J. Biol. Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- 27.Fan X. C., Steitz J. A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savant-Bhonsale S., Cleveland D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 29.Moore M. J. Nuclear RNA turnover. Cell. 2002;108:431–434. doi: 10.1016/s0092-8674(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 30.Chen C. Y., Xu N., Shyu A. B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piechaczyk M., Blanchard J. M., Bonnieu A., Fort P., Mechti N., Rech J., Cuny M., Marty L., Ferre F., Lebleu B., et al. Role of RNA structures in c-myc and c-fos gene regulations. Gene. 1988;72:287–295. doi: 10.1016/0378-1119(88)90154-0. [DOI] [PubMed] [Google Scholar]
- 32.DeMaria C. T., Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]